

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.6 - phase: 0
(437 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
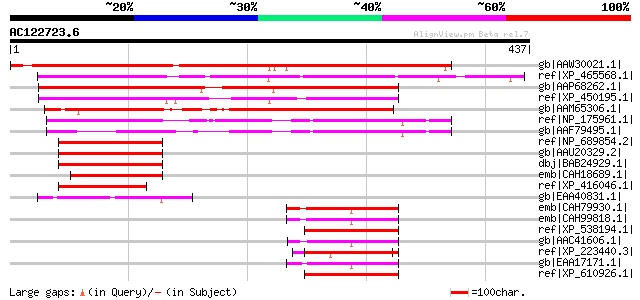
gb|AAW30021.1| At1g04880 [Arabidopsis thaliana] gi|56236040|gb|A... 379 e-104
ref|XP_465568.1| glutathione S-transferase GST16-like protein [O... 288 2e-76
gb|AAP68262.1| At1g76110 [Arabidopsis thaliana] gi|20466328|gb|A... 260 7e-68
ref|XP_450195.1| glutathione S-transferase GST 16 - like protein... 250 7e-65
gb|AAM65306.1| unknown [Arabidopsis thaliana] gi|22137084|gb|AAM... 238 3e-61
ref|NP_175961.1| high mobility group (HMG1/2) family protein / A... 206 2e-51
gb|AAF79495.1| F20N2.8 [Arabidopsis thaliana] gi|25405992|pir||H... 140 1e-31
ref|NP_689854.2| AT rich interactive domain 2 (ARID, RFX-like) [... 77 8e-13
gb|AAU20329.2| ARID2 [Homo sapiens] 77 8e-13
dbj|BAB24929.1| unnamed protein product [Mus musculus] 77 8e-13
emb|CAH18689.1| hypothetical protein [Homo sapiens] 72 3e-11
ref|XP_416046.1| PREDICTED: similar to hypothetical protein [Gal... 70 1e-10
gb|EAA40831.1| GLP_154_23821_22412 [Giardia lamblia ATCC 50803] 69 4e-10
emb|CAH79930.1| high mobility group protein, putative [Plasmodiu... 68 5e-10
emb|CAH99818.1| high mobility group protein, putative [Plasmodiu... 68 6e-10
ref|XP_538194.1| PREDICTED: similar to High mobility group prote... 67 1e-09
gb|AAC41606.1| high mobility group protein 67 1e-09
ref|XP_223440.3| PREDICTED: similar to high mobility group prote... 67 1e-09
gb|EAA17171.1| high mobility group protein [Plasmodium yoelii yo... 66 2e-09
ref|XP_610926.1| PREDICTED: similar to HMGB3 protein, partial [B... 66 2e-09
>gb|AAW30021.1| At1g04880 [Arabidopsis thaliana] gi|56236040|gb|AAV84476.1|
At1g04880 [Arabidopsis thaliana]
gi|15220344|ref|NP_171980.1| high mobility group
(HMG1/2) family protein / ARID/BRIGHT DNA-binding
domain-containing protein [Arabidopsis thaliana]
gi|7211978|gb|AAF40449.1| Contains similarity to the
high mobility group family PF|00505. [Arabidopsis
thaliana] gi|25406818|pir||B86182 hypothetical protein
[imported] - Arabidopsis thaliana
Length = 448
Score = 379 bits (974), Expect = e-104
Identities = 213/394 (54%), Positives = 268/394 (67%), Gaps = 33/394 (8%)
Query: 1 MASTSCFFNNNEFPMKVVAPMSHSYPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVP 60
MAS+SC + PM++ P A Y+ VVA+P+LFM +LE+LH+ +GTKFMVP
Sbjct: 1 MASSSCLKQGS-------VPMNNVCVTPEATYEAVVADPRLFMTSLERLHSLLGTKFMVP 53
Query: 61 IIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLY 120
IIGG++LDL +LF+EVTSRGGI K++ ERRWKEVTA F FP TATNAS+VLRKYY SLL
Sbjct: 54 IIGGRDLDLHKLFVEVTSRGGINKILNERRWKEVTATFVFPPTATNASYVLRKYYFSLLN 113
Query: 121 HYEQIYYFRSKRWTPASSDALQNQSTMSVPASITQFLQPSPGTHPVDFQ-KSGVNASELP 179
+YEQIY+FRS P S QS + P I ++PS + F + +N +E
Sbjct: 114 NYEQIYFFRSNGQIPPDS----MQSPSARPCFIQGAIRPSQELQALTFTPQPKINTAEFL 169
Query: 180 QVSSSGSSLAGVIDGKFESGYLVSVSVGSETLKGVLYE----------SPQS---IKINN 226
S +GS++ GVIDGKFESGYLV+V++GSE LKGVLY+ +PQ + N
Sbjct: 170 GGSLAGSNVVGVIDGKFESGYLVTVTIGSEQLKGVLYQLLPQNTVSYQTPQQSHGVLPNT 229
Query: 227 NNIAS----AALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD 282
NI++ A GV +RRRRRKKSEIKRRDP HPKPNRSGYNFFFAEQHARLK L+ D
Sbjct: 230 LNISANPQGVAGGVTKRRRRRKKSEIKRRDPDHPKPNRSGYNFFFAEQHARLKPLHPGKD 289
Query: 283 KDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTSIVTDNAGPLQQRF 342
+DISRMIGELWN L E EK +YQ KA++DKERY+ EMEDYR+K K + NA PLQQR
Sbjct: 290 RDISRMIGELWNKLNEDEKLIYQGKAMEDKERYRTEMEDYREKKKNGQLISNAVPLQQRL 349
Query: 343 PEGDSALVDVDIKMHDSCQTPEE----SSSGESD 372
PE + + + D+ + + + EE SSGES+
Sbjct: 350 PEQNVDMAEADLPIDEVEEDDEEGDSSGSSGESE 383
>ref|XP_465568.1| glutathione S-transferase GST16-like protein [Oryza sativa
(japonica cultivar-group)] gi|47497529|dbj|BAD19581.1|
glutathione S-transferase GST16-like protein [Oryza
sativa (japonica cultivar-group)]
gi|47497414|dbj|BAD19471.1| glutathione S-transferase
GST16-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 467
Score = 288 bits (737), Expect = 2e-76
Identities = 172/423 (40%), Positives = 248/423 (57%), Gaps = 34/423 (8%)
Query: 24 SYPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIE 83
+YP +A Y DVVA+ +F LE LHA MGTK VPIIGGK+LDL +LF EVTSRGGI+
Sbjct: 63 AYPARVAGYKDVVADAAVFRRALEGLHAQMGTKLKVPIIGGKDLDLHQLFKEVTSRGGID 122
Query: 84 KLMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQN 143
K+ + RW+EVTA+F FP+TATNASF+L+KYY SLLYH+E++Y F ++ W
Sbjct: 123 KVKSDNRWREVTASFIFPATATNASFMLKKYYMSLLYHFERLYLFEAQGW---------Y 173
Query: 144 QSTMSVPASITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVS 203
Q T S S + + +K G N+ +S + + +IDGKFE GY+V+
Sbjct: 174 QETDSRSISCIEMKAEGQASRK---RKRGSNSCSSDLAASLDNDVQVIIDGKFEHGYIVT 230
Query: 204 VSVGSETLKGVLY---ESPQSIKINNNNIASAALGVQRRRRRRKKSEIKRRDPAHPKPNR 260
V +GS++ K VLY E P + +A G++ RRRRR+K ++ DP HPKPNR
Sbjct: 231 VIMGSKSTKAVLYNCTEEPAVPTAVPHVAIDSAEGIRPRRRRRRK-KLSTTDPNHPKPNR 289
Query: 261 SGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEME 320
SGYNFFF +QH +LK D+ IS+MIGE WNNL +K VYQEK ++DK RYQ ++
Sbjct: 290 SGYNFFFQDQHRKLKPEYPGQDRLISKMIGERWNNLGPEDKAVYQEKGVEDKARYQRQLA 349
Query: 321 DYRDKMKTSIVTDNAGPLQQRFPEGDSALVDVDIKMHDS----CQTPEESSSGESDYVAD 376
YR++ +T NA P+QQR P+ + + +VD K+ + SS+ SD AD
Sbjct: 350 LYREQ-RTGQPISNAVPIQQRLPQKEVTIDEVDSKVSEGDILLSNQGYSSSTSSSDETAD 408
Query: 377 DINMDASSGGARVDSEAFVDSDKATKEGVIEVLSHCEGEVNAGG------IQQTQKMNES 430
SG V+ + +++ + + + SH + + +A G ++ K++E
Sbjct: 409 -------SGEKNVEDDEEFNTETSPEPSMETTDSHGQPDPSADGERFELRRRENPKIDEK 461
Query: 431 QNM 433
++M
Sbjct: 462 RDM 464
>gb|AAP68262.1| At1g76110 [Arabidopsis thaliana] gi|20466328|gb|AAM20481.1| unknown
protein [Arabidopsis thaliana]
gi|15222957|ref|NP_177738.1| high mobility group
(HMG1/2) family protein / ARID/BRIGHT DNA-binding
domain-containing protein [Arabidopsis thaliana]
gi|25405003|pir||B96789 protein T23E18.4 [imported] -
Arabidopsis thaliana gi|6573729|gb|AAF17649.1| T23E18.4
[Arabidopsis thaliana]
Length = 338
Score = 260 bits (664), Expect = 7e-68
Identities = 134/313 (42%), Positives = 193/313 (60%), Gaps = 23/313 (7%)
Query: 25 YPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEK 84
YPEPLA ++ VV + +F TL + H+ M TKFM+P+IGGKELDL L++EVT RGG EK
Sbjct: 27 YPEPLALHEVVVKDSSVFWDTLRRFHSIMSTKFMIPVIGGKELDLHVLYVEVTRRGGYEK 86
Query: 85 LMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQ 144
++ E++W+EV F F +T T+ASFVLRK+Y +LL+HYEQ++ F ++
Sbjct: 87 VVVEKKWREVGGVFRFSATTTSASFVLRKHYLNLLFHYEQVHLFTARGPLLHPIATFHAN 146
Query: 145 STMSVPASITQFLQPS---PGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYL 201
+ S ++ ++ PS THP P SS + G I+GKF+ GYL
Sbjct: 147 PSTSKEMALVEYTPPSIRYHNTHP-------------PSQGSSSFTAIGTIEGKFDCGYL 193
Query: 202 VSVSVGSETLKGVLYESPQ-------SIKINNNNIASAALGVQRRRRRRKKSEIKRRDPA 254
V V +GSE L GVLY S Q + +NN + G +RRR +++ +R DP
Sbjct: 194 VKVKLGSEILNGVLYHSAQPGPSSSPTAVLNNAVVPYVETGRRRRRLGKRRRSRRREDPN 253
Query: 255 HPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKER 314
+PKPNRSGYNFFFAE+H +LK L +++ +++IGE W+NL E+ VYQ+ +KDKER
Sbjct: 254 YPKPNRSGYNFFFAEKHCKLKSLYPNKEREFTKLIGESWSNLSTEERMVYQDIGLKDKER 313
Query: 315 YQAEMEDYRDKMK 327
YQ E+ +YR+ ++
Sbjct: 314 YQRELNEYRETLR 326
>ref|XP_450195.1| glutathione S-transferase GST 16 - like protein [Oryza sativa
(japonica cultivar-group)] gi|32490476|dbj|BAC79159.1|
glutathione S-transferase GST 16 - like protein [Oryza
sativa (japonica cultivar-group)]
Length = 306
Score = 250 bits (638), Expect = 7e-65
Identities = 138/317 (43%), Positives = 188/317 (58%), Gaps = 39/317 (12%)
Query: 25 YPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEK 84
YP PL +++V + FM TL + H+ MGTKFM+P+IGGKE+DL L++EVTSRGG+ K
Sbjct: 7 YPPPLLSHEEVANDRAAFMDTLRRFHSLMGTKFMIPVIGGKEMDLHALYVEVTSRGGLAK 66
Query: 85 LMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRS--KRWTPASS---- 138
+M+ER+W+EV A FSFP+T T+AS+VLR+YY SLL+HYEQ+Y+FR+ PA+S
Sbjct: 67 VMEERKWREVMARFSFPATTTSASYVLRRYYLSLLHHYEQVYFFRAHGALLRPAASALTK 126
Query: 139 ------DALQNQSTMSVPASITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVI 192
+QS + A L G P F S+ G I
Sbjct: 127 TPRRKMRGTSDQSPAAAEAGKRMALPERLGGEPCSF------------------SVTGSI 168
Query: 193 DGKFESGYLVSVSVGSETLKGVLYE--SPQSIKINNNNIASAALGVQRRRRRRKKSEIKR 250
DGKFE GYLV+V + +ETL+GVLY P A G +RR RR +
Sbjct: 169 DGKFEHGYLVTVKIAAETLRGVLYRVAPPPPPPAAPPPPPPPARGRRRRGRR-------Q 221
Query: 251 RDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIK 310
RDPA P+PNRS YNFFF E+H LK + +++ SRMIG+ WN L +K VY + +
Sbjct: 222 RDPAQPRPNRSAYNFFFKEKHPELKATHPHREREYSRMIGDAWNRLAADDKMVYYRHSAE 281
Query: 311 DKERYQAEMEDYRDKMK 327
DKERY+ EM++Y +++K
Sbjct: 282 DKERYKREMQEYNERLK 298
>gb|AAM65306.1| unknown [Arabidopsis thaliana] gi|22137084|gb|AAM91387.1|
At3g13350/MDC11_14 [Arabidopsis thaliana]
gi|9294541|dbj|BAB02804.1| high mobility group
protein-like [Arabidopsis thaliana]
gi|13605513|gb|AAK32750.1| AT3g13350/MDC11_14
[Arabidopsis thaliana] gi|18399977|ref|NP_566454.1| high
mobility group (HMG1/2) family protein / ARID/BRIGHT
DNA-binding domain-containing protein [Arabidopsis
thaliana]
Length = 319
Score = 238 bits (607), Expect = 3e-61
Identities = 140/298 (46%), Positives = 184/298 (60%), Gaps = 30/298 (10%)
Query: 30 AKYDDVVANPKLFMLTLEKLHASMGTK---FMVPIIGGKELDLCRLFIEVTSRGGIEKLM 86
AKYDD+V N LF EKL A +G VP +GG LDL RLFIEVTSRGGIE+++
Sbjct: 34 AKYDDLVRNSALFW---EKLRAFLGLTSKTLKVPTVGGNTLDLHRLFIEVTSRGGIERVV 90
Query: 87 KERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQST 146
K+R+WKEV AFSFP+T T+ASFVLRKYY L+ E +YY P S S
Sbjct: 91 KDRKWKEVIGAFSFPTTITSASFVLRKYYLKFLFQLEHVYYLEK----PVS-------SL 139
Query: 147 MSVPASITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSV 206
S ++ SP + G++ PQV G + G IDGKF+SGYLV++ +
Sbjct: 140 QSTDEALKSLANESPN------PEEGIDE---PQV---GYEVQGFIDGKFDSGYLVTMKL 187
Query: 207 GSETLKGVLYESPQSIKINNNNIASAALGVQ-RRRRRRKKSEIKRRDPAHPKPNRSGYNF 265
GS+ LKGVLY PQ+ + + + + VQ +RR RKKS++ D PK +RSGYNF
Sbjct: 188 GSQELKGVLYHIPQTPSQSQQTMETPSAIVQSSQRRHRKKSKLAVVDTQKPKCHRSGYNF 247
Query: 266 FFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYR 323
FFAEQ+ARLK ++ I++ IG +W+NL ESEK VYQ+K +KD ERY+ EM +Y+
Sbjct: 248 FFAEQYARLKPEYHGQERSITKKIGHMWSNLTESEKQVYQDKGVKDVERYRIEMLEYK 305
>ref|NP_175961.1| high mobility group (HMG1/2) family protein / ARID/BRIGHT
DNA-binding domain-containing protein [Arabidopsis
thaliana]
Length = 337
Score = 206 bits (523), Expect = 2e-51
Identities = 127/346 (36%), Positives = 185/346 (52%), Gaps = 46/346 (13%)
Query: 32 YDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRW 91
Y D+V NP+LF L H S KF +PI+GGK LDL RLF EVTSRGG+EK++K+RR
Sbjct: 30 YQDIVRNPELFWEMLRDFHESSDKKFKIPIVGGKSLDLHRLFNEVTSRGGLEKVIKDRRC 89
Query: 92 KEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQSTMSVPA 151
KEV AF+F +T TN++FVLRK Y +L+ +E +YYF+ A
Sbjct: 90 KEVIDAFNFKTTITNSAFVLRKSYLKMLFEFEHLYYFQ---------------------A 128
Query: 152 SITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSETL 211
++ F + + +KS N + Q G+ + G+IDGKFESGYL+S VGSE L
Sbjct: 129 PLSTFWEKEKALKLL-IEKS-ANRDKDSQELKPGTVITGIIDGKFESGYLISTKVGSEKL 186
Query: 212 KGVLYE-SPQSIKINNNNIASAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQ 270
KG+LY SP++ +R +++ K S+ P PK R+GYNFF AEQ
Sbjct: 187 KGMLYHISPET---------------KRGKKKAKSSQGDSHKP--PKRQRTGYNFFVAEQ 229
Query: 271 HARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTS- 329
R+K N + G +W NL ES++ VY EK+ +D +RY+ E+ YR M++
Sbjct: 230 SVRIKAENAGQKVSSPKNFGNMWTNLSESDRKVYYEKSREDGKRYKMEILQYRSLMESRV 289
Query: 330 ---IVTDNAGPLQQRFPEGDSALVDVDIKMHDSCQTPEESSSGESD 372
+ +AG D A + ++ D+C + ++ E +
Sbjct: 290 AEIVAATDAGTSASAAETADEASQE-NLAKTDACTSASSAAETEDE 334
>gb|AAF79495.1| F20N2.8 [Arabidopsis thaliana] gi|25405992|pir||H96598 protein
F20N2.8 [imported] - Arabidopsis thaliana
Length = 315
Score = 140 bits (352), Expect = 1e-31
Identities = 103/346 (29%), Positives = 153/346 (43%), Gaps = 94/346 (27%)
Query: 32 YDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRW 91
Y D+V NP+LF L H S KF +
Sbjct: 56 YQDIVRNPELFWEMLRDFHESSDKKF--------------------------------KC 83
Query: 92 KEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQSTMSVPA 151
KEV AF+F +T TN++FVLRK Y +L+ +E +YYF++ T D+
Sbjct: 84 KEVIDAFNFKTTITNSAFVLRKSYLKMLFEFEHLYYFQAPLSTFWEKDS----------- 132
Query: 152 SITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSETL 211
Q L+P G+ + G+IDGKFESGYL+S VGSE L
Sbjct: 133 ---QELKP-------------------------GTVITGIIDGKFESGYLISTKVGSEKL 164
Query: 212 KGVLYE-SPQSIKINNNNIASAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQ 270
KG+LY SP++ +R +++ K S+ P PK R+GYNFF AEQ
Sbjct: 165 KGMLYHISPET---------------KRGKKKAKSSQGDSHKP--PKRQRTGYNFFVAEQ 207
Query: 271 HARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTS- 329
R+K N + G +W NL ES++ VY EK+ +D +RY+ E+ YR M++
Sbjct: 208 SVRIKAENAGQKVSSPKNFGNMWTNLSESDRKVYYEKSREDGKRYKMEILQYRSLMESRV 267
Query: 330 ---IVTDNAGPLQQRFPEGDSALVDVDIKMHDSCQTPEESSSGESD 372
+ +AG D A + ++ D+C + ++ E +
Sbjct: 268 AEIVAATDAGTSASAAETADEASQE-NLAKTDACTSASSAAETEDE 312
>ref|NP_689854.2| AT rich interactive domain 2 (ARID, RFX-like) [Homo sapiens]
Length = 1835
Score = 77.4 bits (189), Expect = 8e-13
Identities = 35/88 (39%), Positives = 57/88 (64%), Gaps = 1/88 (1%)
Query: 42 FMLTLEKLHASMGTKFM-VPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSF 100
F+ L + H S G+ F +P +GGKELDL L+ VT+ GG K+ ++ +W E+ F+F
Sbjct: 19 FLDELRQFHHSRGSPFKKIPAVGGKELDLHGLYTRVTTLGGFAKVSEKNQWGEIVEEFNF 78
Query: 101 PSTATNASFVLRKYYSSLLYHYEQIYYF 128
P + +NA+F L++YY L YE++++F
Sbjct: 79 PRSCSNAAFALKQYYLRYLEKYEKVHHF 106
>gb|AAU20329.2| ARID2 [Homo sapiens]
Length = 1113
Score = 77.4 bits (189), Expect = 8e-13
Identities = 35/88 (39%), Positives = 57/88 (64%), Gaps = 1/88 (1%)
Query: 42 FMLTLEKLHASMGTKFM-VPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSF 100
F+ L + H S G+ F +P +GGKELDL L+ VT+ GG K+ ++ +W E+ F+F
Sbjct: 19 FLDELRQFHHSRGSPFKKIPAVGGKELDLHGLYTRVTTLGGFAKVSEKNQWGEIVEEFNF 78
Query: 101 PSTATNASFVLRKYYSSLLYHYEQIYYF 128
P + +NA+F L++YY L YE++++F
Sbjct: 79 PRSCSNAAFALKQYYLRYLEKYEKVHHF 106
>dbj|BAB24929.1| unnamed protein product [Mus musculus]
Length = 145
Score = 77.4 bits (189), Expect = 8e-13
Identities = 35/88 (39%), Positives = 57/88 (64%), Gaps = 1/88 (1%)
Query: 42 FMLTLEKLHASMGTKFM-VPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSF 100
F+ L + H S G+ F +P +GGKELDL L+ VT+ GG K+ ++ +W E+ F+F
Sbjct: 19 FLDELRQFHHSRGSPFKKIPAVGGKELDLHGLYTRVTTLGGFAKVSEKNQWGEIVEEFNF 78
Query: 101 PSTATNASFVLRKYYSSLLYHYEQIYYF 128
P + +NA+F L++YY L YE++++F
Sbjct: 79 PRSCSNAAFALKQYYLRYLEKYEKVHHF 106
>emb|CAH18689.1| hypothetical protein [Homo sapiens]
Length = 1756
Score = 72.0 bits (175), Expect = 3e-11
Identities = 32/78 (41%), Positives = 52/78 (66%), Gaps = 1/78 (1%)
Query: 52 SMGTKFM-VPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFPSTATNASFV 110
S G+ F +P +GGKELDL L+ VT+ GG K+ ++ +W E+ F+FP + +NA+F
Sbjct: 2 SRGSPFKKIPAVGGKELDLHGLYTRVTTLGGFAKVSEKNQWGEIVEEFNFPRSCSNAAFA 61
Query: 111 LRKYYSSLLYHYEQIYYF 128
L++YY L YE++++F
Sbjct: 62 LKQYYLRYLEKYEKVHHF 79
>ref|XP_416046.1| PREDICTED: similar to hypothetical protein [Gallus gallus]
Length = 2143
Score = 70.1 bits (170), Expect = 1e-10
Identities = 31/75 (41%), Positives = 50/75 (66%), Gaps = 1/75 (1%)
Query: 42 FMLTLEKLHASMGTKFM-VPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSF 100
F+ L + H S G+ F +P++GGKELDL L+ VT+ GG K+ ++ +W E+ F+F
Sbjct: 19 FLDELRQFHHSRGSPFKKIPVVGGKELDLHALYTRVTTLGGFGKVSEKNQWGEIVEEFNF 78
Query: 101 PSTATNASFVLRKYY 115
P + +NA+F L++YY
Sbjct: 79 PRSCSNAAFALKQYY 93
>gb|EAA40831.1| GLP_154_23821_22412 [Giardia lamblia ATCC 50803]
Length = 469
Score = 68.6 bits (166), Expect = 4e-10
Identities = 42/134 (31%), Positives = 72/134 (53%), Gaps = 8/134 (5%)
Query: 24 SYPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIE 83
+Y + A +D V+ +LF LE+L S + PI+G ++LD+ +LF V +RGG +
Sbjct: 2 NYSQYTALNEDAVS--ELFKRRLEELQRSCRRSYRQPIVGHRQLDMYQLFRAVQARGGAK 59
Query: 84 KLMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIY---YFRSKRWTPASSDA 140
+ +WKE+ P++ TNA + LR Y S + YE+I + + + P S
Sbjct: 60 NV---TQWKEIGKKLGLPASVTNAGYTLRTKYESYILPYEEILCNEFPQMEFADPGLSST 116
Query: 141 LQNQSTMSVPASIT 154
L+ +++S P S+T
Sbjct: 117 LRGVTSISAPISLT 130
>emb|CAH79930.1| high mobility group protein, putative [Plasmodium chabaudi]
Length = 102
Score = 68.2 bits (165), Expect = 5e-10
Identities = 38/98 (38%), Positives = 60/98 (60%), Gaps = 8/98 (8%)
Query: 234 LGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDIS---RMIG 290
+G + ++RRK ++DP PK + S Y FF E+ A + + ++ KD++ +MIG
Sbjct: 9 MGGKEVKKRRKN----KKDPHAPKRSLSAYMFFAKEKRAEIITRDPSLSKDVATVGKMIG 64
Query: 291 ELWNNLKESEKTVYQEKAIKDKERYQAEMEDY-RDKMK 327
E WN L E EK Y++KA +DK RY+ E +Y ++KMK
Sbjct: 65 EAWNKLDEREKAPYEKKAQEDKLRYEKEKMEYAKNKMK 102
>emb|CAH99818.1| high mobility group protein, putative [Plasmodium berghei]
Length = 96
Score = 67.8 bits (164), Expect = 6e-10
Identities = 38/98 (38%), Positives = 59/98 (59%), Gaps = 8/98 (8%)
Query: 234 LGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDIS---RMIG 290
+G + ++RRK ++DP PK + S Y FF E+ A + + ++ KD++ +MIG
Sbjct: 3 MGGKEVKKRRKN----KKDPHAPKRSLSAYMFFAKEKRAEIITRDPSLSKDVATVGKMIG 58
Query: 291 ELWNNLKESEKTVYQEKAIKDKERYQAEMEDY-RDKMK 327
E WN L E EK Y++KA +DK RY+ E +Y + KMK
Sbjct: 59 EAWNKLDEREKAPYEKKAQEDKIRYEKEKMEYAKSKMK 96
>ref|XP_538194.1| PREDICTED: similar to High mobility group protein 4 (HMG-4) (High
mobility group protein 2a) (HMG-2a) [Canis familiaris]
Length = 201
Score = 67.0 bits (162), Expect = 1e-09
Identities = 32/80 (40%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Query: 249 KRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD-KDISRMIGELWNNLKESEKTVYQEK 307
K++DP PK SG+ F +E ++K N + D+++ +GE+WNNL +SEK Y K
Sbjct: 86 KKKDPNAPKRPPSGFFLFCSEFRPKIKSTNPGISIGDVAKKLGEMWNNLSDSEKQPYNNK 145
Query: 308 AIKDKERYQAEMEDYRDKMK 327
A K KE+Y+ ++ DY+ K K
Sbjct: 146 AAKLKEKYEKDVADYKSKGK 165
Score = 44.3 bits (103), Expect = 0.008
Identities = 25/74 (33%), Positives = 33/74 (43%), Gaps = 3/74 (4%)
Query: 252 DPAHPKPNRSGYNFFFA---EQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKA 308
DP PK S Y FF E+H + + S+ E W + EK+ + E A
Sbjct: 5 DPKKPKGKMSAYAFFVQTCREEHKKKNPEVPVNFAEFSKKCSERWKTMSGKEKSKFDEMA 64
Query: 309 IKDKERYQAEMEDY 322
DK RY EM+DY
Sbjct: 65 KADKVRYDREMKDY 78
>gb|AAC41606.1| high mobility group protein
Length = 97
Score = 67.0 bits (162), Expect = 1e-09
Identities = 39/97 (40%), Positives = 57/97 (58%), Gaps = 9/97 (9%)
Query: 235 GVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDIS---RMIGE 291
G + R+RR+KK +DP PK + S Y FF E+ A + + KD++ +MIGE
Sbjct: 5 GKEVRKRRKKK-----KDPHAPKRSLSAYMFFAKEKRAEIISKQPELSKDVATVGKMIGE 59
Query: 292 LWNNLKESEKTVYQEKAIKDKERYQAEMEDYRD-KMK 327
WN L E EK +++KA +DK RY+ E +Y + KMK
Sbjct: 60 AWNKLGEKEKAPFEKKAQEDKLRYEKEKAEYANMKMK 96
>ref|XP_223440.3| PREDICTED: similar to high mobility group protein homolog HMG4
[Rattus norvegicus]
Length = 241
Score = 66.6 bits (161), Expect = 1e-09
Identities = 32/80 (40%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Query: 249 KRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD-KDISRMIGELWNNLKESEKTVYQEK 307
K++DP PK SG+ F +E ++K N + D+++ +GE+WNNL +SEK Y K
Sbjct: 127 KKKDPNAPKRPPSGFFLFCSEFRPKIKSANPGISIGDVAKKLGEMWNNLSDSEKQPYMTK 186
Query: 308 AIKDKERYQAEMEDYRDKMK 327
A K KE+Y+ ++ DY+ K K
Sbjct: 187 AAKLKEKYEKDVADYKSKGK 206
Score = 48.9 bits (115), Expect = 3e-04
Identities = 28/87 (32%), Positives = 38/87 (43%), Gaps = 3/87 (3%)
Query: 239 RRRRRKKSEIKRRDPAHPKPNRSGYNFFFA---EQHARLKLLNQTMDKDISRMIGELWNN 295
R RR + + DP PK S Y FF E+H + + S+ E W
Sbjct: 33 REGRRHSVRMAKGDPKKPKGKMSAYAFFVQTCREEHKKKNPEVPVNFAEFSKKCSERWKT 92
Query: 296 LKESEKTVYQEKAIKDKERYQAEMEDY 322
+ EK+ + E A DK RY EM+DY
Sbjct: 93 MSSKEKSKFDEMAKADKVRYDREMKDY 119
>gb|EAA17171.1| high mobility group protein [Plasmodium yoelii yoelii]
Length = 105
Score = 66.2 bits (160), Expect = 2e-09
Identities = 38/98 (38%), Positives = 58/98 (58%), Gaps = 8/98 (8%)
Query: 234 LGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDIS---RMIG 290
+G + + RRK +DP PK + S Y FF E+ A + + ++ KD++ +MIG
Sbjct: 12 MGGKEVKXRRKNX----KDPHAPKRSLSAYMFFAKEKRAEIITRDPSLSKDVATVGKMIG 67
Query: 291 ELWNNLKESEKTVYQEKAIKDKERYQAEMEDY-RDKMK 327
E WN L E EK Y++KA +DK RY+ E +Y ++KMK
Sbjct: 68 EAWNKLDEREKAPYEKKAQEDKIRYEKEKMEYAKNKMK 105
>ref|XP_610926.1| PREDICTED: similar to HMGB3 protein, partial [Bos taurus]
Length = 179
Score = 65.9 bits (159), Expect = 2e-09
Identities = 32/80 (40%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Query: 249 KRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD-KDISRMIGELWNNLKESEKTVYQEK 307
K++DP PK SG+ F +E ++K N + D+++ +GE+WNNL +SEK Y K
Sbjct: 86 KKKDPNAPKRPPSGFFLFCSEFRPKIKSANPGISIGDVAKKLGEMWNNLSDSEKQPYINK 145
Query: 308 AIKDKERYQAEMEDYRDKMK 327
A K KE+Y+ ++ DY+ K K
Sbjct: 146 AAKLKEKYEKDVADYKSKGK 165
Score = 44.7 bits (104), Expect = 0.006
Identities = 25/74 (33%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query: 252 DPAHPKPNRSGYNFFFA---EQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKA 308
DP PK S Y+FF E+H + + S+ E W + EK+ + E A
Sbjct: 5 DPKKPKGKMSAYSFFVQTCREEHKKKNPEVPVNFAEFSKKCSERWKTMSGKEKSKFDEIA 64
Query: 309 IKDKERYQAEMEDY 322
DK RY EM+DY
Sbjct: 65 KADKVRYDREMKDY 78
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 724,606,319
Number of Sequences: 2540612
Number of extensions: 30313115
Number of successful extensions: 81393
Number of sequences better than 10.0: 1169
Number of HSP's better than 10.0 without gapping: 710
Number of HSP's successfully gapped in prelim test: 460
Number of HSP's that attempted gapping in prelim test: 79973
Number of HSP's gapped (non-prelim): 1608
length of query: 437
length of database: 863,360,394
effective HSP length: 131
effective length of query: 306
effective length of database: 530,540,222
effective search space: 162345307932
effective search space used: 162345307932
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC122723.6


