

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122165.6 + phase: 0
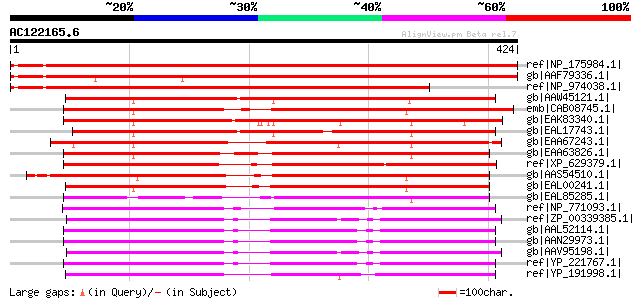
(424 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_175984.1| pyridoxal-5'-phosphate-dependent enzyme, beta f... 638 0.0
gb|AAF79336.1| F14J16.13 [Arabidopsis thaliana] 623 e-177
ref|NP_974038.1| pyridoxal-5'-phosphate-dependent enzyme, beta f... 518 e-145
gb|AAW45121.1| cysteine synthase, putative [Cryptococcus neoform... 365 1e-99
emb|CAB08745.1| SPAC3A12.17c [Schizosaccharomyces pombe] gi|1911... 355 2e-96
gb|EAK83340.1| hypothetical protein UM02218.1 [Ustilago maydis 5... 346 7e-94
gb|EAL17743.1| hypothetical protein CNBL2560 [Cryptococcus neofo... 345 2e-93
gb|EAA67243.1| hypothetical protein FG00903.1 [Gibberella zeae P... 337 4e-91
gb|EAA63826.1| hypothetical protein AN1513.2 [Aspergillus nidula... 334 3e-90
ref|XP_629379.1| cysteine synthase [Dictyostelium discoideum] gi... 330 4e-89
gb|AAS54510.1| AGR021Cp [Ashbya gossypii ATCC 10895] gi|45201116... 314 3e-84
gb|EAL00241.1| hypothetical protein CaO19.5574 [Candida albicans... 308 3e-82
gb|EAL85285.1| cysteine synthase, putative [Aspergillus fumigatu... 283 7e-75
ref|NP_771093.1| cysteine synthase [Bradyrhizobium japonicum USD... 280 5e-74
ref|ZP_00339385.1| COG0031: Cysteine synthase [Silicibacter sp. ... 268 2e-70
gb|AAL52114.1| CYSTEINE SYNTHASE [Brucella melitensis 16M] gi|17... 258 2e-67
gb|AAN29973.1| cysteine synthase [Brucella suis 1330] gi|2350193... 258 3e-67
gb|AAV95198.1| pyridoxal-phosphate dependent enzyme [Silicibacte... 256 1e-66
ref|YP_221767.1| cysteine synthase [Brucella abortus biovar 1 st... 256 1e-66
ref|YP_191998.1| Cysteine synthase A (O-acetylserine sulfhydryla... 248 3e-64
>ref|NP_175984.1| pyridoxal-5'-phosphate-dependent enzyme, beta family protein
[Arabidopsis thaliana] gi|46518441|gb|AAS99702.1|
At1g55880 [Arabidopsis thaliana]
Length = 421
Score = 638 bits (1646), Expect = 0.0
Identities = 321/425 (75%), Positives = 364/425 (85%), Gaps = 5/425 (1%)
Query: 1 MAPVAARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRI 60
MAPV GAV A+ + ++FF RL K+K+K +NG++DAIGNTPLIRI
Sbjct: 1 MAPV--NMTGAVVAAAALLMLTSYSFFF--RLSEKKKRKEKLTMRNGLVDAIGNTPLIRI 56
Query: 61 NSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGSTAISI 120
NSLS+ATGCEILGKCEFLNPGGSVKDRVAV+II+EALESG+L GGIVTEGSAGSTAIS+
Sbjct: 57 NSLSEATGCEILGKCEFLNPGGSVKDRVAVKIIQEALESGKLFPGGIVTEGSAGSTAISL 116
Query: 121 ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEAN 180
ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGA+VERVRPVSITHKDH+VNIARRRA EAN
Sbjct: 117 ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGASVERVRPVSITHKDHYVNIARRRADEAN 176
Query: 181 EFAFKHRK-SQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFENLANFRAHYEGT 239
E A K R S+ G ++ NGC + K +LF + GGFFADQFENLAN+RAHYEGT
Sbjct: 177 ELASKRRLGSETNGIHQEKTNGCTVEEVKEPSLFSDSVTGGFFADQFENLANYRAHYEGT 236
Query: 240 GPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVM 299
GPEIW QT GN+DAFVAAAGTGGT+AGVS+FLQ+KN +KC+LIDPPGSGL+NKV RGVM
Sbjct: 237 GPEIWHQTQGNIDAFVAAAGTGGTLAGVSRFLQDKNERVKCFLIDPPGSGLYNKVTRGVM 296
Query: 300 YTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGL 359
YT+EEAEGRRLKNPFDTITEGIGINR+TKNF AKLDG FRGTD EAVEM+RFL+KNDGL
Sbjct: 297 YTREEAEGRRLKNPFDTITEGIGINRLTKNFLMAKLDGGFRGTDKEAVEMSRFLLKNDGL 356
Query: 360 FLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEYLSQLGLTPKATGLE 419
F+GSSSAMNCVGAVR+AQ++GPGHTIVTILCDSGMRHLSKF++ +YL+ GL+P A GLE
Sbjct: 357 FVGSSSAMNCVGAVRVAQTLGPGHTIVTILCDSGMRHLSKFHDPKYLNLYGLSPTAIGLE 416
Query: 420 FLGIK 424
FLGIK
Sbjct: 417 FLGIK 421
>gb|AAF79336.1| F14J16.13 [Arabidopsis thaliana]
Length = 439
Score = 623 bits (1606), Expect = e-177
Identities = 321/443 (72%), Positives = 364/443 (81%), Gaps = 23/443 (5%)
Query: 1 MAPVAARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRI 60
MAPV GAV A+ + ++FF RL K+K+K +NG++DAIGNTPLIRI
Sbjct: 1 MAPV--NMTGAVVAAAALLMLTSYSFFF--RLSEKKKRKEKLTMRNGLVDAIGNTPLIRI 56
Query: 61 NSLSDATGCE----ILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGST 116
NSLS+ATGCE ILGKCEFLNPGGSVKDRVAV+II+EALESG+L GGIVTEGSAGST
Sbjct: 57 NSLSEATGCEVFLDILGKCEFLNPGGSVKDRVAVKIIQEALESGKLFPGGIVTEGSAGST 116
Query: 117 AISIATVAPAYGCKCHVVIPDDAAIEK--------------SQIIEALGATVERVRPVSI 162
AIS+ATVAPAYGCKCHVVIPDDAAIEK SQIIEALGA+VERVRPVSI
Sbjct: 117 AISLATVAPAYGCKCHVVIPDDAAIEKVISFKYLDDDSNSLSQIIEALGASVERVRPVSI 176
Query: 163 THKDHFVNIARRRASEANEFAFKHRK-SQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGF 221
THKDH+VNIARRRA EANE A K R S+ G ++ NGC + K +LF + GGF
Sbjct: 177 THKDHYVNIARRRADEANELASKRRLGSETNGIHQEKTNGCTVEEVKEPSLFSDSVTGGF 236
Query: 222 FADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCY 281
FADQFENLAN+RAHYEGTGPEIW QT GN+DAFVAAAGTGGT+AGVS+FLQ+KN +KC+
Sbjct: 237 FADQFENLANYRAHYEGTGPEIWHQTQGNIDAFVAAAGTGGTLAGVSRFLQDKNERVKCF 296
Query: 282 LIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRG 341
LIDPPGSGL+NKV RGVMYT+EEAEGRRLKNPFDTITEGIGINR+TKNF AKLDG FRG
Sbjct: 297 LIDPPGSGLYNKVTRGVMYTREEAEGRRLKNPFDTITEGIGINRLTKNFLMAKLDGGFRG 356
Query: 342 TDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFY 401
TD EAVEM+RFL+KNDGLF+GSSSAMNCVGAVR+AQ++GPGHTIVTILCDSGMRHLSKF+
Sbjct: 357 TDKEAVEMSRFLLKNDGLFVGSSSAMNCVGAVRVAQTLGPGHTIVTILCDSGMRHLSKFH 416
Query: 402 NAEYLSQLGLTPKATGLEFLGIK 424
+ +YL+ GL+P A GLEFLGIK
Sbjct: 417 DPKYLNLYGLSPTAIGLEFLGIK 439
>ref|NP_974038.1| pyridoxal-5'-phosphate-dependent enzyme, beta family protein
[Arabidopsis thaliana]
Length = 348
Score = 518 bits (1334), Expect = e-145
Identities = 263/352 (74%), Positives = 296/352 (83%), Gaps = 5/352 (1%)
Query: 1 MAPVAARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRI 60
MAPV GAV A+ + ++FF RL K+K+K +NG++DAIGNTPLIRI
Sbjct: 1 MAPV--NMTGAVVAAAALLMLTSYSFFF--RLSEKKKRKEKLTMRNGLVDAIGNTPLIRI 56
Query: 61 NSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGSTAISI 120
NSLS+ATGCEILGKCEFLNPGGSVKDRVAV+II+EALESG+L GGIVTEGSAGSTAIS+
Sbjct: 57 NSLSEATGCEILGKCEFLNPGGSVKDRVAVKIIQEALESGKLFPGGIVTEGSAGSTAISL 116
Query: 121 ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEAN 180
ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGA+VERVRPVSITHKDH+VNIARRRA EAN
Sbjct: 117 ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGASVERVRPVSITHKDHYVNIARRRADEAN 176
Query: 181 EFAFKHR-KSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFENLANFRAHYEGT 239
E A K R S+ G ++ NGC + K +LF + GGFFADQFENLAN+RAHYEGT
Sbjct: 177 ELASKRRLGSETNGIHQEKTNGCTVEEVKEPSLFSDSVTGGFFADQFENLANYRAHYEGT 236
Query: 240 GPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVM 299
GPEIW QT GN+DAFVAAAGTGGT+AGVS+FLQ+KN +KC+LIDPPGSGL+NKV RGVM
Sbjct: 237 GPEIWHQTQGNIDAFVAAAGTGGTLAGVSRFLQDKNERVKCFLIDPPGSGLYNKVTRGVM 296
Query: 300 YTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAVEMAR 351
YT+EEAEGRRLKNPFDTITEGIGINR+TKNF AKLDG FRGTD EAVEM+R
Sbjct: 297 YTREEAEGRRLKNPFDTITEGIGINRLTKNFLMAKLDGGFRGTDKEAVEMSR 348
>gb|AAW45121.1| cysteine synthase, putative [Cryptococcus neoformans var.
neoformans JEC21] gi|58270544|ref|XP_572428.1| cysteine
synthase, putative [Cryptococcus neoformans var.
neoformans JEC21]
Length = 458
Score = 365 bits (937), Expect = 1e-99
Identities = 197/367 (53%), Positives = 254/367 (68%), Gaps = 9/367 (2%)
Query: 47 GIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQL--RR 104
G+ IGNTPLIRINSLSDA G EILGK EFLNPGGSVKDRVA+QIIE+A G L
Sbjct: 76 GVTGLIGNTPLIRINSLSDALGVEILGKAEFLNPGGSVKDRVALQIIEDAEAQGLLYPNT 135
Query: 105 GGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITH 164
G ++ EG+ GST IS+ATV A G + +++PDD AIEK QI+E LGA VERVRP SI
Sbjct: 136 GSVLFEGTVGSTGISLATVGKAKGYESCIIMPDDVAIEKVQILEKLGARVERVRPASIVD 195
Query: 165 KDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQG---GF 221
+ FVN+AR+RA E + + G + + H LFP+ + GF
Sbjct: 196 QKQFVNLARKRALEFGKSELTNIPGH--GEEIAVSTAAEPSEINHHHLFPSAFEHKPRGF 253
Query: 222 FADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCY 281
FADQFEN +NF AHY+GTGPEI QT+G+LD FV+ AGTGGT+AG FL++ P++K
Sbjct: 254 FADQFENDSNFMAHYKGTGPEILRQTSGHLDGFVSGAGTGGTIAGTGCFLKKALPDLKIV 313
Query: 282 LIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEA--KLDGAF 339
L DP GSGLFNKV VM+ +E+EG++ ++ DT+ EGIGINR+T NF+ +D A+
Sbjct: 314 LSDPEGSGLFNKVRFNVMFDPKESEGKKRRHQVDTVVEGIGINRITYNFSLGLPVIDDAY 373
Query: 340 RGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSK 399
R +D EAV M+R+LV++DGLFLGSSSA N + +R+A+++ G I TILCDSG RH SK
Sbjct: 374 RVSDEEAVAMSRYLVQHDGLFLGSSSACNLIACIRLAKTMSRGSRIATILCDSGSRHQSK 433
Query: 400 FYNAEYL 406
F++ +YL
Sbjct: 434 FWSDDYL 440
>emb|CAB08745.1| SPAC3A12.17c [Schizosaccharomyces pombe]
gi|19114255|ref|NP_593343.1| hypothetical protein
SPAC3A12.17c [Schizosaccharomyces pombe 972h-]
gi|3219785|sp|P87131|CYSK2_SCHPO Cysteine synthase 2
(O-acetylserine sulfhydrylase 2) (O-acetylserine
(Thiol)-lyase 2) (CSase 2)
Length = 395
Score = 355 bits (910), Expect = 2e-96
Identities = 190/381 (49%), Positives = 251/381 (65%), Gaps = 36/381 (9%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQL--R 103
NG+ IGNT ++RI SLS ATGC+IL K EFLNPG S KDRVA+Q+I A E+G L
Sbjct: 44 NGVEGLIGNTKMVRIKSLSQATGCDILAKAEFLNPGNSPKDRVALQMIRTAEENGDLVPY 103
Query: 104 RGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
+ V EG+AGST ISIA + + G + +P D + EKS I+E LGA V+RV P I
Sbjct: 104 QSNAVYEGTAGSTGISIAMLCCSLGYDSRIYMPSDQSKEKSDILELLGAHVQRVTPAPIV 163
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
+HFVN ARR A+ T + I G G+FA
Sbjct: 164 DPNHFVNTARRNAANH--------------TVDESIPG-----------------KGYFA 192
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
+QFEN AN++AH+ TGPEIW Q G LDAF+A +GTGGT+AG+S++L+ K+P+I L
Sbjct: 193 NQFENPANWQAHFNSTGPEIWRQCAGKLDAFIAGSGTGGTIAGISRYLKSKDPSITVCLA 252
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNF--AEAKLDGAFRG 341
DPPGSGL++KV+ GVM+ E EG R ++ DTI EG+GINR+T+NF AE +D A+R
Sbjct: 253 DPPGSGLYHKVLHGVMFDLAEREGTRRRHQVDTIVEGVGINRMTRNFSIAEPLIDMAYRV 312
Query: 342 TDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFY 401
TD +AV M+R+LV +DGLF+GSSSA+NCV AVR+A+ +GPGH IVT+LCD G RH SK Y
Sbjct: 313 TDEQAVAMSRYLVTHDGLFVGSSSAVNCVAAVRLAKKLGPGHRIVTLLCDPGSRHFSKLY 372
Query: 402 NAEYLSQLGLTPKA-TGLEFL 421
N E+L + + P+ + L+F+
Sbjct: 373 NEEFLRKKNIVPQVPSSLDFV 393
>gb|EAK83340.1| hypothetical protein UM02218.1 [Ustilago maydis 521]
gi|49071088|ref|XP_399833.1| hypothetical protein
UM02218.1 [Ustilago maydis 521]
Length = 537
Score = 346 bits (888), Expect = 7e-94
Identities = 204/416 (49%), Positives = 254/416 (61%), Gaps = 51/416 (12%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQL--R 103
+G+ IGNTPL+RINSLS+ATGCEILGK EFLNP GS KDRVA+QI+++A E G L
Sbjct: 109 SGVEGLIGNTPLMRINSLSEATGCEILGKAEFLNPAGSPKDRVALQILKDAEEEGLLYPH 168
Query: 104 RGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
G + EG+ GST IS+AT+A A G +C +VIPDD A EK +++E LGA +E VRP I
Sbjct: 169 TGSCIFEGTVGSTGISLATLARAKGYRCSIVIPDDVAREKVELLEKLGAEIESVRPRGIV 228
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDG-----HKH-----STLF 213
HFVN AR RA E P G + + G + H+H S
Sbjct: 229 DPRHFVNEARARAEAFGEVELVG--PHPNGLGTSRYAGSDAGQGEEYVHRHDLVVSSRRV 286
Query: 214 PN--DCQG--------GFFADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGT 263
PN D G GFFADQFEN +NF AHY GTGPEIW QT G +DAFVA AGTGGT
Sbjct: 287 PNTDDASGIVNETQARGFFADQFENPSNFWAHYNGTGPEIWRQTGGLIDAFVAGAGTGGT 346
Query: 264 VAGVSKFLQEKN-------------------PNIKCYLIDPPGSGLFNKVMRGVMYTKEE 304
++G + FL+ + +K L DP GSGL+NKV GVMY+ E
Sbjct: 347 LSGCAAFLKRVSCDPDESQQGGGFIRRPGSAQEVKVVLADPQGSGLYNKVKYGVMYSATE 406
Query: 305 AEGRRLKNPFDTITEGIGINRVTKNFAEAK--LDGAFRGTDMEAVEMARFLVKNDGLFLG 362
AEG+R ++ D++ EGIGINR+T+N +D A R +D EA M R L+ NDGLFLG
Sbjct: 407 AEGKRRRHQVDSVVEGIGINRITRNLQMGLQCIDDAERVSDDEAARMGRHLILNDGLFLG 466
Query: 363 SSSAMNCVGAVRIAQSI------GPGHTIVTILCDSGMRHLSKFYNAEYLSQLGLT 412
SSSA+NCV AVR A I P +VT+LCDSG RHLSKF+N + L +LG++
Sbjct: 467 SSSAVNCVAAVRTALKIKRQRGDDPPPVVVTVLCDSGSRHLSKFHNDDALVRLGVS 522
>gb|EAL17743.1| hypothetical protein CNBL2560 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 453
Score = 345 bits (885), Expect = 2e-93
Identities = 191/361 (52%), Positives = 246/361 (67%), Gaps = 16/361 (4%)
Query: 53 GNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQL--RRGGIVTE 110
GNTPLIRINSLSDA G EILGK EFLNPGGSVKDRVA+QIIE+A G L G ++ E
Sbjct: 84 GNTPLIRINSLSDALGVEILGKAEFLNPGGSVKDRVALQIIEDAEAQGLLYPNTGSVLFE 143
Query: 111 GSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVN 170
G+ GST IS+ATV A G + +++PDD AIEK QI+E LGA VERVRP SI + FVN
Sbjct: 144 GTVGSTGISLATVGKAKGYESCIIMPDDVAIEKVQILEKLGARVERVRPASIVDQKQFVN 203
Query: 171 IARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQG---GFFADQFE 227
+AR+RA E + + G + + H LFP+ + GFFADQFE
Sbjct: 204 LARKRALEFGKSELTNIPGH--GEEIAVSTAAEPSEINHHHLFPSAFEHKPRGFFADQFE 261
Query: 228 NLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPG 287
N +NF AHY+GTGPEI QT+G+LD FV+ AGTG FL++ P++K L DP G
Sbjct: 262 NDSNFMAHYKGTGPEILRQTSGHLDGFVSGAGTGC-------FLKKALPDLKIVLSDPEG 314
Query: 288 SGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAK--LDGAFRGTDME 345
SGLFNKV VM+ +E+EG++ ++ DT+ EGIGINR+T NF+ +D A+R +D E
Sbjct: 315 SGLFNKVRFNVMFDPKESEGKKRRHQVDTVVEGIGINRITYNFSLGLPVIDDAYRVSDEE 374
Query: 346 AVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEY 405
AV M+R+LV++DGLFLGSSSA N + +R+A+++ G I TILCDSG RH SKF++ +Y
Sbjct: 375 AVAMSRYLVQHDGLFLGSSSACNLIACIRLAKTMSRGSRIATILCDSGSRHQSKFWSDDY 434
Query: 406 L 406
L
Sbjct: 435 L 435
>gb|EAA67243.1| hypothetical protein FG00903.1 [Gibberella zeae PH-1]
gi|46108042|ref|XP_381079.1| hypothetical protein
FG00903.1 [Gibberella zeae PH-1]
Length = 445
Score = 337 bits (864), Expect = 4e-91
Identities = 191/388 (49%), Positives = 248/388 (63%), Gaps = 44/388 (11%)
Query: 35 PSKKKKKKKSKNGIIDAI----GNTPLIRINSLSDATGCEILGKCEFLN-PGGSVKDRVA 89
PS ++NG+ D I GNTPLI++ SLS+ATG I+ K EFLN G S KDRVA
Sbjct: 71 PSSPTFYDGARNGLADGIEATIGNTPLIKVQSLSEATGRTIMAKAEFLNGAGNSPKDRVA 130
Query: 90 VQIIEEALESGQL--RRGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQII 147
+ +I EA G L +G + EG+ GST IS+AT+A A G + H+ +P D A+EKS ++
Sbjct: 131 LNMIREAEAQGLLTPHKGDTIYEGTVGSTGISLATLARAMGYRAHICMPSDMALEKSDLL 190
Query: 148 EALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGH 207
LGATVERV P IT DHFVN+ARRRA E A
Sbjct: 191 LHLGATVERVTPAPITSPDHFVNLARRRAEEHASKA------------------------ 226
Query: 208 KHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGV 267
ND GFFA+QFE+ AN++AH+ TGPEIW QTNG LDAFVA AGTGGTV+GV
Sbjct: 227 -------NDGSKGFFANQFESEANWKAHFNTTGPEIWGQTNGELDAFVAGAGTGGTVSGV 279
Query: 268 SKFLQE--KNPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINR 325
+K+L+E + +IK L DP GSGL+NKV GVMY+ E EG R + DT+ EGIGI R
Sbjct: 280 AKYLKEEKEKSDIKVVLADPQGSGLYNKVRHGVMYSSTEREGTRRRQQVDTMVEGIGITR 339
Query: 326 VTKNFAEAK--LDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGH 383
+T+NF + +D A R TD +A MAR+LV++DG+F+GSSS++NCV AV A + G
Sbjct: 340 LTENFEAGRELIDDAVRVTDEQACRMARWLVEHDGIFVGSSSSVNCVAAVETAMRLPKGS 399
Query: 384 TIVTILCDSGMRHLSKFYNAEYLSQLGL 411
+VTILCDSG RHLSKF+ +++ ++G+
Sbjct: 400 RVVTILCDSGTRHLSKFW--KHIKEMGI 425
>gb|EAA63826.1| hypothetical protein AN1513.2 [Aspergillus nidulans FGSC A4]
gi|67522112|ref|XP_659117.1| hypothetical protein
AN1513_2 [Aspergillus nidulans FGSC A4]
gi|49087428|ref|XP_405650.1| hypothetical protein
AN1513.2 [Aspergillus nidulans FGSC A4]
Length = 428
Score = 334 bits (857), Expect = 3e-90
Identities = 187/361 (51%), Positives = 238/361 (65%), Gaps = 27/361 (7%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGG-SVKDRVAVQIIEEALESGQL-- 102
+GI IGNTPL+RI SLS+ATGCEIL K EFLN G S KDRVA+ +IE A E G +
Sbjct: 66 DGIEGCIGNTPLLRIKSLSEATGCEILAKAEFLNGAGQSSKDRVALSMIELAEERGLMTP 125
Query: 103 RRGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSI 162
G + EG++GST IS+AT+A A G H+ +P D AIEKS ++ LGA V+RV P I
Sbjct: 126 HSGDTIYEGTSGSTGISLATLARAKGYLAHICMPSDQAIEKSNLLLKLGAIVDRVPPAPI 185
Query: 163 THKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFF 222
KD+FVN AR A ++ T S+ G S + G+F
Sbjct: 186 VEKDNFVNRARALA-----------QAHTNSTASESSVGTASQRGR-----------GYF 223
Query: 223 ADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYL 282
ADQFEN AN+RAHY GTGPEI+ Q NG+LDAFVA AGTGGT++GV+ +L+ + PN+ +
Sbjct: 224 ADQFENEANWRAHYNGTGPEIYAQCNGSLDAFVAGAGTGGTISGVALYLKPRIPNMTVVV 283
Query: 283 IDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAK--LDGAFR 340
DP GSGL+N+V GVM+ +E EG R + DTI EGIGINRVT NF + +D A R
Sbjct: 284 ADPQGSGLYNRVRYGVMFDTKEKEGTRRRRQVDTIVEGIGINRVTANFEAGRELVDDAVR 343
Query: 341 GTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKF 400
TD +A+ MAR+LV+ DG+F GSSSA+NC AV+ A +GPGH IVT+L DSG RHLS+F
Sbjct: 344 VTDAQALAMARWLVEKDGIFAGSSSAVNCFAAVKTALKLGPGHRIVTMLSDSGSRHLSRF 403
Query: 401 Y 401
+
Sbjct: 404 W 404
>ref|XP_629379.1| cysteine synthase [Dictyostelium discoideum]
gi|60462769|gb|EAL60969.1| cysteine synthase
[Dictyostelium discoideum]
Length = 378
Score = 330 bits (847), Expect = 4e-89
Identities = 172/362 (47%), Positives = 238/362 (65%), Gaps = 39/362 (10%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG 105
NGII+ +GNTPLIRI SLS+ATGCEI GK EF+NPGGS KDRVA +II + + G L++G
Sbjct: 40 NGIIETVGNTPLIRIKSLSEATGCEIYGKAEFMNPGGSPKDRVAREIILDGEKKGLLKKG 99
Query: 106 GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHK 165
+ E +AGST IS+ + + G + IPD+ + EK ++E LGA + V V + +
Sbjct: 100 STIVEATAGSTGISLTMLGKSRGYNVQLFIPDNVSKEKVDLLEMLGAETKIVPIVGMNNA 159
Query: 166 DHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQ 225
+HF++ A +R C D F+A+Q
Sbjct: 160 NHFMHCAYQR--------------------------CLGDDM------------AFYANQ 181
Query: 226 FENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
F+NL+NF AHY GT EIWEQT G++D FVAAAGTGGTVAG+S +L+E NPNI+ +LIDP
Sbjct: 182 FDNLSNFNAHYNGTAKEIWEQTKGDVDGFVAAAGTGGTVAGISSYLKEVNPNIQNWLIDP 241
Query: 286 PGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDME 345
PGSGL++ V GV++ ++ P + EG+G+N+ T+NF +A L+GAFRGT+ E
Sbjct: 242 PGSGLYSLVNTGVIFHPKDRLVVEKLGP-RSFYEGVGVNKKTENFNKASLNGAFRGTEEE 300
Query: 346 AVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEY 405
V+MA +L+K+DGLFLG SSA+NCVGAV++A+ +GPG TIVT+LCDSG R+ S+ Y+ +
Sbjct: 301 GVDMAHYLLKHDGLFLGGSSALNCVGAVKLARKLGPGKTIVTVLCDSGHRYTSRLYSKSW 360
Query: 406 LS 407
L+
Sbjct: 361 LN 362
>gb|AAS54510.1| AGR021Cp [Ashbya gossypii ATCC 10895] gi|45201116|ref|NP_986686.1|
AGR021Cp [Eremothecium gossypii]
Length = 387
Score = 314 bits (805), Expect = 3e-84
Identities = 172/392 (43%), Positives = 240/392 (60%), Gaps = 40/392 (10%)
Query: 15 GTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGCEILGK 74
G ++ VS V + F+ + S+ KNG+ + IGNTP++ + SLS TGC+I K
Sbjct: 15 GVSVIVS-VASLFYAYK--TSSRLTDLAPPKNGVEELIGNTPMVLLPSLSKFTGCKIYAK 71
Query: 75 CEFLNPGGSVKDRVAVQIIEEALESGQLRRG--GIVTEGSAGSTAISIATVAPAYGCKCH 132
E NP GS KDRVA+ I+ A G L RG +V EG++GST ISIAT+ A G + H
Sbjct: 72 LELANPAGSAKDRVALNIVRTAETQGLLTRGHRDVVFEGTSGSTGISIATICNALGYQSH 131
Query: 133 VVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPK 192
+ +PDD + EK ++E LGA V++V+P SI D +VN A++ NE
Sbjct: 132 ISLPDDTSPEKLALLECLGAHVKKVKPASIVDPDQYVNAAKKACKLLNE----------- 180
Query: 193 GTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPEIWEQTNGNLD 252
+GH + G FADQFEN AN+ HY+ TGPEI++Q G ++
Sbjct: 181 ------------EGHGNK---------GIFADQFENEANWTIHYQNTGPEIYQQLGGRIN 219
Query: 253 AFVAAAGTGGTVAGVSKFLQEK-NPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRLK 311
AF+A GTGGT+ GVS++L+E+ ++ L DP GSG +N++ GVMY E EG R +
Sbjct: 220 AFIAGCGTGGTIGGVSRYLKERLGDSVHTILADPQGSGFYNRINYGVMYDHVEKEGSRRR 279
Query: 312 NPFDTITEGIGINRVTKNF--AEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNC 369
+ DTI EGIG+NR+T+NF E +D + R TD +A++MA++L NDGLF+GSS+A+N
Sbjct: 280 HQVDTIVEGIGLNRITRNFKCCEEYIDSSIRVTDEQAIKMAKYLCVNDGLFIGSSTAVNA 339
Query: 370 VGAVRIAQSIGPGHTIVTILCDSGMRHLSKFY 401
V AVR+AQ + G T+V I CDSG RHLSKF+
Sbjct: 340 VAAVRVAQEMPAGSTLVLIACDSGSRHLSKFW 371
>gb|EAL00241.1| hypothetical protein CaO19.5574 [Candida albicans SC5314]
Length = 398
Score = 308 bits (788), Expect = 3e-82
Identities = 173/360 (48%), Positives = 221/360 (61%), Gaps = 34/360 (9%)
Query: 47 GIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQL--RR 104
GI IGNTPLI I SLS GC+I K E NPGGS KDRVA+ II A SGQL
Sbjct: 51 GIESLIGNTPLIEIKSLSRQLGCKIYAKLELCNPGGSAKDRVALAIIRAAESSGQLIPNA 110
Query: 105 GGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITH 164
I+ EG++GST IS+A + A G CH+ +PDD ++EK Q++++LGA +E V+ SI
Sbjct: 111 NNIIFEGTSGSTGISLAILCNALGYICHICLPDDTSLEKLQLLKSLGAELEPVKSASIVD 170
Query: 165 KDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFAD 224
+ + N ARR A N+ DGH + FAD
Sbjct: 171 PNQYTNAARRGALAVNQ---------------------NRDGHSQ--------RRAIFAD 201
Query: 225 QFENLANFRAHYEGTGPEIWEQTNGN-LDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
QFEN N+R HYE TGPE+ +Q + +D F+ +GTGGT+AGV + L+E NP K L
Sbjct: 202 QFENDFNWRIHYETTGPELLQQMGQDKIDVFINGSGTGGTIAGVGRCLKEYNPRTKIVLA 261
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNF--AEAKLDGAFRG 341
DP GSGL N++ GVMY E EG R ++ DT+ EGIG+NR+T NF AE +D A R
Sbjct: 262 DPQGSGLANRINYGVMYDSVEKEGTRRRHQVDTLVEGIGLNRLTWNFKQAEPYIDEAIRV 321
Query: 342 TDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFY 401
TD +A++MA++L NDGLFLGSSSA+NCV AV++A GPG IV I CDSG RHLSKF+
Sbjct: 322 TDDQALKMAKYLSINDGLFLGSSSAINCVAAVKMALKNGPGQKIVVIACDSGARHLSKFW 381
>gb|EAL85285.1| cysteine synthase, putative [Aspergillus fumigatus Af293]
Length = 401
Score = 283 bits (724), Expect = 7e-75
Identities = 173/360 (48%), Positives = 216/360 (59%), Gaps = 51/360 (14%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGG-SVKDRVAVQIIEEALESGQLRR 104
+GI IGNTPL+RI SLSDATGCEILGK EFLN G S KDRVA+ +IE +
Sbjct: 65 DGIEGCIGNTPLLRIKSLSDATGCEILGKAEFLNGAGQSSKDRVALSMIEIVCD------ 118
Query: 105 GGIVTEGSAGSTAISIATVAPAYGCKCH-VVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
+ S I++ + + H +V+P+ A + I V+RV P I
Sbjct: 119 ---MWPRSLLMPKINLQRPSKKVRPEFHWLVLPEQRATSLTCAI------VDRVPPAPIV 169
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
KD+FVN AR A TL P+ +G FFA
Sbjct: 170 EKDNFVNRARALAQA-------------------------------QTLSPSGGRG-FFA 197
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
DQFEN AN+RAHY GTGPEI+ Q NG LDAFVA AGTGGT++GV+ FL+ + PN+ L
Sbjct: 198 DQFENEANWRAHYNGTGPEIYAQCNGKLDAFVAGAGTGGTISGVALFLKPRIPNLSVVLA 257
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAK--LDGAFRG 341
DP GSGL+N+V GVM+ +E EG R + DTI EGIGINRVT NF K +D A R
Sbjct: 258 DPQGSGLYNRVRFGVMFDIKEREGTRRRRQVDTIVEGIGINRVTANFEAGKELVDDAVRV 317
Query: 342 TDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFY 401
TD +A+ MAR+LV+ DG+F+GSSSA+NC AV+ A +GPGH IVTIL DSG RHLS+F+
Sbjct: 318 TDAQALAMARWLVEKDGIFVGSSSAVNCFAAVKTAMKLGPGHRIVTILSDSGSRHLSRFW 377
>ref|NP_771093.1| cysteine synthase [Bradyrhizobium japonicum USDA 110]
gi|27352716|dbj|BAC49718.1| cysteine synthase
[Bradyrhizobium japonicum USDA 110]
Length = 344
Score = 280 bits (717), Expect = 5e-74
Identities = 152/362 (41%), Positives = 219/362 (59%), Gaps = 43/362 (11%)
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
+N +++AIGNTPLI++ S+ATGC ILGK EF+NPG SVKDR +I EA + G+L+
Sbjct: 4 RNDVVEAIGNTPLIKLKRASEATGCTILGKAEFMNPGQSVKDRAGKWMILEAEKRGELKP 63
Query: 105 GGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITH 164
GG+V E +AG+T I +A VA A G + +VIP+ + EK ++ GA + V + +
Sbjct: 64 GGLVVEATAGNTGIGLAVVASARGYRTLIVIPETQSQEKKDFLKLCGAELIEVPALPYAN 123
Query: 165 KDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFAD 224
+++ ++ RR A E RK++P G FAD
Sbjct: 124 PNNYQHVGRRLADEL-------RKTEPNGV--------------------------LFAD 150
Query: 225 QFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLID 284
Q+ NL N +AHYE TGPEIWEQT G +D FV + G+GGT+AGVS+ L+EKN N++ D
Sbjct: 151 QWNNLDNAKAHYESTGPEIWEQTGGKVDGFVCSVGSGGTLAGVSRLLKEKNKNVRIACAD 210
Query: 285 PPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDM 344
P G+G+F+ G EA+ P +ITEGIG+NR T AK+D A+ D
Sbjct: 211 PHGAGMFDYFRTG------EAKA----TPGGSITEGIGLNRATAIVESAKVDDAYLIPDT 260
Query: 345 EAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAE 404
+AV L++++GL LG S+ +N GA+R+A+ +GPG TIVT+LCDSG R+ SK +NA+
Sbjct: 261 DAVTEIYDLLQHEGLCLGGSTGINIAGAIRLAKQLGPGKTIVTVLCDSGSRYQSKLFNAD 320
Query: 405 YL 406
++
Sbjct: 321 FM 322
>ref|ZP_00339385.1| COG0031: Cysteine synthase [Silicibacter sp. TM1040]
Length = 394
Score = 268 bits (685), Expect = 2e-70
Identities = 152/364 (41%), Positives = 215/364 (58%), Gaps = 46/364 (12%)
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGI 107
+ DA+G+TPLIR+N +S+ TGCEILGK EFLNPG SVKDR A+ II++A+ G+L+ GG
Sbjct: 57 LADAVGHTPLIRLNKVSEETGCEILGKAEFLNPGQSVKDRAALYIIKDAIARGELKPGGT 116
Query: 108 VTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDH 167
+ EG+AG+T I +A V + G K +VIP+ + EK ++ GA + +V + ++
Sbjct: 117 IVEGTAGNTGIGLALVGASMGFKTVIVIPETQSEEKKDMLRLAGAQLVQVPAAPYKNPNN 176
Query: 168 FVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFE 227
+V + R A+E K++P G +A+QF+
Sbjct: 177 YVRYSERLANEL-------AKTEP--------------------------NGAIWANQFD 203
Query: 228 NLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPG 287
N+AN +AH E TGPEIWEQT G +D F+ A G+GGT+AG+++ LQ K +K L DP G
Sbjct: 204 NVANRQAHVETTGPEIWEQTGGKVDGFICAVGSGGTLAGIAEALQPK--GVKIGLADPNG 261
Query: 288 SGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAV 347
+ L++ YT E LK+ +ITEGIG R+TKN D ++ D EA+
Sbjct: 262 AALYS------YYTTGE-----LKSEGSSITEGIGQGRITKNLEGLTPDFNYQIPDAEAL 310
Query: 348 EMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEYLS 407
L+ +GL LG SS +N GAVR+A+ +GPGHTIVTILCD G R+ SK +N E+L
Sbjct: 311 PYVFDLLHEEGLVLGGSSGVNIAGAVRLAKELGPGHTIVTILCDYGTRYQSKLFNPEFLK 370
Query: 408 QLGL 411
GL
Sbjct: 371 DKGL 374
>gb|AAL52114.1| CYSTEINE SYNTHASE [Brucella melitensis 16M]
gi|17987216|ref|NP_539850.1| CYSTEINE SYNTHASE [Brucella
melitensis 16M] gi|25291729|pir||AG3368 cysteine
synthase (EC 4.2.99.8) [imported] - Brucella melitensis
(strain 16M)
Length = 342
Score = 258 bits (659), Expect = 2e-67
Identities = 146/361 (40%), Positives = 208/361 (57%), Gaps = 44/361 (12%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG 105
N ++D IGNTPLIR++ S+ TGC+I GK EFLNPG SVKDR A+ II +A + G LR G
Sbjct: 3 NSVLDTIGNTPLIRLSKASELTGCDIYGKAEFLNPGQSVKDRAALYIIRDAEKRGLLRPG 62
Query: 106 GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHK 165
G++ EG+AG+T I + VA A G + +VIP+ + EK +++ LGA + V +
Sbjct: 63 GVIVEGTAGNTGIGLTMVAKALGYRTAIVIPETQSQEKKEVLRLLGAELIEVPAAPYRNP 122
Query: 166 DHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQ 225
+++V ++ R A + K++P G +A+Q
Sbjct: 123 NNYVRLSGRLAEQL-------AKTEP--------------------------NGAIWANQ 149
Query: 226 FENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
F+N N +AH E T EIW T+ +D FVAA G+GGT+AG + L+E+N NIK L DP
Sbjct: 150 FDNTVNRQAHIETTAQEIWRDTSDQIDGFVAAVGSGGTLAGTAIGLKERNHNIKIALADP 209
Query: 286 PGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDME 345
G+ L YT E LK D+ITEGIG R+T N D +++ D E
Sbjct: 210 HGAALH------AFYTTGE-----LKAEGDSITEGIGQGRITANLEGFTPDFSYQIPDAE 258
Query: 346 AVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEY 405
A+++ LV+ +GL LG SS +N GA+R+A+ +GPGHTIVT+LCD G R+ SK +N +
Sbjct: 259 ALDILFALVEEEGLCLGGSSGINIAGAIRLAKDLGPGHTIVTVLCDYGNRYQSKLFNPAF 318
Query: 406 L 406
L
Sbjct: 319 L 319
>gb|AAN29973.1| cysteine synthase [Brucella suis 1330] gi|23501931|ref|NP_698058.1|
cysteine synthase [Brucella suis 1330]
Length = 342
Score = 258 bits (658), Expect = 3e-67
Identities = 147/361 (40%), Positives = 206/361 (56%), Gaps = 44/361 (12%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG 105
N ++D IGNTPLIR++ S+ TGC+I GK EFLNPG SVKDR A+ II +A + G LR G
Sbjct: 3 NSVLDTIGNTPLIRLSKASELTGCDIYGKAEFLNPGQSVKDRAALYIIRDAEKRGLLRPG 62
Query: 106 GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHK 165
G++ EG+AG+T I + VA A G + +VIP+ + EK + LGA + V +
Sbjct: 63 GVIVEGTAGNTGIGLTMVAKALGYRTAIVIPETQSQEKKDALRLLGAELIEVPAAPYRNP 122
Query: 166 DHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQ 225
+++V ++ R A + K++P G +A+Q
Sbjct: 123 NNYVRLSGRLAEQL-------AKTEP--------------------------NGAIWANQ 149
Query: 226 FENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
F+N N +AH E T EIW TN +D FVAA G+GGT+AG + L+E+N NIK L DP
Sbjct: 150 FDNTVNRQAHIETTAQEIWRDTNDQIDGFVAAVGSGGTLAGTAIGLKERNHNIKIALADP 209
Query: 286 PGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDME 345
G+ L YT E LK D+ITEGIG R+T N D +++ D E
Sbjct: 210 HGAALH------AFYTTGE-----LKAEGDSITEGIGQGRITANLEGFTPDFSYQIPDAE 258
Query: 346 AVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEY 405
A+++ LV+ +GL LG SS +N GA+R+A+ +GPGHTIVT+LCD G R+ SK +N +
Sbjct: 259 ALDILFALVEEEGLCLGGSSGINIAGAIRLAKDLGPGHTIVTVLCDYGNRYQSKLFNPAF 318
Query: 406 L 406
L
Sbjct: 319 L 319
>gb|AAV95198.1| pyridoxal-phosphate dependent enzyme [Silicibacter pomeroyi DSS-3]
gi|56696795|ref|YP_167156.1| pyridoxal-phosphate
dependent enzyme [Silicibacter pomeroyi DSS-3]
Length = 344
Score = 256 bits (653), Expect = 1e-66
Identities = 148/364 (40%), Positives = 212/364 (57%), Gaps = 46/364 (12%)
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGI 107
+ +A+G+TPLIR+ +S+ TGCEI GK EF+NPG SVKDR A+ II +A+ G+L+ GG
Sbjct: 7 LAEAVGHTPLIRLRRVSEETGCEIYGKAEFMNPGQSVKDRAALYIIRDAIARGELQPGGT 66
Query: 108 VTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDH 167
+ EG+AG+T I +A V + G K +VIP+ + EK ++ GA + +V + ++
Sbjct: 67 IVEGTAGNTGIGLALVGASMGFKTVIVIPETQSEEKKDMLRLAGAELVQVPAAPYRNPNN 126
Query: 168 FVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFE 227
FV + R A E K++P G +A+QF+
Sbjct: 127 FVRYSERLAREL-------AKTEP--------------------------NGAIWANQFD 153
Query: 228 NLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPG 287
N AN +AH E TGPEIWEQT G +D FVAA G+GGT+AGV++ LQ K +K L DP G
Sbjct: 154 NTANRQAHVETTGPEIWEQTGGKVDGFVAAVGSGGTLAGVAEALQPK--GVKIGLADPMG 211
Query: 288 SGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAV 347
+ LF+ YT E L +I EGIG R+T+N K D A++ D EA+
Sbjct: 212 AALFS------YYTTGE-----LAMEGGSIAEGIGQVRITRNLEGFKPDFAYQVPDAEAL 260
Query: 348 EMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEYLS 407
L++ +GL +G S+A+N GAVR+A+ +GPGHTIVT+LCD G R+ SK +N ++L
Sbjct: 261 PYIFDLLRQEGLVMGGSTAINIAGAVRMAREMGPGHTIVTVLCDYGTRYQSKLFNPDFLR 320
Query: 408 QLGL 411
+ L
Sbjct: 321 EKDL 324
>ref|YP_221767.1| cysteine synthase [Brucella abortus biovar 1 str. 9-941]
gi|62196106|gb|AAX74406.1| cysteine synthase [Brucella
abortus biovar 1 str. 9-941]
Length = 342
Score = 256 bits (653), Expect = 1e-66
Identities = 146/361 (40%), Positives = 206/361 (56%), Gaps = 44/361 (12%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG 105
N ++D IGNTPLIR++ S+ TGC+I GK EFLNPG SVKDR A+ II +A + G LR G
Sbjct: 3 NSVLDTIGNTPLIRLSKASELTGCDIYGKAEFLNPGQSVKDRAALYIIRDAEKRGLLRPG 62
Query: 106 GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHK 165
G++ EG+AG+T I + VA A G + +VIP+ + EK + LGA + V +
Sbjct: 63 GVIVEGTAGNTGIGLTMVAKALGYRTAIVIPETQSQEKKDALRLLGAELIEVPAAPYRNP 122
Query: 166 DHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQ 225
+++V ++ R A + K++P G +A+Q
Sbjct: 123 NNYVRLSGRLAEQL-------AKTEP--------------------------NGAIWANQ 149
Query: 226 FENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
F+N N +AH E T EIW T+ +D FVAA G+GGT+AG + L+E+N NIK L DP
Sbjct: 150 FDNTVNRQAHIETTAQEIWRDTSDQIDGFVAAVGSGGTLAGTAIGLKERNHNIKIALADP 209
Query: 286 PGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDME 345
G+ L YT E LK D+ITEGIG R+T N D +++ D E
Sbjct: 210 HGAALH------AFYTTGE-----LKAEGDSITEGIGQGRITANLEGFTPDFSYQIPDAE 258
Query: 346 AVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEY 405
A+++ LV+ +GL LG SS +N GA+R+A+ +GPGHTIVT+LCD G R+ SK +N +
Sbjct: 259 ALDILFALVEEEGLCLGGSSGINIAGAIRLAKDLGPGHTIVTVLCDYGNRYQSKLFNPAF 318
Query: 406 L 406
L
Sbjct: 319 L 319
>ref|YP_191998.1| Cysteine synthase A (O-acetylserine sulfhydrylase A) protein
[Gluconobacter oxydans 621H] gi|58002448|gb|AAW61342.1|
Cysteine synthase A (O-acetylserine sulfhydrylase A)
protein [Gluconobacter oxydans 621H]
Length = 351
Score = 248 bits (633), Expect = 3e-64
Identities = 147/365 (40%), Positives = 196/365 (53%), Gaps = 55/365 (15%)
Query: 47 GIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGG 106
G+ AIG TPLIR+ S+ TGC+I GK EF+NPGGSVKDR A+ IIE+AL+SG L+ GG
Sbjct: 25 GMTAAIGGTPLIRLRRASEETGCDIYGKAEFMNPGGSVKDRAALAIIEDALKSGALKPGG 84
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
V EG+AG+T I + VA A GCK +V+P+ + EK + +GA + V
Sbjct: 85 TVVEGTAGNTGIGLTLVAHAVGCKAVIVVPETQSREKLDFLRMIGADLRLVPAKPYRDPG 144
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
++V+++RR A E GGF+A+QF
Sbjct: 145 NYVHVSRRLAEEI---------------------------------------GGFWANQF 165
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEK-----NPNIKCY 281
+N AN H T EIWEQT G +DAF A GTGGT+AG++ L+++ +
Sbjct: 166 DNTANREGHRATTAREIWEQTGGKVDAFTCACGTGGTLAGIALGLRDQAQATGKTAPRIV 225
Query: 282 LIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRG 341
L DP GS L+ V + L +ITEGIG RVT N + D A R
Sbjct: 226 LADPEGSALYGWV-----------KSNDLTITGGSITEGIGQGRVTANLEGSHPDDAERI 274
Query: 342 TDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFY 401
D EA+E L +GL +G SS +N A+R A+ +GPGHTIVTILCD G R+ SK +
Sbjct: 275 PDAEALEQIFSLTSEEGLSVGGSSGINVAAAIRTARRLGPGHTIVTILCDGGARYQSKLF 334
Query: 402 NAEYL 406
N E+L
Sbjct: 335 NPEFL 339
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 732,211,132
Number of Sequences: 2540612
Number of extensions: 32093464
Number of successful extensions: 81164
Number of sequences better than 10.0: 1261
Number of HSP's better than 10.0 without gapping: 878
Number of HSP's successfully gapped in prelim test: 383
Number of HSP's that attempted gapping in prelim test: 77866
Number of HSP's gapped (non-prelim): 2326
length of query: 424
length of database: 863,360,394
effective HSP length: 131
effective length of query: 293
effective length of database: 530,540,222
effective search space: 155448285046
effective search space used: 155448285046
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122165.6


