

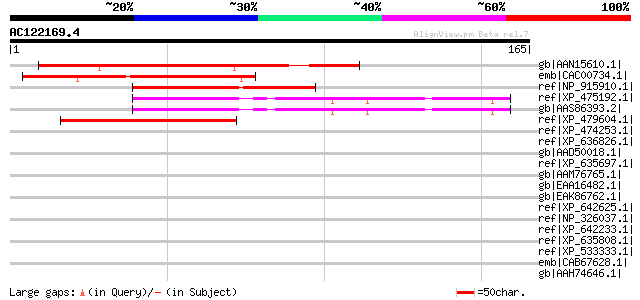
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.4 - phase: 0
(165 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN15610.1| unknown protein [Arabidopsis thaliana] gi|2046663... 75 9e-13
emb|CAC00734.1| SigA binding protein [Arabidopsis thaliana] gi|2... 67 2e-10
ref|NP_915910.1| P0468B07.6 [Oryza sativa (japonica cultivar-gro... 62 4e-09
ref|XP_475192.1| hypothetical protein [Oryza sativa (japonica cu... 61 1e-08
gb|AAS86393.2| hypothetical protein [Oryza sativa (japonica cult... 61 1e-08
ref|XP_479604.1| unknown protein [Oryza sativa (japonica cultiva... 45 7e-04
ref|XP_474253.1| OSJNBa0087O24.11 [Oryza sativa (japonica cultiv... 40 0.018
ref|XP_636826.1| hypothetical protein DDB0187816 [Dictyostelium ... 39 0.052
gb|AAD50018.1| Hypothetical Protein [Arabidopsis thaliana] gi|25... 39 0.068
ref|XP_635697.1| hypothetical protein DDB0189000 [Dictyostelium ... 38 0.12
gb|AAM76765.1| hypothetical protein [Arabidopsis thaliana] gi|61... 37 0.15
gb|EAA16482.1| hypothetical protein [Plasmodium yoelii yoelii] 37 0.26
gb|EAK86762.1| hypothetical protein UM05817.1 [Ustilago maydis 5... 36 0.34
ref|XP_642625.1| hypothetical protein DDB0169228 [Dictyostelium ... 36 0.44
ref|NP_326037.1| LIPOPROTEIN [Mycoplasma pulmonis UAB CTIP] gi|1... 35 0.75
ref|XP_642233.1| RNA polymerase II elongation factor [Dictyostel... 35 0.75
ref|XP_635808.1| hypothetical protein DDB0219551 [Dictyostelium ... 35 0.75
ref|XP_533333.1| PREDICTED: hypothetical protein XP_533333 [Cani... 35 0.98
emb|CAB67628.1| putative protein [Arabidopsis thaliana] gi|15230... 35 0.98
gb|AAH74646.1| Serine/arginine repetitive matrix 1 [Xenopus trop... 35 0.98
>gb|AAN15610.1| unknown protein [Arabidopsis thaliana] gi|20466638|gb|AAM20636.1|
unknown protein [Arabidopsis thaliana]
gi|3402700|gb|AAD11994.1| expressed protein [Arabidopsis
thaliana] gi|16323165|gb|AAL15317.1| At2g41180/T3K9.5
[Arabidopsis thaliana] gi|18405627|ref|NP_030663.1|
sigA-binding protein-related [Arabidopsis thaliana]
gi|7487632|pir||T02101 hypothetical protein At2g41180
[imported] - Arabidopsis thaliana
Length = 141
Score = 74.7 bits (182), Expect = 9e-13
Identities = 46/104 (44%), Positives = 65/104 (62%), Gaps = 8/104 (7%)
Query: 10 SSSVISTVQQKTPTKLTK-PKKKKNNNNTYNKPIKVVYISNPMKVKTSASEFRALVQELT 68
SSS + Q+K+ + T+ P K+K + T +KPIKV YISNPM+V+T S+FR LVQELT
Sbjct: 4 SSSTLLINQRKSSSSPTRIPPKQKRKSTTTHKPIKVRYISNPMRVETCPSKFRELVQELT 63
Query: 69 GQ-YAESPPNPSRFQEFDVNDSGTDQGGCENMMDCDKSDQTVVG 111
GQ A+ PP+P+ F D++ CE+ M+ + D V G
Sbjct: 64 GQDAADLPPSPTTFTAVDLHRP------CESEMNLEPLDGEVRG 101
>emb|CAC00734.1| SigA binding protein [Arabidopsis thaliana]
gi|27312003|gb|AAO00967.1| SigA binding protein
[Arabidopsis thaliana] gi|6980074|gb|AAF34713.1| SigA
binding protein [Arabidopsis thaliana]
gi|14596087|gb|AAK68771.1| SigA binding protein
[Arabidopsis thaliana] gi|15228994|ref|NP_191230.1|
sigA-binding protein [Arabidopsis thaliana]
gi|11289616|pir||T51259 SigA binding protein -
Arabidopsis thaliana
Length = 151
Score = 67.0 bits (162), Expect = 2e-10
Identities = 41/76 (53%), Positives = 52/76 (67%), Gaps = 3/76 (3%)
Query: 5 STSSTSSSVISTVQQK-TPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEFRAL 63
S+SST + S ++K +P PK+KK +T NKPIKV YISNPM+V+T AS+FR L
Sbjct: 3 SSSSTFLTTTSLDKKKPSPVSRKSPKQKKKTTST-NKPIKVRYISNPMRVQTCASKFREL 61
Query: 64 VQELTGQYA-ESPPNP 78
VQELTGQ A + P P
Sbjct: 62 VQELTGQDAVDLQPEP 77
>ref|NP_915910.1| P0468B07.6 [Oryza sativa (japonica cultivar-group)]
gi|20160684|dbj|BAB89627.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 184
Score = 62.4 bits (150), Expect = 4e-09
Identities = 36/58 (62%), Positives = 41/58 (70%), Gaps = 1/58 (1%)
Query: 40 KPIKVVYISNPMKVKTSASEFRALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCE 97
KPIKVVYISNPM+VKTSA+ FRALVQELTG+ A+ P S D +D G GG E
Sbjct: 37 KPIKVVYISNPMRVKTSAAGFRALVQELTGRNAD-PSKYSPRASADDDDGGGGGGGGE 93
>ref|XP_475192.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 216
Score = 60.8 bits (146), Expect = 1e-08
Identities = 45/129 (34%), Positives = 68/129 (51%), Gaps = 17/129 (13%)
Query: 40 KPIKVVYISNPMKVKTSASEFRALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCENM 99
KPIKVVYISNPM+V+TSA+ FRALVQELTG+ A+ PS++ +G D GG +
Sbjct: 89 KPIKVVYISNPMRVRTSAAGFRALVQELTGRNAD----PSKYS--PRASAGDDGGGATAL 142
Query: 100 MD---CDKSDQTVVGV-----PSLVDPDDKGKPSEAGSSNESFDEDVLLMPEMMDNIWDL 151
D +D G P+ D+G G ++ D+D + +++D + +
Sbjct: 143 PDTGAASDADALEAGAAPGRHPAETATFDEGGGGGGGGYDD--DDDDVFRSQLLDTSYSV 200
Query: 152 L-PTSAFYE 159
P + Y+
Sbjct: 201 FSPPTLLYD 209
>gb|AAS86393.2| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 168
Score = 60.8 bits (146), Expect = 1e-08
Identities = 45/129 (34%), Positives = 68/129 (51%), Gaps = 17/129 (13%)
Query: 40 KPIKVVYISNPMKVKTSASEFRALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCENM 99
KPIKVVYISNPM+V+TSA+ FRALVQELTG+ A+ PS++ +G D GG +
Sbjct: 41 KPIKVVYISNPMRVRTSAAGFRALVQELTGRNAD----PSKYS--PRASAGDDGGGATAL 94
Query: 100 MD---CDKSDQTVVGV-----PSLVDPDDKGKPSEAGSSNESFDEDVLLMPEMMDNIWDL 151
D +D G P+ D+G G ++ D+D + +++D + +
Sbjct: 95 PDTGAASDADALEAGAAPGRHPAETATFDEGGGGGGGGYDD--DDDDVFRSQLLDTSYSV 152
Query: 152 L-PTSAFYE 159
P + Y+
Sbjct: 153 FSPPTLLYD 161
>ref|XP_479604.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|50509155|dbj|BAD30295.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|22324437|dbj|BAC10354.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 137
Score = 45.1 bits (105), Expect = 7e-04
Identities = 22/56 (39%), Positives = 34/56 (60%)
Query: 17 VQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEFRALVQELTGQYA 72
+ + P K K +K ++VVYI++PMK+ S EFRA+VQELTG+++
Sbjct: 14 ISKPPPAKKGKAAARKYKPPQCPGAVRVVYIASPMKLTASPEEFRAVVQELTGRHS 69
>ref|XP_474253.1| OSJNBa0087O24.11 [Oryza sativa (japonica cultivar-group)]
gi|32488661|emb|CAE03588.1| OSJNBa0087O24.11 [Oryza
sativa (japonica cultivar-group)]
Length = 151
Score = 40.4 bits (93), Expect = 0.018
Identities = 23/56 (41%), Positives = 32/56 (57%), Gaps = 1/56 (1%)
Query: 40 KPIKVVYISNPMKVKTSASEFRALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGG 95
+ IK+V++ P +KT A FR LVQ LTG+ A P + Q D D+ D+GG
Sbjct: 26 RKIKIVHVLAPEVIKTDARHFRDLVQRLTGKPAADGPAAASSQP-DPCDTAGDEGG 80
>ref|XP_636826.1| hypothetical protein DDB0187816 [Dictyostelium discoideum]
gi|60465230|gb|EAL63324.1| hypothetical protein
DDB0187816 [Dictyostelium discoideum]
Length = 450
Score = 38.9 bits (89), Expect = 0.052
Identities = 25/70 (35%), Positives = 35/70 (49%), Gaps = 5/70 (7%)
Query: 3 SISTSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEFRA 62
S ST+ST+++ +T +TPT T P NNNN N + I++P KT +
Sbjct: 348 SPSTTSTTTTTTTTTSPQTPTNATSPVNNNNNNNNNNNNTRPP-INSPPVAKTP----QP 402
Query: 63 LVQELTGQYA 72
Q T QYA
Sbjct: 403 TPQPTTNQYA 412
>gb|AAD50018.1| Hypothetical Protein [Arabidopsis thaliana] gi|25518484|pir||D86307
hypothetical protein F20D23.16 - Arabidopsis thaliana
Length = 89
Score = 38.5 bits (88), Expect = 0.068
Identities = 36/110 (32%), Positives = 47/110 (42%), Gaps = 26/110 (23%)
Query: 42 IKVVYISNPMKVKTSASEFRALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCENMMD 101
+KVV+I N V+T A F+ +VQELTG+ A P F SG GG +
Sbjct: 1 MKVVFI-NTQYVQTDARSFKTIVQELTGKNAVVADGPYEF-------SGQGYGGKD---- 48
Query: 102 CDKSDQTVVGVPSLVDPDDKGKPSEAGSSNESFDEDVLLMPEM--MDNIW 149
S Q GV GK +E G FD MP + + N+W
Sbjct: 49 ---SSQRFCGV---------GKEAERGVETTEFDSFFREMPPVGELYNLW 86
>ref|XP_635697.1| hypothetical protein DDB0189000 [Dictyostelium discoideum]
gi|60464007|gb|EAL62170.1| hypothetical protein
DDB0189000 [Dictyostelium discoideum]
Length = 720
Score = 37.7 bits (86), Expect = 0.12
Identities = 34/136 (25%), Positives = 56/136 (41%), Gaps = 17/136 (12%)
Query: 3 SISTSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEFRA 62
S+S+SS+SSS S+ +KTP+ LT K K N ++T N SNP+ +S
Sbjct: 62 SLSSSSSSSSTTSSTVKKTPSTLTANKSKTNLSSTSNN--STTLKSNPLMSDRLSST--- 116
Query: 63 LVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCENMMDCDK-----SDQTVVGVPSLVD 117
+ + P P + + + + + + K S TV + D
Sbjct: 117 -------SLSSTFPGPRKSTTGGTSSTSSTSNSASSTLSSKKLTDKLSTPTVASMMKKTD 169
Query: 118 PDDKGKPSEAGSSNES 133
PS++ SS+ S
Sbjct: 170 SHHHTLPSKSSSSSSS 185
>gb|AAM76765.1| hypothetical protein [Arabidopsis thaliana]
gi|61742669|gb|AAX55155.1| hypothetical protein
At2g42140 [Arabidopsis thaliana]
gi|2673905|gb|AAB88639.1| hypothetical protein
[Arabidopsis thaliana] gi|18491279|gb|AAL69464.1|
At2g42140/T24P15.5 [Arabidopsis thaliana]
gi|15227891|ref|NP_181744.1| VQ motif-containing protein
[Arabidopsis thaliana] gi|7487417|pir||T00924
hypothetical protein At2g42140 [imported] - Arabidopsis
thaliana
Length = 172
Score = 37.4 bits (85), Expect = 0.15
Identities = 36/137 (26%), Positives = 57/137 (41%), Gaps = 21/137 (15%)
Query: 15 STVQQKTPTKLTKPKKKKNNNNTYNKP-IKVVYISNPMKVKTSASEFRALVQELTGQ--- 70
+T QK + T K++ T +KP I++++I P +KT + FR +VQ LTG+
Sbjct: 5 ATTVQKRRSLPTIAMHKQSRTLTKSKPKIRIIHIFAPEIIKTDVANFREIVQNLTGKQDH 64
Query: 71 ----------YAESPPNPSRFQEFDVNDSGTDQGGCENMMDCDKSDQTVVGVPSLVDPDD 120
+P + +V+D G C N SD+ G+ S+ +
Sbjct: 65 HHHDLPHQKGLKRNPRSRRSHDHHEVHDMNKSHGFCIN------SDEEEEGMVSMT-WNG 117
Query: 121 KGKPSEAGSSNESFDED 137
G S G N D D
Sbjct: 118 NGDESSGGFLNGLGDLD 134
>gb|EAA16482.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1048
Score = 36.6 bits (83), Expect = 0.26
Identities = 33/129 (25%), Positives = 57/129 (43%), Gaps = 19/129 (14%)
Query: 3 SISTSST---SSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNP-------MK 52
SIS++S+ SS + + K + + + NNN YNK KV+ I +
Sbjct: 783 SISSNSSFLSDSSFFTNGDNENERKRKRSENESPNNNFYNKKNKVIEIKKRRPTVIDFLN 842
Query: 53 VKTSASEFRALVQELTGQYAESPPN--------PSRFQEFDVNDSGTDQGGC-ENMMDCD 103
K + + V+ + +Y E+ P+ S E D+NDS + C N+++ +
Sbjct: 843 EKWNKNSETQNVRSYSKEYEENEPHENSCNTNLESIDSETDINDSEIESSECSSNIIENE 902
Query: 104 KSDQTVVGV 112
K D+T V
Sbjct: 903 KEDKTEFSV 911
>gb|EAK86762.1| hypothetical protein UM05817.1 [Ustilago maydis 521]
gi|49079612|ref|XP_403432.1| hypothetical protein
UM05817.1 [Ustilago maydis 521]
Length = 439
Score = 36.2 bits (82), Expect = 0.34
Identities = 31/133 (23%), Positives = 49/133 (36%), Gaps = 3/133 (2%)
Query: 1 MDSISTSSTSSSVISTVQQKTPTKLT--KPKKKK-NNNNTYNKPIKVVYISNPMKVKTSA 57
+D +S+ + S S+ S V P+ L KP K+K + T KP K S T A
Sbjct: 113 LDDLSSIAPSPSIASEVSTSKPSSLKAKKPSKQKADGTETKPKPKKPKKKSTTGAATTDA 172
Query: 58 SEFRALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCENMMDCDKSDQTVVGVPSLVD 117
+ F + Q A + P P + + +D + + P
Sbjct: 173 TAFSSSAQPSNVTAASTAPKPKKTSKNKASDKDKSKEEAHQQPTSTAASPAPFATPVSSS 232
Query: 118 PDDKGKPSEAGSS 130
+ K K +AG S
Sbjct: 233 KESKPKAKKAGGS 245
>ref|XP_642625.1| hypothetical protein DDB0169228 [Dictyostelium discoideum]
gi|21281373|gb|AAM45260.1| similar to Plasmodium
falciparum. Hypothetical protein [Dictyostelium
discoideum] gi|60470698|gb|EAL68672.1| hypothetical
protein DDB0169228 [Dictyostelium discoideum]
Length = 2625
Score = 35.8 bits (81), Expect = 0.44
Identities = 18/87 (20%), Positives = 39/87 (44%)
Query: 1 MDSISTSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEF 60
+ S S +S+SSS++S + + KL N+N +Y+ P+++ + + +
Sbjct: 515 ISSNSPNSSSSSIVSNLSSNSLNKLVNSNTNNNSNQSYSPPLQIQQQQQQQQQQQQQQQQ 574
Query: 61 RALVQELTGQYAESPPNPSRFQEFDVN 87
+ Q+ Q + + EF+ N
Sbjct: 575 QQQQQQQQQQQQQQQQQQQQINEFNCN 601
>ref|NP_326037.1| LIPOPROTEIN [Mycoplasma pulmonis UAB CTIP]
gi|14089619|emb|CAC13379.1| LIPOPROTEIN [Mycoplasma
pulmonis] gi|25393284|pir||F90537 lipoprotein [imported]
- Mycoplasma pulmonis (strain UAB CTIP)
Length = 773
Score = 35.0 bits (79), Expect = 0.75
Identities = 34/139 (24%), Positives = 55/139 (39%), Gaps = 10/139 (7%)
Query: 1 MDSISTSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEF 60
M +S +T S+ K TK T P+ NNT ++ NP KT ++
Sbjct: 21 MALVSCVTTKSNNQIDPSAKQNTKQTSPQSAPKENNTNTNRNSIISPQNPDSSKTPETQV 80
Query: 61 RALVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCE--NMMDCDKSDQTVVGVPSLVDP 118
E + +SP +P ++ D+G G + + D + DQ VP P
Sbjct: 81 PPTKPEDQNKEPQSPKDP------EIKDNGQKNEGSKAPEIKDMSQKDQ-APQVPQ-KQP 132
Query: 119 DDKGKPSEAGSSNESFDED 137
+D KP +S D++
Sbjct: 133 EDPKKPETQKPPVKSEDQN 151
>ref|XP_642233.1| RNA polymerase II elongation factor [Dictyostelium discoideum]
gi|60470313|gb|EAL68293.1| RNA polymerase II elongation
factor [Dictyostelium discoideum]
Length = 319
Score = 35.0 bits (79), Expect = 0.75
Identities = 24/75 (32%), Positives = 37/75 (49%), Gaps = 6/75 (8%)
Query: 5 STSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVY------ISNPMKVKTSAS 58
S+SS+SSS ST KT + K+K + +T ++P IS P KTS+
Sbjct: 85 SSSSSSSSTTSTTTTKTASPSESLKRKSISEDTSDRPTSKPLLQENKKISPPTTPKTSSP 144
Query: 59 EFRALVQELTGQYAE 73
+L+ +TG A+
Sbjct: 145 PIASLIAPITGANAD 159
>ref|XP_635808.1| hypothetical protein DDB0219551 [Dictyostelium discoideum]
gi|60464150|gb|EAL62311.1| hypothetical protein
DDB0219551 [Dictyostelium discoideum]
Length = 1886
Score = 35.0 bits (79), Expect = 0.75
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query: 3 SISTSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMK 52
S +SS+SSS +S++QQ +T P NNNN N PI + P K
Sbjct: 1381 SSPSSSSSSSSLSSLQQ-----ITSPLTNNNNNNNNNIPISAITPETPKK 1425
>ref|XP_533333.1| PREDICTED: hypothetical protein XP_533333 [Canis familiaris]
Length = 558
Score = 34.7 bits (78), Expect = 0.98
Identities = 44/158 (27%), Positives = 64/158 (39%), Gaps = 38/158 (24%)
Query: 5 STSSTSSSVISTVQQKTP-TKLTKP------KKKKNNNNTYNKPIKVVYISN-----PMK 52
S S++SSSV S ++ K P T KP KK+K+ + PI S MK
Sbjct: 295 SQSASSSSVTSPIKMKIPITNTEKPEKYMADKKEKSGSLKLRIPIPPTDKSASKEELKMK 354
Query: 53 VKTSASEFRALVQELTGQYAESPPNPSRFQEFDVND-----------------SGTDQGG 95
+K S+SE + E +G+ S P+ SR + D SG+ GG
Sbjct: 355 IKVSSSERHSSSDEGSGKSKHSSPHISRDHKEKHKDHPSNRHHPSSHKHSHSHSGSSSGG 414
Query: 96 CENMMDCDKSDQTVVGVPSLVDPDDKGKPSEAGSSNES 133
++ D G+P V G S+ SS+ S
Sbjct: 415 SKHSAD---------GIPPTVLRSPVGLSSDGISSSSS 443
>emb|CAB67628.1| putative protein [Arabidopsis thaliana]
gi|15230913|ref|NP_191359.1| VQ motif-containing
protein [Arabidopsis thaliana] gi|11281932|pir||T46022
hypothetical protein T10K17.210 - Arabidopsis thaliana
Length = 175
Score = 34.7 bits (78), Expect = 0.98
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 10/70 (14%)
Query: 1 MDSISTSSTSSSVISTVQQKTPTKLTKPKKKKNNNNTYNKPIKVVYISNPMKVKTSASEF 60
M++ SS+ S K +TK K K I++++I P +KT + F
Sbjct: 1 MEATIFEKRRSSLSSIAVHKQSYSITKSKPK----------IRIIHIFAPEIIKTDVANF 50
Query: 61 RALVQELTGQ 70
R LVQ LTG+
Sbjct: 51 RELVQSLTGK 60
>gb|AAH74646.1| Serine/arginine repetitive matrix 1 [Xenopus tropicalis]
gi|54020799|ref|NP_001005645.1| serine/arginine
repetitive matrix 1 [Xenopus tropicalis]
Length = 874
Score = 34.7 bits (78), Expect = 0.98
Identities = 38/152 (25%), Positives = 69/152 (45%), Gaps = 27/152 (17%)
Query: 5 STSSTSSSVISTVQQKTPTKLTK--PKKKKNNNNTYNKPIKVVY---------------- 46
S+SS+SSS +K P +++ P+K + ++ + + P + Y
Sbjct: 351 SSSSSSSSRSPPPPKKQPKRISSSPPRKPRRSSPSSSPPPRRRYRPSPTPPPKRRSPSPT 410
Query: 47 -ISNPMKVKTSASEFRA-LVQELTGQYAESPPNPSRFQEFDVNDSGTDQGGCENMMDCDK 104
S P K++ S S R +++ + +SP PS+ ++ D+++S D+GG M D
Sbjct: 411 QSSRPRKIRGSVSPARPPVIKHKVIEKRKSPSPPSKPRKRDLSESDDDKGG--RMAPPDS 468
Query: 105 SDQTVVGVPSLVDPDDKGKPSEAGSSNESFDE 136
Q ++ SE+GSS+ S DE
Sbjct: 469 VQQR-----RQYRRQNQQSSSESGSSSSSEDE 495
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.306 0.125 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 295,892,515
Number of Sequences: 2540612
Number of extensions: 12881116
Number of successful extensions: 58150
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 97
Number of HSP's that attempted gapping in prelim test: 57995
Number of HSP's gapped (non-prelim): 205
length of query: 165
length of database: 863,360,394
effective HSP length: 118
effective length of query: 47
effective length of database: 563,568,178
effective search space: 26487704366
effective search space used: 26487704366
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC122169.4


