

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122165.12 + phase: 1 /pseudo
(150 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
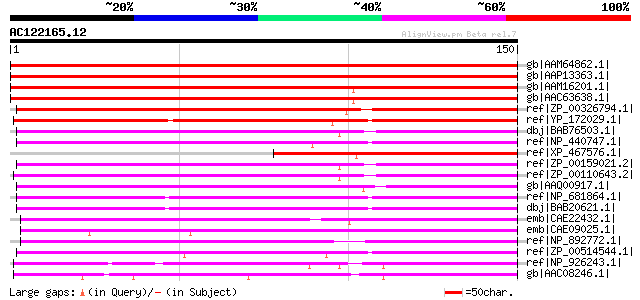
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM64862.1| unknown [Arabidopsis thaliana] 260 5e-69
gb|AAP13363.1| At5g55710 [Arabidopsis thaliana] gi|9758605|dbj|B... 259 1e-68
gb|AAM16201.1| At2g47840/F17A22.23 [Arabidopsis thaliana] gi|152... 139 1e-32
gb|AAC63638.1| expressed protein [Arabidopsis thaliana] gi|25409... 139 1e-32
ref|ZP_00326794.1| hypothetical protein Tery02002129 [Trichodesm... 119 2e-26
ref|YP_172029.1| hypothetical protein YCF60 [Synechococcus elong... 109 1e-23
dbj|BAB76503.1| all4804 [Nostoc sp. PCC 7120] gi|25531819|pir||A... 104 5e-22
ref|NP_440747.1| hypothetical protein sll1737 [Synechocystis sp.... 103 6e-22
ref|XP_467576.1| unknown protein [Oryza sativa (japonica cultiva... 102 1e-21
ref|ZP_00159021.2| hypothetical protein Avar03004993 [Anabaena v... 102 1e-21
ref|ZP_00110643.2| hypothetical protein Npun02002017 [Nostoc pun... 102 2e-21
gb|AAQ00917.1| Uncharacterized membrane protein [Prochlorococcus... 93 1e-18
ref|NP_681864.1| hypothetical protein tlr1073 [Thermosynechococc... 90 1e-17
dbj|BAB20621.1| hypothetical protein [Thermosynechococcus elonga... 90 1e-17
emb|CAE22432.1| conserved hypothetical protein [Prochlorococcus ... 87 1e-16
emb|CAE09025.1| conserved hypothetical protein [Synechococcus sp... 79 2e-14
ref|NP_892772.1| hypothetical protein PMM0654 [Prochlorococcus m... 75 3e-13
ref|ZP_00514544.1| conserved hypothetical protein [Crocosphaera ... 69 2e-11
ref|NP_926243.1| hypothetical protein glr3297 [Gloeobacter viola... 66 2e-10
gb|AAC08246.1| ORF203 [Porphyra purpurea] gi|11465826|ref|NP_053... 64 1e-09
>gb|AAM64862.1| unknown [Arabidopsis thaliana]
Length = 209
Score = 260 bits (664), Expect = 5e-69
Identities = 122/150 (81%), Positives = 138/150 (91%)
Query: 1 ASDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTL 60
ASDR+ISA+CYFYPFFDGIQYGKF+ITQ++P Q ++QPL PAIR FKSFPFNGFL+F+TL
Sbjct: 59 ASDRIISAVCYFYPFFDGIQYGKFIITQYHPFQILIQPLFPAIRAFKSFPFNGFLIFITL 118
Query: 61 YFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFLFL 120
YF VVRNPNFS+YVRFNTMQAIVLDVLLIFPDLLER FNPR GFGLD++MSLDSTVFLFL
Sbjct: 119 YFVVVRNPNFSRYVRFNTMQAIVLDVLLIFPDLLERSFNPRDGFGLDVVMSLDSTVFLFL 178
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
LV LIYG S+C+ G PRLP+VA+AADRQV
Sbjct: 179 LVSLIYGFSACLFGLTPRLPLVAEAADRQV 208
>gb|AAP13363.1| At5g55710 [Arabidopsis thaliana] gi|9758605|dbj|BAB09238.1| unnamed
protein product [Arabidopsis thaliana]
gi|15240567|ref|NP_200382.1| expressed protein
[Arabidopsis thaliana] gi|25082851|gb|AAN72007.1|
putative protein [Arabidopsis thaliana]
Length = 209
Score = 259 bits (661), Expect = 1e-68
Identities = 122/150 (81%), Positives = 137/150 (91%)
Query: 1 ASDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTL 60
ASDR+ISA+CYFYPFFDGIQYGKF+ITQ+ P Q ++QPL PAIR FKSFPFNGFL+F+TL
Sbjct: 59 ASDRIISAVCYFYPFFDGIQYGKFIITQYQPFQILIQPLFPAIRAFKSFPFNGFLIFITL 118
Query: 61 YFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFLFL 120
YF VVRNPNFS+YVRFNTMQAIVLDVLLIFPDLLER FNPR GFGLD++MSLDSTVFLFL
Sbjct: 119 YFVVVRNPNFSRYVRFNTMQAIVLDVLLIFPDLLERSFNPRDGFGLDVVMSLDSTVFLFL 178
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
LV LIYG S+C+ G PRLP+VA+AADRQV
Sbjct: 179 LVSLIYGFSACLFGLTPRLPLVAEAADRQV 208
>gb|AAM16201.1| At2g47840/F17A22.23 [Arabidopsis thaliana]
gi|15215636|gb|AAK91363.1| At2g47840/F17A22.23
[Arabidopsis thaliana]
Length = 208
Score = 139 bits (350), Expect = 1e-32
Identities = 68/152 (44%), Positives = 100/152 (65%), Gaps = 2/152 (1%)
Query: 1 ASDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTL 60
A++R+IS Y PFF+ +QYG+F+ Q+ + + +P+ P + +++S P+ F+ F L
Sbjct: 57 ATERVISIASYALPFFNSLQYGRFLFAQYPRLGLLFEPIFPILNLYRSVPYASFVAFFGL 116
Query: 61 YFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNP--RGGFGLDLLMSLDSTVFL 118
Y VVRN +FS+YVRFN MQA+ LDVLL P LL R +P GGFG+ +M + VF+
Sbjct: 117 YLGVVRNTSFSRYVRFNAMQAVTLDVLLAVPVLLTRILDPGQGGGFGMKAMMWGHTGVFV 176
Query: 119 FLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
F +C +YG S +LG+ P +P VADAA RQ+
Sbjct: 177 FSFMCFVYGVVSSLLGKTPYIPFVADAAGRQL 208
>gb|AAC63638.1| expressed protein [Arabidopsis thaliana] gi|25409066|pir||B84920
hypothetical protein At2g47840 [imported] - Arabidopsis
thaliana gi|18407462|ref|NP_566112.1| tic20
protein-related [Arabidopsis thaliana]
Length = 208
Score = 139 bits (350), Expect = 1e-32
Identities = 68/152 (44%), Positives = 100/152 (65%), Gaps = 2/152 (1%)
Query: 1 ASDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTL 60
A++R+IS Y PFF+ +QYG+F+ Q+ + + +P+ P + +++S P+ F+ F L
Sbjct: 57 ATERVISIASYALPFFNSLQYGRFLFAQYPRLGLLFEPIFPILNLYRSVPYASFVAFFGL 116
Query: 61 YFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNP--RGGFGLDLLMSLDSTVFL 118
Y VVRN +FS+YVRFN MQA+ LDVLL P LL R +P GGFG+ +M + VF+
Sbjct: 117 YLGVVRNTSFSRYVRFNAMQAVTLDVLLAVPVLLTRILDPGQGGGFGMKAMMWGHTGVFV 176
Query: 119 FLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
F +C +YG S +LG+ P +P VADAA RQ+
Sbjct: 177 FSFMCFVYGVVSSLLGKTPYIPFVADAAGRQL 208
>ref|ZP_00326794.1| hypothetical protein Tery02002129 [Trichodesmium erythraeum IMS101]
Length = 157
Score = 119 bits (297), Expect = 2e-26
Identities = 58/149 (38%), Positives = 95/149 (62%), Gaps = 4/149 (2%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
DR+ + L Y P DG++YG+++ QF P++ I+ PL+P +++++ PF+GF++FL L+
Sbjct: 11 DRIFACLPYLLPLLDGLEYGRYLFQQFPPLKLILIPLLPLLQIYQGIPFSGFIIFLALFL 70
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGF-NPRGGFGLDLLMSLDSTVFLFLL 121
VVRN N S ++RFNTMQAI+LD++LI L+ + GGF +L +L + +FL +L
Sbjct: 71 LVVRNNNISHFIRFNTMQAILLDIVLILCSLILSVLGSALGGF---ILETLSNMIFLGIL 127
Query: 122 VCLIYGSSSCILGQLPRLPIVADAADRQV 150
IY +G+ +P ++DA QV
Sbjct: 128 GSFIYTVIQSAIGRYAEIPTISDAVYMQV 156
>ref|YP_172029.1| hypothetical protein YCF60 [Synechococcus elongatus PCC 6301]
gi|56686287|dbj|BAD79509.1| hypothetical protein YCF60
[Synechococcus elongatus PCC 6301]
gi|45512142|ref|ZP_00163709.1| hypothetical protein
Selo03002389 [Synechococcus elongatus PCC 7942]
Length = 158
Score = 109 bits (273), Expect = 1e-23
Identities = 56/150 (37%), Positives = 93/150 (61%), Gaps = 3/150 (2%)
Query: 2 SDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLY 61
+DR + L Y P +G+ +G + QF +Q ++ PL+P +++F+ PF G ++F L+
Sbjct: 10 ADRFFACLPYLLPLAEGVSFGFALFNQFPVLQYLILPLVPVLQLFQ-IPFAGLIIFFLLF 68
Query: 62 FFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLL-ERGFNPRGGFGLDLLMSLDSTVFLFL 120
F VVRN N S ++RFN MQAI++D+LLI +++ + P G G +L +L++T+FL +
Sbjct: 69 FLVVRNENISHFIRFNAMQAILIDILLILANIIFQMVLRPTLGGGF-ILETLNNTIFLAM 127
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
LV +Y ILG+ +P ++DA QV
Sbjct: 128 LVGCLYSIIQSILGRYAEIPTISDAVYMQV 157
>dbj|BAB76503.1| all4804 [Nostoc sp. PCC 7120] gi|25531819|pir||AD2406 hypothetical
protein all4804 [imported] - Nostoc sp. (strain PCC
7120) gi|17232296|ref|NP_488844.1| hypothetical protein
all4804 [Nostoc sp. PCC 7120]
Length = 158
Score = 104 bits (259), Expect = 5e-22
Identities = 49/150 (32%), Positives = 89/150 (58%), Gaps = 5/150 (3%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
DR+ + L Y P + +G+F++ QF +Q + PLIP +R++ + G ++F L+
Sbjct: 11 DRIFACLPYLLPLIEVFAFGEFLLRQFPVLQLLFLPLIPLLRIYYGVRYAGLIIFFVLWL 70
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLER--GFNPRGGFGLDLLMSLDSTVFLFL 120
VVRN + ++RFNTMQAI+LD+++ +L G P GGF + +L +T+F+ +
Sbjct: 71 LVVRNEKINHFIRFNTMQAILLDIIIFLFSILTDIVGLIPTGGFA---IQTLYTTIFIGI 127
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
+ ++Y ++ +LG+ +P ++DA QV
Sbjct: 128 IAAVVYSVANSLLGRYAEIPAISDAVYMQV 157
>ref|NP_440747.1| hypothetical protein sll1737 [Synechocystis sp. PCC 6803]
gi|1652506|dbj|BAA17427.1| sll1737 [Synechocystis sp.
PCC 6803] gi|7470141|pir||S77324 hypothetical protein
sll1737 - Synechocystis sp. (strain PCC 6803)
Length = 160
Score = 103 bits (258), Expect = 6e-22
Identities = 56/149 (37%), Positives = 86/149 (57%), Gaps = 2/149 (1%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
DR SAL Y P D +G F++ QF +Q I P++P ++ + FPF F++F+ L+
Sbjct: 11 DRFFSALIYVIPLIDAFMFGGFLLQQFPVLQIIYLPIMPLLQFYYQFPFASFIIFIVLFM 70
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLL-IFPDLLERGFNPRGGFGLDLLMSLDSTVFLFLL 121
VVRN N S ++RFN MQAI++ +LL +F ++ P G GL + +L + FL L
Sbjct: 71 AVVRNNNISHFIRFNAMQAILIGILLSLFGLIVAYVIQPVFGQGL-VTETLYNFAFLGAL 129
Query: 122 VCLIYGSSSCILGQLPRLPIVADAADRQV 150
C +G +LG+ +P ++DAA QV
Sbjct: 130 ACGFFGIVQSVLGRYAEIPTISDAAYSQV 158
>ref|XP_467576.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|46390600|dbj|BAD16084.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 75
Score = 102 bits (255), Expect = 1e-21
Identities = 50/74 (67%), Positives = 60/74 (80%), Gaps = 2/74 (2%)
Query: 79 MQAIVLDVLLIFPDLLERGFNPR--GGFGLDLLMSLDSTVFLFLLVCLIYGSSSCILGQL 136
MQA+ LDVLLIFPDLL + F P GG G +L S++STVFLFLLVCL+YG +C+LG+
Sbjct: 1 MQAVALDVLLIFPDLLVQSFAPSTGGGIGFELFQSMESTVFLFLLVCLVYGGGACLLGKT 60
Query: 137 PRLPIVADAADRQV 150
PRLPIVADAA+RQV
Sbjct: 61 PRLPIVADAAERQV 74
>ref|ZP_00159021.2| hypothetical protein Avar03004993 [Anabaena variabilis ATCC 29413]
Length = 158
Score = 102 bits (255), Expect = 1e-21
Identities = 49/150 (32%), Positives = 89/150 (58%), Gaps = 5/150 (3%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
DR+ + L Y P + +G+F++ QF +Q + PLIP +R++ + G ++F L+
Sbjct: 11 DRIFACLPYLLPLIEVFVFGEFLLRQFPVLQLLFLPLIPLLRIYYGVRYAGIIIFFALWL 70
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLER--GFNPRGGFGLDLLMSLDSTVFLFL 120
VVRN + ++RFNTMQAI+LD+++ +L G P GGF + +L +T+F+ +
Sbjct: 71 LVVRNEKINHFIRFNTMQAILLDIIIFLFSILTDVVGLIPTGGFA---IQTLYTTIFIGI 127
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
+ ++Y ++ +LG+ +P ++DA QV
Sbjct: 128 IAAVVYCVANSLLGRYAEIPAISDAVYMQV 157
>ref|ZP_00110643.2| hypothetical protein Npun02002017 [Nostoc punctiforme PCC 73102]
Length = 158
Score = 102 bits (253), Expect = 2e-21
Identities = 48/151 (31%), Positives = 88/151 (57%), Gaps = 5/151 (3%)
Query: 2 SDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLY 61
SDR+ + L Y P + +G+++++ F P+Q + PL+P +R++ + G ++F L+
Sbjct: 10 SDRIFACLPYLLPLIEVFVFGRYLLSDFPPLQLLFLPLLPLLRIYYGVRYAGMIIFFALF 69
Query: 62 FFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLER--GFNPRGGFGLDLLMSLDSTVFLF 119
VVRN S ++RFNTMQAI+LD+++ ++L G P GGF + +L +T+F+
Sbjct: 70 LLVVRNEKISHFIRFNTMQAILLDIIIFLFNILTDVVGLVPVGGFA---IQTLSTTIFIG 126
Query: 120 LLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
++ + Y + G+ +P ++DA QV
Sbjct: 127 IVGAVAYSVIQSVSGRYAEIPAISDAVYMQV 157
>gb|AAQ00917.1| Uncharacterized membrane protein [Prochlorococcus marinus subsp.
marinus str. CCMP1375] gi|33241322|ref|NP_876264.1|
Uncharacterized membrane protein [Prochlorococcus
marinus subsp. marinus str. CCMP1375]
Length = 152
Score = 92.8 bits (229), Expect = 1e-18
Identities = 51/150 (34%), Positives = 83/150 (55%), Gaps = 5/150 (3%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
D++IS Y P+ D ++YG + F + + P +P + +S P FLVFL L+
Sbjct: 6 DKIISVFLYMLPWSDALKYGNNIFQNFPLSKIFIIPTLPIFIIERSLPIGSFLVFLLLFI 65
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGG--FGLDLLMSLDSTVFLFL 120
+ +NP S ++RFN MQA++L ++LI + LE F G F LD+ L+ +F+
Sbjct: 66 GIAKNPRVSYFIRFNAMQALLLKLILIIFNYLEILFIQLSGSFFRLDI---LEIIIFISS 122
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
L +IY S+ CI G P +P ++ +A Q+
Sbjct: 123 LAIVIYASTQCIRGIEPDIPGISASAKMQI 152
>ref|NP_681864.1| hypothetical protein tlr1073 [Thermosynechococcus elongatus BP-1]
gi|22294797|dbj|BAC08626.1| ycf60 [Thermosynechococcus
elongatus BP-1]
Length = 157
Score = 90.1 bits (222), Expect = 1e-17
Identities = 49/148 (33%), Positives = 84/148 (56%), Gaps = 2/148 (1%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
DR+ ++L Y P F + +G F+ F P+QA+V ++P ++ S PF G LVF+ LY
Sbjct: 11 DRIFASLAYLLPLFYVMPFGGFLFELFPPLQALVWVVLPVALIY-SIPFAGLLVFMLLYL 69
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFLFLLV 122
VVRN S ++R+NTMQA+++ ++L LL + F L L+ L + +F+ ++
Sbjct: 70 LVVRNNRVSLFIRYNTMQALLMGIVLFIVQLLVQLLLRIASFDL-LIKVLFNFIFIATVI 128
Query: 123 CLIYGSSSCILGQLPRLPIVADAADRQV 150
++YG + G +P +++A QV
Sbjct: 129 GVVYGVVQSVRGIYAEIPTLSEATKSQV 156
>dbj|BAB20621.1| hypothetical protein [Thermosynechococcus elongatus]
Length = 153
Score = 90.1 bits (222), Expect = 1e-17
Identities = 49/148 (33%), Positives = 84/148 (56%), Gaps = 2/148 (1%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYF 62
DR+ ++L Y P F + +G F+ F P+QA+V ++P ++ S PF G LVF+ LY
Sbjct: 7 DRIFASLAYLLPLFYVMPFGGFLFELFPPLQALVWVVLPVALIY-SIPFAGLLVFMLLYL 65
Query: 63 FVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFLFLLV 122
VVRN S ++R+NTMQA+++ ++L LL + F L L+ L + +F+ ++
Sbjct: 66 LVVRNNRVSLFIRYNTMQALLMGIVLFIVQLLVQLLLRIASFDL-LIKVLFNFIFIATVI 124
Query: 123 CLIYGSSSCILGQLPRLPIVADAADRQV 150
++YG + G +P +++A QV
Sbjct: 125 GVVYGVVQSVRGIYAEIPTLSEATKSQV 152
>emb|CAE22432.1| conserved hypothetical protein [Prochlorococcus marinus str. MIT
9313] gi|33864522|ref|NP_896082.1| hypothetical protein
PMT2258 [Prochlorococcus marinus str. MIT 9313]
Length = 154
Score = 86.7 bits (213), Expect = 1e-16
Identities = 48/149 (32%), Positives = 83/149 (55%), Gaps = 5/149 (3%)
Query: 4 RLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYFF 63
RL+ L Y P+ D I +G+ + QF ++ + P +P I + + PF L+F L+
Sbjct: 8 RLLGVLIYMLPWSDTIPFGRSLFVQFPLLELLALPALPLIILEQGIPFGSLLIFFLLFLA 67
Query: 64 VVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFN--PRGGFGLDLLMSLDSTVFLFLL 121
VVRNP ++RFNT+QA+++D++++ LL F + G +L +L STV + +L
Sbjct: 68 VVRNPKVPYFLRFNTLQALLVDIVVV---LLGYAFQILLQPLSGAFMLRTLTSTVLVAML 124
Query: 122 VCLIYGSSSCILGQLPRLPIVADAADRQV 150
+I+ C+ G+ P LP ++ A Q+
Sbjct: 125 AIVIFALIECLRGREPDLPGISQAVRMQL 153
>emb|CAE09025.1| conserved hypothetical protein [Synechococcus sp. WH 8102]
gi|33867040|ref|NP_898599.1| hypothetical protein
SYNW2510 [Synechococcus sp. WH 8102]
Length = 160
Score = 79.0 bits (193), Expect = 2e-14
Identities = 49/152 (32%), Positives = 81/152 (53%), Gaps = 5/152 (3%)
Query: 4 RLISALCYFYPFFDGIQYG---KFVITQFYPIQAIVQPLIPAIRVFKSFPFN--GFLVFL 58
RL++ L Y P+ D I G + Q+ ++ +V P +P +++ +S PF G L+F
Sbjct: 8 RLVAPLMYLLPWSDAIPLGFGPDGLFLQYPVLRPLVLPALPLMQLERSIPFGLGGLLLFF 67
Query: 59 TLYFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFL 118
L+ VVRNPN ++RFN +QA++ D+ LI + R G LL +L S V +
Sbjct: 68 VLFLAVVRNPNVPYFLRFNALQALLTDIALIVLSIGFRLLLQPIAAGSLLLGTLSSAVVV 127
Query: 119 FLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
+L L++ C+ G+ P LP ++ A Q+
Sbjct: 128 AVLAILLFSLVECLRGREPDLPGISQAVRMQL 159
>ref|NP_892772.1| hypothetical protein PMM0654 [Prochlorococcus marinus subsp.
pastoris str. CCMP1986] gi|33639943|emb|CAE19113.1|
conserved hypothetical protein [Prochlorococcus marinus
subsp. pastoris str. CCMP1986]
Length = 144
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/147 (29%), Positives = 73/147 (49%), Gaps = 9/147 (6%)
Query: 4 RLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLYFF 63
R S + Y P + +G +++ +F ++ ++ P + +S PF G L F+ L+
Sbjct: 7 RATSVIFYTLPLKASLPFGYYLLYKFSFLKVLLFLTFPVAIIERSLPFGGLLFFIILFAG 66
Query: 64 VVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFLFLLVC 123
VVRNPN ++R+N QA++LD+ LI L R + L+ S VF+F L
Sbjct: 67 VVRNPNVPYFIRYNACQALLLDIALIIISYLLR---------ILPLVEFGSIVFVFSLSI 117
Query: 124 LIYGSSSCILGQLPRLPIVADAADRQV 150
I+ CI G P +P ++ + Q+
Sbjct: 118 FIFSIFQCINGVEPEIPFISKSVRMQI 144
>ref|ZP_00514544.1| conserved hypothetical protein [Crocosphaera watsonii WH 8501]
gi|67857142|gb|EAM52382.1| conserved hypothetical
protein [Crocosphaera watsonii WH 8501]
Length = 168
Score = 68.9 bits (167), Expect = 2e-11
Identities = 45/157 (28%), Positives = 77/157 (48%), Gaps = 10/157 (6%)
Query: 3 DRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFP--------FNGF 54
DR+ L Y + +D + +G F++ QF +V P +P + F F
Sbjct: 11 DRIFGTLVYCFAVYDTLFFGTFLLQQFPAFNFLVLPAVPVGITYGLVSAILGPLGRFGSF 70
Query: 55 LVFLTLYFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPD-LLERGFNPRGGFGLDLLMSLD 113
LVF+ L+ VVRN S ++R+N MQ+I++ +LL +++ P G G+ L+ +L
Sbjct: 71 LVFILLFAAVVRNDKISHFIRYNAMQSILIGILLALGGIIMQYVLVPALGGGI-LIETLF 129
Query: 114 STVFLFLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
+ +FL L + LG +P +++AA QV
Sbjct: 130 NVLFLGGLAASYFSMIQSALGNYAEIPTISEAAYSQV 166
>ref|NP_926243.1| hypothetical protein glr3297 [Gloeobacter violaceus PCC 7421]
gi|35213868|dbj|BAC91238.1| glr3297 [Gloeobacter
violaceus PCC 7421]
Length = 159
Score = 65.9 bits (159), Expect = 2e-10
Identities = 46/156 (29%), Positives = 83/156 (52%), Gaps = 14/156 (8%)
Query: 2 SDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTLY 61
SDR+ ++L Y P + YG +++Q + A++Q L P F + F G VF+ L+
Sbjct: 10 SDRIFASLPYLLPMAASLAYGVTLLSQL-GLGAVIQILAPIN--FLNSGFIGLAVFIALF 66
Query: 62 FFVVRNPNFSKYVRFNTMQAIVLDVL-----LIFPDLLER-GFNPRGGFGLDLL-MSLDS 114
VV N + S ++RFN QA+++ ++ L+ P L+ F P G+D++ +L +
Sbjct: 67 ALVVNNTSISHFIRFNVFQALLVGIILSLFSLVLPLLVNMFAFLP----GIDVIQQTLGN 122
Query: 115 TVFLFLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
T+FL + IYG +LG+ +P +++ Q+
Sbjct: 123 TLFLGVFAFGIYGIVQSLLGRYAEVPTISEVVYSQL 158
>gb|AAC08246.1| ORF203 [Porphyra purpurea] gi|11465826|ref|NP_053970.1|
hypothetical protein PopuCp175 [Porphyra purpurea]
gi|2147522|pir||S73281 hypothetical protein 203 - red
alga (Porphyra purpurea) chloroplast
gi|1723409|sp|P51360|YCXM_PORPU Hypothetical 23.3 kDa
protein in RPS6-THIG intergenic region (ORF203)
Length = 203
Score = 63.5 bits (153), Expect = 1e-09
Identities = 44/157 (28%), Positives = 86/157 (54%), Gaps = 11/157 (7%)
Query: 2 SDRLISALCYFYPFFDGIQ-YGKFVITQFYPIQAI---VQPLIPAIRVFKSFPFNGFLVF 57
S RL+S + Y+ P F+G+Q +G++V+ YP+ AI + L+P + + + G + F
Sbjct: 45 SIRLVSIIPYYLPLFEGLQNFGQYVLPD-YPVGAIPLYKKILLPMLIFYMNHAILGLVTF 103
Query: 58 LTLYFFVVRNPN---FSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLL-MSLD 113
LY+ +VRN + + VRFN+MQ+I+L ++ + R F F + + +++
Sbjct: 104 FALYYVLVRNKSPITVHQLVRFNSMQSILLFLVGSLFGAIFRAFPIE--FRISFIGLTVC 161
Query: 114 STVFLFLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
+ +F F+L + Y I G+ +P++++A Q+
Sbjct: 162 NMMFWFILSTITYSIVKAIQGKYSNIPVISEAVRIQI 198
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.335 0.153 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 252,628,428
Number of Sequences: 2540612
Number of extensions: 10107774
Number of successful extensions: 37683
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 37616
Number of HSP's gapped (non-prelim): 59
length of query: 150
length of database: 863,360,394
effective HSP length: 126
effective length of query: 24
effective length of database: 543,243,282
effective search space: 13037838768
effective search space used: 13037838768
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122165.12


