

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.9 - phase: 0
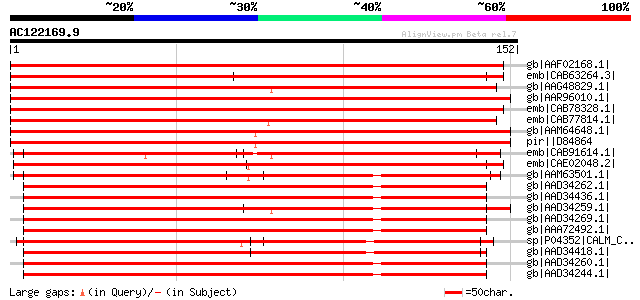
(152 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF02168.1| putative calmodulin [Arabidopsis thaliana] gi|152... 246 1e-64
emb|CAB63264.3| calcium-binding protein [Lotus corniculatus var.... 237 5e-62
gb|AAG48829.1| putative calcium-binding protein [Arabidopsis tha... 230 5e-60
gb|AAR96010.1| calmodulin-like protein [Musa acuminata] 228 2e-59
emb|CAB78328.1| putative calmodulin [Arabidopsis thaliana] gi|45... 220 5e-57
emb|CAB77814.1| putative calmodulin [Arabidopsis thaliana] gi|28... 210 5e-54
gb|AAM64648.1| putative calcium binding protein [Arabidopsis tha... 209 1e-53
pir||D84864 probable calcium binding protein [imported] - Arabid... 209 1e-53
emb|CAB91614.1| calmodulin-like protein [Arabidopsis thaliana] g... 199 9e-51
emb|CAE02048.2| OJ990528_30.6 [Oryza sativa (japonica cultivar-g... 182 1e-45
gb|AAM63501.1| touch-induced calmodulin-related protein TCH2 [Ar... 128 2e-29
gb|AAD34262.1| calmodulin mutant SYNCAM57D [synthetic construct] 127 7e-29
gb|AAD34436.1| calmodulin mutant SYNCAM33 [synthetic construct] 126 1e-28
gb|AAD34259.1| calmodulin mutant SYNCAM57A [synthetic construct] 125 2e-28
gb|AAD34269.1| calmodulin mutant SYNCAM71A [synthetic construct] 125 3e-28
gb|AAA72492.1| VU1 calmodulin [synthetic construct] gi|208412|gb... 124 4e-28
sp|P04352|CALM_CHLRE Calmodulin (CaM) gi|167411|gb|AAA33083.1| c... 124 4e-28
gb|AAD34418.1| calmodulin mutant SYNCAM24 [synthetic construct] 124 4e-28
gb|AAD34260.1| calmodulin mutant SYNCAM57B [synthetic construct] 124 4e-28
gb|AAD34244.1| calmodulin mutant SYNCAM30 [synthetic construct] 124 4e-28
>gb|AAF02168.1| putative calmodulin [Arabidopsis thaliana]
gi|15231470|ref|NP_187405.1| calcium-binding protein,
putative [Arabidopsis thaliana]
Length = 153
Score = 246 bits (627), Expect = 1e-64
Identities = 119/148 (80%), Positives = 136/148 (91%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MDQ ELARIFQMFD+NGDG+ITK+EL+DSL+NLGI I ++DLVQMIEKID+NGDGYVDI+
Sbjct: 1 MDQAELARIFQMFDRNGDGKITKQELNDSLENLGIYIPDKDLVQMIEKIDLNGDGYVDIE 60
Query: 61 EFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
EFG LYQTIM+E+DEEEDM+EAFNVFDQN DGFI+ EEL +VL+SLGLK G+TLEDCK M
Sbjct: 61 EFGGLYQTIMEERDEEEDMREAFNVFDQNRDGFITVEELRSVLASLGLKQGRTLEDCKRM 120
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAGAFAA 148
I KVDVDGDGMVNFKEF+QMMK G FAA
Sbjct: 121 ISKVDVDGDGMVNFKEFKQMMKGGGFAA 148
>emb|CAB63264.3| calcium-binding protein [Lotus corniculatus var. japonicus]
Length = 230
Score = 237 bits (604), Expect = 5e-62
Identities = 113/148 (76%), Positives = 133/148 (89%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MD EL R+FQMFD+NGDGRITKKEL+DSL+NLGI I +++L QMIE+IDVNGDG VDID
Sbjct: 81 MDPTELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPDKELTQMIERIDVNGDGCVDID 140
Query: 61 EFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
EFGELYQ+IMDE+DEEEDM+EAFNVFDQNGDGFI+ EEL VL+SLG+K G+T+EDCK M
Sbjct: 141 EFGELYQSIMDERDEEEDMREAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKM 200
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAGAFAA 148
I KVDVDGDGMV++KEF+QMMK G F+A
Sbjct: 201 IMKVDVDGDGMVDYKEFKQMMKGGGFSA 228
Score = 50.8 bits (120), Expect = 6e-06
Identities = 26/76 (34%), Positives = 48/76 (62%), Gaps = 2/76 (2%)
Query: 68 TIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVD 127
T + K + ++K F +FD+NGDG I+ +EL+ L +LG+ ++ MI+++DV+
Sbjct: 75 TSLIPKMDPTELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPD--KELTQMIERIDVN 132
Query: 128 GDGMVNFKEFQQMMKA 143
GDG V+ EF ++ ++
Sbjct: 133 GDGCVDIDEFGELYQS 148
>gb|AAG48829.1| putative calcium-binding protein [Arabidopsis thaliana]
gi|8810461|gb|AAF80122.1| Contains similarity to a
calcium-binding protein from Lotus japonicus gi|6580549
and contains a EF hand PF|00036 domain. EST gb|T46471
comes from this gene. [Arabidopsis thaliana]
gi|15221358|ref|NP_172089.1| calcium-binding protein,
putative [Arabidopsis thaliana] gi|25295760|pir||H86194
hypothetical protein [imported] - Arabidopsis thaliana
Length = 150
Score = 230 bits (587), Expect = 5e-60
Identities = 115/147 (78%), Positives = 130/147 (88%), Gaps = 1/147 (0%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MD EL R+FQMFDKNGDG IT KELS++L++LGI I +++L QMIEKIDVNGDG VDID
Sbjct: 1 MDPTELKRVFQMFDKNGDGTITGKELSETLRSLGIYIPDKELTQMIEKIDVNGDGCVDID 60
Query: 61 EFGELYQTIMDEKDEEE-DMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKN 119
EFGELY+TIMDE+DEEE DMKEAFNVFDQNGDGFI+ +EL AVLSSLGLK GKTL+DCK
Sbjct: 61 EFGELYKTIMDEEDEEEEDMKEAFNVFDQNGDGFITVDELKAVLSSLGLKQGKTLDDCKK 120
Query: 120 MIKKVDVDGDGMVNFKEFQQMMKAGAF 146
MIKKVDVDGDG VN+KEF+QMMK G F
Sbjct: 121 MIKKVDVDGDGRVNYKEFRQMMKGGGF 147
>gb|AAR96010.1| calmodulin-like protein [Musa acuminata]
Length = 210
Score = 228 bits (582), Expect = 2e-59
Identities = 108/150 (72%), Positives = 129/150 (86%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MD EL R+FQMFD+NGDGRITK EL+DSL+NLGI + E +L MIE+ID NGDG VD++
Sbjct: 61 MDPSELKRVFQMFDRNGDGRITKAELTDSLENLGILVPEAELASMIERIDANGDGCVDVE 120
Query: 61 EFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
EFG LY+TIMDE+DEEEDM+EAFNVFD+NGDGFI+ EEL +VL+SLGLK G+T EDC+ M
Sbjct: 121 EFGTLYRTIMDERDEEEDMREAFNVFDRNGDGFITVEELRSVLASLGLKQGRTAEDCRKM 180
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAGAFAADS 150
I +VDVDGDG+VNFKEF+QMMK G FAA S
Sbjct: 181 INEVDVDGDGVVNFKEFKQMMKGGGFAAPS 210
>emb|CAB78328.1| putative calmodulin [Arabidopsis thaliana]
gi|4586262|emb|CAB41003.1| putative calmodulin
[Arabidopsis thaliana] gi|67633742|gb|AAY78795.1|
putative calcium-binding protein [Arabidopsis thaliana]
gi|15235516|ref|NP_193022.1| calcium-binding protein,
putative [Arabidopsis thaliana] gi|7441488|pir||T06644
calmodulin homolog T20K18.210 - Arabidopsis thaliana
Length = 152
Score = 220 bits (561), Expect = 5e-57
Identities = 102/148 (68%), Positives = 127/148 (84%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MD+GEL+R+FQMFDKNGDG+I K EL D +++GI + E ++ +MI K+DVNGDG +DID
Sbjct: 1 MDRGELSRVFQMFDKNGDGKIAKNELKDFFKSVGIMVPENEINEMIAKMDVNGDGAMDID 60
Query: 61 EFGELYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
EFG LYQ +++EK+EEEDM+EAF VFDQNGDGFI+ EEL +VL+S+GLK G+TLEDCK M
Sbjct: 61 EFGSLYQEMVEEKEEEEDMREAFRVFDQNGDGFITDEELRSVLASMGLKQGRTLEDCKKM 120
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAGAFAA 148
I KVDVDGDGMVNFKEF+QMM+ G FAA
Sbjct: 121 ISKVDVDGDGMVNFKEFKQMMRGGGFAA 148
>emb|CAB77814.1| putative calmodulin [Arabidopsis thaliana]
gi|28827616|gb|AAO50652.1| putative calmodulin
[Arabidopsis thaliana] gi|28393101|gb|AAO41984.1|
putative calmodulin [Arabidopsis thaliana]
gi|15236276|ref|NP_192238.1| calcium-binding protein,
putative [Arabidopsis thaliana]
gi|4262157|gb|AAD14457.1| putative calmodulin
[Arabidopsis thaliana] gi|25295759|pir||G85041 probable
calmodulin [imported] - Arabidopsis thaliana
Length = 154
Score = 210 bits (535), Expect = 5e-54
Identities = 104/149 (69%), Positives = 126/149 (83%), Gaps = 3/149 (2%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
MD EL R+FQMFDK+GDG+IT KEL++S +NLGI I E++L Q+I+KIDVNGDG VDI+
Sbjct: 1 MDSTELNRVFQMFDKDGDGKITTKELNESFKNLGIIIPEDELTQIIQKIDVNGDGCVDIE 60
Query: 61 EFGELYQTIMDEKDEE---EDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDC 117
EFGELY+TIM E ++E EDMKEAFNVFD+NGDGFI+ +EL AVLSSLGLK GKTLE+C
Sbjct: 61 EFGELYKTIMVEDEDEVGEEDMKEAFNVFDRNGDGFITVDELKAVLSSLGLKQGKTLEEC 120
Query: 118 KNMIKKVDVDGDGMVNFKEFQQMMKAGAF 146
+ MI +VDVDGDG VN+ EF+QMMK G F
Sbjct: 121 RKMIMQVDVDGDGRVNYMEFRQMMKKGRF 149
>gb|AAM64648.1| putative calcium binding protein [Arabidopsis thaliana]
gi|20197147|gb|AAM14938.1| putative calcium binding
protein [Arabidopsis thaliana]
gi|20196862|gb|AAB64310.2| putative calcium binding
protein [Arabidopsis thaliana]
gi|20148491|gb|AAM10136.1| putative Ca2+-binding protein
[Arabidopsis thaliana] gi|17065478|gb|AAL32893.1|
putative Ca2+-binding protein [Arabidopsis thaliana]
gi|18406202|ref|NP_565996.1| calmodulin-like protein
(MSS3) [Arabidopsis thaliana] gi|9965747|gb|AAG10150.1|
calmodulin-like MSS3 [Arabidopsis thaliana]
Length = 215
Score = 209 bits (532), Expect = 1e-53
Identities = 103/155 (66%), Positives = 125/155 (80%), Gaps = 5/155 (3%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
+D EL R+FQMFDKNGDGRITK+EL+DSL+NLGI I ++DL QMI KID NGDG VDID
Sbjct: 61 IDPSELKRVFQMFDKNGDGRITKEELNDSLENLGIYIPDKDLTQMIHKIDANGDGCVDID 120
Query: 61 EFGELYQTIMDE-----KDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLE 115
EF LY +I+DE + EEEDMK+AFNVFDQ+GDGFI+ EEL +V++SLGLK GKTL+
Sbjct: 121 EFESLYSSIVDEHHNDGETEEEDMKDAFNVFDQDGDGFITVEELKSVMASLGLKQGKTLD 180
Query: 116 DCKNMIKKVDVDGDGMVNFKEFQQMMKAGAFAADS 150
CK MI +VD DGDG VN+KEF QMMK G F++ +
Sbjct: 181 GCKKMIMQVDADGDGRVNYKEFLQMMKGGGFSSSN 215
>pir||D84864 probable calcium binding protein [imported] - Arabidopsis thaliana
Length = 169
Score = 209 bits (532), Expect = 1e-53
Identities = 103/155 (66%), Positives = 125/155 (80%), Gaps = 5/155 (3%)
Query: 1 MDQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDID 60
+D EL R+FQMFDKNGDGRITK+EL+DSL+NLGI I ++DL QMI KID NGDG VDID
Sbjct: 15 IDPSELKRVFQMFDKNGDGRITKEELNDSLENLGIYIPDKDLTQMIHKIDANGDGCVDID 74
Query: 61 EFGELYQTIMDE-----KDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLE 115
EF LY +I+DE + EEEDMK+AFNVFDQ+GDGFI+ EEL +V++SLGLK GKTL+
Sbjct: 75 EFESLYSSIVDEHHNDGETEEEDMKDAFNVFDQDGDGFITVEELKSVMASLGLKQGKTLD 134
Query: 116 DCKNMIKKVDVDGDGMVNFKEFQQMMKAGAFAADS 150
CK MI +VD DGDG VN+KEF QMMK G F++ +
Sbjct: 135 GCKKMIMQVDADGDGRVNYKEFLQMMKGGGFSSSN 169
>emb|CAB91614.1| calmodulin-like protein [Arabidopsis thaliana]
gi|15231685|ref|NP_191503.1| calcium-binding protein,
putative [Arabidopsis thaliana] gi|11276990|pir||T49012
calmodulin-like protein - Arabidopsis thaliana
Length = 195
Score = 199 bits (507), Expect = 9e-51
Identities = 97/143 (67%), Positives = 124/143 (85%), Gaps = 1/143 (0%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
+L R+FQMFDKNGDGRITK+EL+DSL+NLGI + ++DL+QMI+K+D NGDG VDI+EF
Sbjct: 51 DLKRVFQMFDKNGDGRITKEELNDSLENLGIFMPDKDLIQMIQKMDANGDGCVDINEFES 110
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
LY +I++EK EE DM++AFNVFDQ+GDGFI+ EEL++V++SLGLK GKTLE CK MI +V
Sbjct: 111 LYGSIVEEK-EEGDMRDAFNVFDQDGDGFITVEELNSVMTSLGLKQGKTLECCKEMIMQV 169
Query: 125 DVDGDGMVNFKEFQQMMKAGAFA 147
D DGDG VN+KEF QMMK+G F+
Sbjct: 170 DEDGDGRVNYKEFLQMMKSGDFS 192
Score = 55.1 bits (131), Expect = 3e-07
Identities = 30/72 (41%), Positives = 45/72 (61%), Gaps = 4/72 (5%)
Query: 71 DEKDEEE--DMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDG 128
DE + E D+K F +FD+NGDG I+ EEL+ L +LG+ +D MI+K+D +G
Sbjct: 42 DESETESPVDLKRVFQMFDKNGDGRITKEELNDSLENLGIFMPD--KDLIQMIQKMDANG 99
Query: 129 DGMVNFKEFQQM 140
DG V+ EF+ +
Sbjct: 100 DGCVDINEFESL 111
Score = 45.4 bits (106), Expect = 3e-04
Identities = 23/69 (33%), Positives = 45/69 (64%), Gaps = 2/69 (2%)
Query: 2 DQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISE--EDLVQMIEKIDVNGDGYVDI 59
++G++ F +FD++GDG IT +EL+ + +LG+ + E +MI ++D +GDG V+
Sbjct: 120 EEGDMRDAFNVFDQDGDGFITVEELNSVMTSLGLKQGKTLECCKEMIMQVDEDGDGRVNY 179
Query: 60 DEFGELYQT 68
EF ++ ++
Sbjct: 180 KEFLQMMKS 188
>emb|CAE02048.2| OJ990528_30.6 [Oryza sativa (japonica cultivar-group)]
gi|21740787|emb|CAD41532.1| OSJNBb0091E11.1 [Oryza
sativa (japonica cultivar-group)]
gi|50925781|ref|XP_473002.1| OJ990528_30.6 [Oryza sativa
(japonica cultivar-group)]
Length = 196
Score = 182 bits (462), Expect = 1e-45
Identities = 89/160 (55%), Positives = 119/160 (73%), Gaps = 13/160 (8%)
Query: 2 DQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDE 61
D ELAR+F++FD+NGDGRIT++EL DSL LGI + ++L +I +ID NGDG VD++E
Sbjct: 35 DAAELARVFELFDRNGDGRITREELEDSLGKLGIPVPADELAAVIARIDANGDGCVDVEE 94
Query: 62 FGELYQTIM-------------DEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGL 108
FGELY++IM +E++E+ DM+EAF VFD NGDG+I+ +EL AVL+SLGL
Sbjct: 95 FGELYRSIMAGGDDSKDGRAKEEEEEEDGDMREAFRVFDANGDGYITVDELGAVLASLGL 154
Query: 109 KHGKTLEDCKNMIKKVDVDGDGMVNFKEFQQMMKAGAFAA 148
K G+T E+C+ MI +VD DGDG V+F EF QMM+ G FAA
Sbjct: 155 KQGRTAEECRRMIGQVDRDGDGRVDFHEFLQMMRGGGFAA 194
Score = 46.6 bits (109), Expect = 1e-04
Identities = 22/72 (30%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Query: 72 EKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGM 131
++ + ++ F +FD+NGDG I+ EEL L LG+ ++ +I ++D +GDG
Sbjct: 32 QQADAAELARVFELFDRNGDGRITREELEDSLGKLGIP--VPADELAAVIARIDANGDGC 89
Query: 132 VNFKEFQQMMKA 143
V+ +EF ++ ++
Sbjct: 90 VDVEEFGELYRS 101
>gb|AAM63501.1| touch-induced calmodulin-related protein TCH2 [Arabidopsis
thaliana] gi|10177164|dbj|BAB10353.1| calmodulin-related
protein 2, touch-induced [Arabidopsis thaliana]
gi|15240340|ref|NP_198593.1| touch-responsive protein /
calmodulin-related protein 2, touch-induced (TCH2)
[Arabidopsis thaliana] gi|3123295|sp|P25070|TCH2_ARATH
Calmodulin-related protein 2, touch-induced
gi|2583169|gb|AAB82713.1| calmodulin-related protein
[Arabidopsis thaliana]
Length = 161
Score = 128 bits (322), Expect = 2e-29
Identities = 63/147 (42%), Positives = 103/147 (69%), Gaps = 6/147 (4%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
++ ++FQ FDKNGDG+I+ EL + ++ L S E+ V M+++ D++G+G++D+DEF
Sbjct: 17 DIKKVFQRFDKNGDGKISVDELKEVIRALSPTASPEETVTMMKQFDLDGNGFIDLDEFVA 76
Query: 65 LYQTIM----DEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNM 120
L+Q + + +++ D+KEAF ++D +G+G IS +EL +V+ +LG K +++DCK M
Sbjct: 77 LFQIGIGGGGNNRNDVSDLKEAFELYDLDGNGRISAKELHSVMKNLGEK--CSVQDCKKM 134
Query: 121 IKKVDVDGDGMVNFKEFQQMMKAGAFA 147
I KVD+DGDG VNF EF++MM G A
Sbjct: 135 ISKVDIDGDGCVNFDEFKKMMSNGGGA 161
Score = 60.5 bits (145), Expect = 8e-09
Identities = 27/64 (42%), Positives = 44/64 (68%)
Query: 2 DQGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDE 61
D +L F+++D +G+GRI+ KEL ++NLG S +D +MI K+D++GDG V+ DE
Sbjct: 91 DVSDLKEAFELYDLDGNGRISAKELHSVMKNLGEKCSVQDCKKMISKVDIDGDGCVNFDE 150
Query: 62 FGEL 65
F ++
Sbjct: 151 FKKM 154
Score = 49.7 bits (117), Expect = 1e-05
Identities = 25/68 (36%), Positives = 42/68 (61%), Gaps = 2/68 (2%)
Query: 77 EDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMVNFKE 136
+D+K+ F FD+NGDG IS +EL V+ + L + E+ M+K+ D+DG+G ++ E
Sbjct: 16 DDIKKVFQRFDKNGDGKISVDELKEVIRA--LSPTASPEETVTMMKQFDLDGNGFIDLDE 73
Query: 137 FQQMMKAG 144
F + + G
Sbjct: 74 FVALFQIG 81
>gb|AAD34262.1| calmodulin mutant SYNCAM57D [synthetic construct]
Length = 149
Score = 127 bits (318), Expect = 7e-29
Identities = 65/139 (46%), Positives = 93/139 (66%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L +M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARVMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
>gb|AAD34436.1| calmodulin mutant SYNCAM33 [synthetic construct]
Length = 149
Score = 126 bits (316), Expect = 1e-28
Identities = 65/139 (46%), Positives = 93/139 (66%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G++D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGWIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARKMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
>gb|AAD34259.1| calmodulin mutant SYNCAM57A [synthetic construct]
Length = 149
Score = 125 bits (314), Expect = 2e-28
Identities = 65/139 (46%), Positives = 92/139 (65%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARAMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
Score = 59.3 bits (142), Expect = 2e-08
Identities = 32/82 (39%), Positives = 51/82 (62%), Gaps = 4/82 (4%)
Query: 71 DEKDEEE--DMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDG 128
D+ +E+ + KEAF++FD++GDG I+ +EL V+ SLG T + ++MI +VD DG
Sbjct: 3 DQLTDEQIAEFKEAFSLFDKDGDGTITTKELGTVMRSLG--QNPTEAELQDMINEVDADG 60
Query: 129 DGMVNFKEFQQMMKAGAFAADS 150
+G ++F EF +M DS
Sbjct: 61 NGTIDFPEFLNLMARAMKDTDS 82
>gb|AAD34269.1| calmodulin mutant SYNCAM71A [synthetic construct]
Length = 149
Score = 125 bits (313), Expect = 3e-28
Identities = 65/139 (46%), Positives = 92/139 (65%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARKMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMSA 148
>gb|AAA72492.1| VU1 calmodulin [synthetic construct] gi|208412|gb|AAA72766.1|
camodulin
Length = 149
Score = 124 bits (312), Expect = 4e-28
Identities = 65/139 (46%), Positives = 92/139 (65%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARKMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
>sp|P04352|CALM_CHLRE Calmodulin (CaM) gi|167411|gb|AAA33083.1| calmodulin
Length = 163
Score = 124 bits (312), Expect = 4e-28
Identities = 64/141 (45%), Positives = 94/141 (66%), Gaps = 2/141 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 15 EFKEAFALFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMISEVDADGNGTIDFPEFLM 74
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M E D E++++EAF VFD++G+GFIS EL V+++LG K + E+ MI++
Sbjct: 75 LMARKMKETDHEDELREAFKVFDKDGNGFISAAELRHVMTNLGEKLSE--EEVDEMIREA 132
Query: 125 DVDGDGMVNFKEFQQMMKAGA 145
DVDGDG VN++EF +MM +GA
Sbjct: 133 DVDGDGQVNYEEFVRMMTSGA 153
Score = 58.5 bits (140), Expect = 3e-08
Identities = 30/75 (40%), Positives = 50/75 (66%), Gaps = 1/75 (1%)
Query: 3 QGELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEF 62
+ EL F++FDK+G+G I+ EL + NLG +SEE++ +MI + DV+GDG V+ +EF
Sbjct: 86 EDELREAFKVFDKDGNGFISAAELRHVMTNLGEKLSEEEVDEMIREADVDGDGQVNYEEF 145
Query: 63 GELYQT-IMDEKDEE 76
+ + D+KD++
Sbjct: 146 VRMMTSGATDDKDKK 160
Score = 57.4 bits (137), Expect = 7e-08
Identities = 28/69 (40%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 73 KDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMV 132
+++ + KEAF +FD++GDG I+ +EL V+ SLG T + ++MI +VD DG+G +
Sbjct: 10 EEQIAEFKEAFALFDKDGDGTITTKELGTVMRSLG--QNPTEAELQDMISEVDADGNGTI 67
Query: 133 NFKEFQQMM 141
+F EF +M
Sbjct: 68 DFPEFLMLM 76
>gb|AAD34418.1| calmodulin mutant SYNCAM24 [synthetic construct]
Length = 149
Score = 124 bits (312), Expect = 4e-28
Identities = 65/139 (46%), Positives = 92/139 (65%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARKMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
Score = 58.5 bits (140), Expect = 3e-08
Identities = 29/69 (42%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 73 KDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKVDVDGDGMV 132
K + + KEAF++FD++GDG I+ +EL V+ SLG T + ++MI +VD DG+G +
Sbjct: 7 KKKIAEFKEAFSLFDKDGDGTITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 133 NFKEFQQMM 141
+F EF +M
Sbjct: 65 DFPEFLNLM 73
>gb|AAD34260.1| calmodulin mutant SYNCAM57B [synthetic construct]
Length = 149
Score = 124 bits (312), Expect = 4e-28
Identities = 65/139 (46%), Positives = 92/139 (65%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMAREMKDTDSEEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
>gb|AAD34244.1| calmodulin mutant SYNCAM30 [synthetic construct]
Length = 149
Score = 124 bits (312), Expect = 4e-28
Identities = 65/139 (46%), Positives = 92/139 (65%), Gaps = 2/139 (1%)
Query: 5 ELARIFQMFDKNGDGRITKKELSDSLQNLGICISEEDLVQMIEKIDVNGDGYVDIDEFGE 64
E F +FDK+GDG IT KEL +++LG +E +L MI ++D +G+G +D EF
Sbjct: 12 EFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 71
Query: 65 LYQTIMDEKDEEEDMKEAFNVFDQNGDGFISGEELSAVLSSLGLKHGKTLEDCKNMIKKV 124
L M + D EE++KEAF VFD++G+GFIS EL V+++LG K T E+ MI++
Sbjct: 72 LMARKMKDTDREEELKEAFRVFDKDGNGFISAAELRHVMTNLGEK--LTDEEVDEMIREA 129
Query: 125 DVDGDGMVNFKEFQQMMKA 143
DVDGDG VN++EF Q+M A
Sbjct: 130 DVDGDGQVNYEEFVQVMMA 148
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.137 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 253,805,708
Number of Sequences: 2540612
Number of extensions: 11055859
Number of successful extensions: 55067
Number of sequences better than 10.0: 4518
Number of HSP's better than 10.0 without gapping: 2777
Number of HSP's successfully gapped in prelim test: 1753
Number of HSP's that attempted gapping in prelim test: 37830
Number of HSP's gapped (non-prelim): 11544
length of query: 152
length of database: 863,360,394
effective HSP length: 128
effective length of query: 24
effective length of database: 538,162,058
effective search space: 12915889392
effective search space used: 12915889392
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122169.9


