

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.2 + phase: 0
(428 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
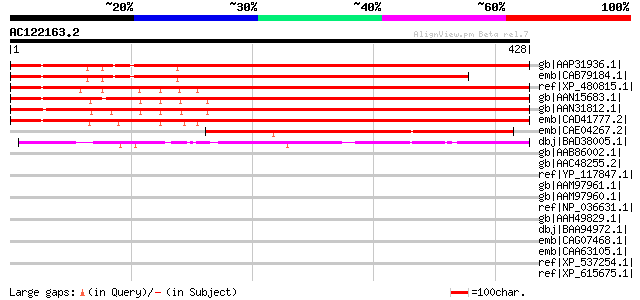
gb|AAP31936.1| At4g22290 [Arabidopsis thaliana] gi|22655310|gb|A... 555 e-156
emb|CAB79184.1| putative protein [Arabidopsis thaliana] gi|28326... 475 e-132
ref|XP_480815.1| unknown protein [Oryza sativa (japonica cultiva... 445 e-123
gb|AAN15683.1| unknown protein [Arabidopsis thaliana] gi|2046570... 433 e-120
gb|AAN31812.1| unknown protein [Arabidopsis thaliana] gi|2143633... 432 e-120
emb|CAD41777.2| OSJNBa0035M09.9 [Oryza sativa (japonica cultivar... 424 e-117
emb|CAE04267.2| OSJNBb0103I08.7 [Oryza sativa (japonica cultivar... 192 1e-47
dbj|BAD38005.1| hypothetical protein [Oryza sativa (japonica cul... 189 2e-46
gb|AAB86002.1| coenzyme F390 synthetase I [Methanothermobacter t... 44 0.010
gb|AAC48255.2| Prion-like-(q/n-rich)-domain-bearing protein prot... 42 0.037
ref|YP_117847.1| hypothetical protein nfa16370 [Nocardia farcini... 42 0.037
gb|AAM97961.1| Prion-like-(q/n-rich)-domain-bearing protein prot... 42 0.037
gb|AAM97960.1| Prion-like-(q/n-rich)-domain-bearing protein prot... 42 0.037
ref|NP_036631.1| adenomatosis polyposis coli [Rattus norvegicus]... 42 0.048
gb|AAH49829.1| Col1a1-prov protein [Xenopus laevis] 41 0.062
dbj|BAA94972.1| type I collagen alpha 1 [Xenopus laevis] 41 0.062
emb|CAG07468.1| unnamed protein product [Tetraodon nigroviridis] 41 0.081
emb|CAA63105.1| coenzyme F390 synthetase [Methanothermobacter th... 40 0.11
ref|XP_537254.1| PREDICTED: similar to Thrombospondin 3 precurso... 40 0.11
ref|XP_615675.1| PREDICTED: similar to KIAA0774 protein, partial... 40 0.11
>gb|AAP31936.1| At4g22290 [Arabidopsis thaliana] gi|22655310|gb|AAM98245.1| unknown
protein [Arabidopsis thaliana]
Length = 445
Score = 555 bits (1429), Expect = e-156
Identities = 289/450 (64%), Positives = 355/450 (78%), Gaps = 27/450 (6%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
M RI SHQL +GLYVSG+ EQPKER PPTMA+R+VPYTGGD KKSGELG+M DI V+D
Sbjct: 1 MAGRIQSHQLPNGLYVSGKLEQPKER-PPTMAARAVPYTGGDIKKSGELGRMFDISVVDS 59
Query: 61 KS-----------HPSSSSSQLSTGP----ARSRPNSGQVGKNITGSGTLSRKSTGSGPI 105
S + S +S+L P + S PNSG V ++ SG++ + S GP+
Sbjct: 60 ASFQGPPPLIVGGNSSGGTSRLQAPPRVSGSSSNPNSGSV-RSGPNSGSVKKFS---GPL 115
Query: 106 A-LQPTGLITSGPVGS-GPV-GASRRSGQLEQSGS---MGKAVYGSAVTSLG-EEVKVGF 158
+ LQPTGLITSG +GS GP+ SRRSGQL+ S K YGS+VTSL + V+VGF
Sbjct: 116 SQLQPTGLITSGSLGSSGPILSGSRRSGQLDHQLSNLASSKPKYGSSVTSLNVDPVRVGF 175
Query: 159 RVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGGVIVPVLVLIIWNCVLGRKGLL 218
+V +++VW ++V AM LLVG FL VAVKK V++ A+ + P +V+++WNCV RKGLL
Sbjct: 176 KVPKAMVWAVLIVAAMGLLVGAFLTVAVKKPVVIAAVLAAVCPAIVVLVWNCVWRRKGLL 235
Query: 219 GFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESSYQRIPRCVYVSSELYEYKGWGGKS 278
F+K+YPDAELRGAIDGQ+VKVTGVVTCGSIPLESS+QR PRCVYVS+ELYEYKG+GGKS
Sbjct: 236 SFIKKYPDAELRGAIDGQFVKVTGVVTCGSIPLESSFQRTPRCVYVSTELYEYKGFGGKS 295
Query: 279 AHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVKAGYGNKVAPFVKPTTVVDVTKENR 338
A+PKHRCF+WGSR++EKY++DFYISDFQ+GLRALVKAGYG+KV+PFVKP TV +VT +N+
Sbjct: 296 ANPKHRCFSWGSRHAEKYVSDFYISDFQSGLRALVKAGYGSKVSPFVKPATVANVTTQNK 355
Query: 339 ELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTG 398
+LSP+FL WL+DR LS DDR+MRLKEG+IKEGSTVSVMG+VRRH+NVLMIVPP E VS+G
Sbjct: 356 DLSPSFLKWLSDRNLSADDRVMRLKEGYIKEGSTVSVMGMVRRHDNVLMIVPPAEAVSSG 415
Query: 399 CQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
C+W CL PT +GLIITC+DNQNADVI V
Sbjct: 416 CRWWHCLFPTYADGLIITCDDNQNADVIPV 445
>emb|CAB79184.1| putative protein [Arabidopsis thaliana] gi|2832679|emb|CAA16779.1|
putative protein [Arabidopsis thaliana]
gi|15235599|ref|NP_193960.1| ubiquitin carboxyl-terminal
hydrolase family protein [Arabidopsis thaliana]
gi|7486824|pir||T04910 hypothetical protein T10I14.120 -
Arabidopsis thaliana
Length = 974
Score = 475 bits (1223), Expect = e-132
Identities = 252/400 (63%), Positives = 313/400 (78%), Gaps = 27/400 (6%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
M RI SHQL +GLYVSG+ EQPKER PPTMA+R+VPYTGGD KKSGELG+M DI V+D
Sbjct: 1 MAGRIQSHQLPNGLYVSGKLEQPKER-PPTMAARAVPYTGGDIKKSGELGRMFDISVVDS 59
Query: 61 KS-----------HPSSSSSQLSTGP----ARSRPNSGQVGKNITGSGTLSRKSTGSGPI 105
S + S +S+L P + S PNSG V ++ SG++ + S GP+
Sbjct: 60 ASFQGPPPLIVGGNSSGGTSRLQAPPRVSGSSSNPNSGSV-RSGPNSGSVKKFS---GPL 115
Query: 106 A-LQPTGLITSGPVGS-GPV-GASRRSGQLEQSGS---MGKAVYGSAVTSLG-EEVKVGF 158
+ LQPTGLITSG +GS GP+ SRRSGQL+ S K YGS+VTSL + V+VGF
Sbjct: 116 SQLQPTGLITSGSLGSSGPILSGSRRSGQLDHQLSNLASSKPKYGSSVTSLNVDPVRVGF 175
Query: 159 RVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGGVIVPVLVLIIWNCVLGRKGLL 218
+V +++VW ++V AM LLVG FL VAVKK V++ A+ + P +V+++WNCV RKGLL
Sbjct: 176 KVPKAMVWAVLIVAAMGLLVGAFLTVAVKKPVVIAAVLAAVCPAIVVLVWNCVWRRKGLL 235
Query: 219 GFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESSYQRIPRCVYVSSELYEYKGWGGKS 278
F+K+YPDAELRGAIDGQ+VKVTGVVTCGSIPLESS+QR PRCVYVS+ELYEYKG+GGKS
Sbjct: 236 SFIKKYPDAELRGAIDGQFVKVTGVVTCGSIPLESSFQRTPRCVYVSTELYEYKGFGGKS 295
Query: 279 AHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVKAGYGNKVAPFVKPTTVVDVTKENR 338
A+PKHRCF+WGSR++EKY++DFYISDFQ+GLRALVKAGYG+KV+PFVKP TV +VT +N+
Sbjct: 296 ANPKHRCFSWGSRHAEKYVSDFYISDFQSGLRALVKAGYGSKVSPFVKPATVANVTTQNK 355
Query: 339 ELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVSVMGV 378
+LSP+FL WL+DR LS DDR+MRLKEG+IKEGSTVSVMG+
Sbjct: 356 DLSPSFLKWLSDRNLSADDRVMRLKEGYIKEGSTVSVMGM 395
>ref|XP_480815.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|51964774|ref|XP_507171.1| PREDICTED OSJNBa0038P10.32
gene product [Oryza sativa (japonica cultivar-group)]
gi|38175699|dbj|BAD01408.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|38175441|dbj|BAD01247.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 482
Score = 445 bits (1144), Expect = e-123
Identities = 236/483 (48%), Positives = 316/483 (64%), Gaps = 56/483 (11%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPV--- 57
MG+R PSHQLS+GLYVSGRPEQPKE+ P + S ++PYTGGD KKSGELGKM D+ V
Sbjct: 1 MGSRFPSHQLSNGLYVSGRPEQPKEK-APVICSTAMPYTGGDIKKSGELGKMFDLHVEKS 59
Query: 58 -------LDPKSHPSSSSSQLSTGP---ARSRPN-SGQVGKNITGSGTLSRKSTGSGPI- 105
P + S + ++GP A R N SG + ++ G+G +R + SGP+
Sbjct: 60 RKSGPLGNQPSRNTSFGGAGSNSGPVSNALGRSNYSGSISSSVPGAGGSARAKSNSGPLN 119
Query: 106 ------------------------------ALQPTGLITSGPVGSGPV---GASRR-SGQ 131
L TGLITSGP+ SGP+ GA R+ SG
Sbjct: 120 KHGEPGKKSSGPQSGGVTPMARQNSGPLPPVLPTTGLITSGPISSGPLNSSGAPRKVSGP 179
Query: 132 LEQSGSM----GKAVYGSAVTSLGEE--VKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVA 185
L+ S SM + AVT+L + + + ++++W+ +++ M + G F++ A
Sbjct: 180 LDPSVSMKMRATSFAHNPAVTNLNADDGYSIKGSIPKTILWMVILLFLMGFIAGGFILGA 239
Query: 186 VKKNVILFALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVT 245
V ++L + + V L+IWN G +G+ GFV RYPDA+LR A DGQYVKVTGVVT
Sbjct: 240 VHNPILLVVVVVIFCFVAALVIWNICWGTRGVTGFVSRYPDADLRTAKDGQYVKVTGVVT 299
Query: 246 CGSIPLESSYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDF 305
CG+ PLESS+QR+PRCVY S+ LYEY+GW K+A+ +HR FTWG R E++ DFYISDF
Sbjct: 300 CGNFPLESSFQRVPRCVYTSTCLYEYRGWDSKAANTEHRQFTWGLRSMERHAVDFYISDF 359
Query: 306 QTGLRALVKAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEG 365
Q+GLRALVK GYG +V P+V + V+D+ +N+++SP FL WL +R LS+DDRIMRLKEG
Sbjct: 360 QSGLRALVKTGYGARVTPYVDESVVIDINPDNKDMSPEFLRWLRERNLSSDDRIMRLKEG 419
Query: 366 HIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADV 425
+IKEGSTVSVMGVV+R++NVLMIVPP+EP+STGCQW +C+LPT ++GL++ CED N DV
Sbjct: 420 YIKEGSTVSVMGVVQRNDNVLMIVPPSEPISTGCQWAKCILPTSLDGLVLRCEDTSNIDV 479
Query: 426 IAV 428
I V
Sbjct: 480 IPV 482
>gb|AAN15683.1| unknown protein [Arabidopsis thaliana] gi|20465703|gb|AAM20320.1|
unknown protein [Arabidopsis thaliana]
gi|18176178|gb|AAL59998.1| unknown protein [Arabidopsis
thaliana] gi|21539479|gb|AAM53292.1| unknown protein
[Arabidopsis thaliana] gi|15219232|ref|NP_178009.1|
balbiani ring 1-related / BR1-related [Arabidopsis
thaliana] gi|3834307|gb|AAC83023.1| Strong similarity to
gene T10I14.120 gi|2832679 putative protein from
Arabidopsis thaliana BAC gb|AL021712. ESTs gb|N65887
and gb|N65627 come from this gene
gi|25372814|pir||C96818 hypothetical protein F9K20.7
[imported] - Arabidopsis thaliana
Length = 468
Score = 433 bits (1113), Expect = e-120
Identities = 235/472 (49%), Positives = 306/472 (64%), Gaps = 48/472 (10%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+R SHQLS+GL+VSGRPEQPKE+ PPTM+S ++PYTGGD KKSGELGKM DIP
Sbjct: 1 MGSRYASHQLSNGLFVSGRPEQPKEK-PPTMSSVAMPYTGGDIKKSGELGKMFDIPTDGT 59
Query: 61 KSHPS------SSSSQLSTGPARSRPNS-GQVGKNITGSGTLSRKSTGSGPIA------- 106
KS S SS S +GP PN+ G++ N+ +G+ S K T SGP++
Sbjct: 60 KSRKSGPITGGSSRSGAQSGPV---PNATGRMSGNLASAGSNSMKKTNSGPLSKHGEPLK 116
Query: 107 --------------------LQPTGLITSGPVGSGPV---GASRR-SGQLEQSGSMG--- 139
L TGLITSGP+ SGP+ GA R+ SG L+ SGSM
Sbjct: 117 KSSGPQSGGVTRQNSGPIPILPTTGLITSGPITSGPLNSSGAPRKISGPLDYSGSMKTHM 176
Query: 140 -KAVYGSAVTSLGEEVKVGFRVS--RSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALG 196
V+ AVT+L E S + V+W+ +++ M L G F++ AV ++L +
Sbjct: 177 PSVVHNQAVTTLAPEDDFSCMKSFPKPVLWLVILIFVMGFLAGGFILGAVHNAILLIVVA 236
Query: 197 GVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESSYQ 256
+ V L IWN R+G+ F+ RYPDA+LR A +GQYVKVTGVVTCG++PLESS+
Sbjct: 237 VLFTVVAALFIWNISCERRGITDFIARYPDADLRTAKNGQYVKVTGVVTCGNVPLESSFH 296
Query: 257 RIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVKAG 316
R+PRCVY S+ LYEY+GWG K A+ HR FTWG R +E+++ DFYISDFQ+GLRALVK G
Sbjct: 297 RVPRCVYTSTCLYEYRGWGSKPANASHRRFTWGLRSAERHVVDFYISDFQSGLRALVKTG 356
Query: 317 YGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVSVM 376
G KV P V + V+D N + SP+F+ WL + L+ DDRIMRLKEG+IKEGSTVSV+
Sbjct: 357 NGAKVTPLVDDSVVIDFKPGNEQASPDFVRWLGKKNLTNDDRIMRLKEGYIKEGSTVSVI 416
Query: 377 GVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
GVV+R++NVLMIVP TEP++ G QW +C P +EG+++ CED+ N D I V
Sbjct: 417 GVVQRNDNVLMIVPTTEPLAAGWQWSKCTFPASLEGIVLRCEDSSNVDAIPV 468
>gb|AAN31812.1| unknown protein [Arabidopsis thaliana] gi|21436333|gb|AAM51336.1|
unknown protein [Arabidopsis thaliana]
gi|15292907|gb|AAK92824.1| unknown protein [Arabidopsis
thaliana] gi|18394411|ref|NP_564009.1| merozoite surface
protein-related [Arabidopsis thaliana]
gi|25372815|pir||H86303 hypothetical protein F6I1.14
[imported] - Arabidopsis thaliana
gi|9802778|gb|AAF99847.1| Unknown protein [Arabidopsis
thaliana]
Length = 474
Score = 432 bits (1112), Expect = e-120
Identities = 233/475 (49%), Positives = 311/475 (65%), Gaps = 48/475 (10%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+R PSHQLS+GL+VSGRPEQPKER P TM++ ++PYTGGD K+SGELGKM DIP
Sbjct: 1 MGSRYPSHQLSNGLFVSGRPEQPKERAP-TMSAVAMPYTGGDIKRSGELGKMFDIPADGT 59
Query: 61 KSHPSS------SSSQLSTGPARSRPNS----GQVGKNITGSGTLSRKSTGSGPIA---- 106
KS S S S G A+S P + G++ ++ +G++S K T SGP++
Sbjct: 60 KSRKSGPIPGAPSRSGSFAGTAQSGPGAPMATGRMSGSLASAGSVSMKKTNSGPLSKHGE 119
Query: 107 -----------------------LQPTGLITSGPVGSGPV---GASRR-SGQLEQSGSMG 139
L TGLITSGP+ SGP+ GA R+ SG L+ SG M
Sbjct: 120 PLKKSSGPQSGGVTRQNSGSIPILPATGLITSGPITSGPLNSSGAPRKVSGPLDSSGLMK 179
Query: 140 K----AVYGSAVTSLGEEVKVGFRVS--RSVVWVFMVVVAMCLLVGVFLMVAVKKNVILF 193
V+ AVT+LG E S + V+W+ +++ M L G F++ AV ++L
Sbjct: 180 SHMPTVVHNQAVTTLGPEDDFSCLKSFPKPVLWLVVLIFIMGFLAGGFILGAVHNPILLV 239
Query: 194 ALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLES 253
+ + V L IWN GR+G+ F+ RYPDA+LR A +GQ+VKVTGVVTCG++PLES
Sbjct: 240 VVAILFTVVAALFIWNICWGRRGITDFIARYPDADLRTAKNGQHVKVTGVVTCGNVPLES 299
Query: 254 SYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALV 313
S+ R+PRCVY S+ LYEY+GWG K A+ HR FTWG R SE+++ DFYISDFQ+GLRALV
Sbjct: 300 SFHRVPRCVYTSTCLYEYRGWGSKPANSSHRHFTWGLRSSERHVVDFYISDFQSGLRALV 359
Query: 314 KAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTV 373
K G G KV P V + V+D + + ++SP+F+ WL + L++DDRIMRLKEG+IKEGSTV
Sbjct: 360 KTGSGAKVTPLVDDSVVIDFKQGSEQVSPDFVRWLGKKNLTSDDRIMRLKEGYIKEGSTV 419
Query: 374 SVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
SV+GVV+R++NVLMIVP +EP++ G QW RC PT +EG+++ CED+ N D I V
Sbjct: 420 SVIGVVQRNDNVLMIVPSSEPLAAGWQWRRCTFPTSLEGIVLRCEDSSNVDAIPV 474
>emb|CAD41777.2| OSJNBa0035M09.9 [Oryza sativa (japonica cultivar-group)]
gi|50928559|ref|XP_473807.1| OSJNBa0035M09.9 [Oryza
sativa (japonica cultivar-group)]
Length = 482
Score = 424 bits (1089), Expect = e-117
Identities = 231/483 (47%), Positives = 311/483 (63%), Gaps = 56/483 (11%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDI-PVLD 59
MG+R PSHQLS+GLYVSGRPEQPKE+ PT+ S ++PYTGGD KKSGELGKM ++ V
Sbjct: 1 MGSRFPSHQLSNGLYVSGRPEQPKEK-APTICSTAMPYTGGDIKKSGELGKMFELHAVKS 59
Query: 60 PKSHP---------------------------------SSSSSQLSTGPARSRPNSGQVG 86
KS P SSS ++G AR++ +SG +
Sbjct: 60 RKSGPLSNAPSRNASFGGAASNSGPVPNAGDRSNYSGSLSSSVPGASGSARAKSSSGPLN 119
Query: 87 KN-----------ITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPV---GASRR-SGQ 131
K+ G ++R+++G P L TGLITSGP+ SGP+ GA R+ SG
Sbjct: 120 KHGEPVKRSSGPQSGGVTPMARQNSGPLPPMLPTTGLITSGPITSGPLNSSGAQRKVSGP 179
Query: 132 LEQSGSMGKAV----YGSAVTSLGEE--VKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVA 185
L+ + S + AVT + E + +S+ ++ V+ + L+ G+ ++ A
Sbjct: 180 LDSAASKKTRATSFSHNQAVTKITTEDSYSITGSLSKLILGAVGVLFVLGLIAGILILSA 239
Query: 186 VKKNVILFALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVT 245
V ++L + + V L IWN R+G++GFV RY DA+LR A DGQY+KVTGVVT
Sbjct: 240 VHNAILLIVVLVLFGFVAALFIWNACWARRGVIGFVDRYSDADLRTAKDGQYIKVTGVVT 299
Query: 246 CGSIPLESSYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDF 305
CG+ PLESSYQR+PRCVY S+ L+EY+GW K+A+ +H FTWG R E++ DFYISDF
Sbjct: 300 CGNFPLESSYQRVPRCVYTSTTLHEYRGWDSKAANTQHHRFTWGLRSMEQHAVDFYISDF 359
Query: 306 QTGLRALVKAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEG 365
Q+GLRALVKAGYG +V PFV + ++D+ +N+++SP F WL +R LS+DDRIMRLKEG
Sbjct: 360 QSGLRALVKAGYGARVTPFVDESVIIDIDPDNKDMSPEFRRWLRERNLSSDDRIMRLKEG 419
Query: 366 HIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADV 425
+IKEGSTVSVMGVV++++NVLMIVPP EP+STGCQW +C+LP + GL++ CED N DV
Sbjct: 420 YIKEGSTVSVMGVVQKNDNVLMIVPPPEPISTGCQWAKCVLPRDLYGLVLRCEDTSNIDV 479
Query: 426 IAV 428
IAV
Sbjct: 480 IAV 482
>emb|CAE04267.2| OSJNBb0103I08.7 [Oryza sativa (japonica cultivar-group)]
gi|50927160|ref|XP_473366.1| OSJNBb0103I08.7 [Oryza
sativa (japonica cultivar-group)]
Length = 465
Score = 192 bits (489), Expect = 1e-47
Identities = 98/257 (38%), Positives = 157/257 (60%), Gaps = 4/257 (1%)
Query: 162 RSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGGVIVPVLVLIIWNCVLGRKG--LLG 219
R+ VW ++A+ L +G ++ V +L + V+ ++WN G L
Sbjct: 207 RAAVWAVAALLAVGLGLGALVLAVVHSAALLVVAVLLSAAVVAFLLWNAAASASGRALRR 266
Query: 220 FVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESSYQRIPRCVYVSSELYEYKGWGGKSA 279
FV P + LR A D Q VK+TG+V CG I L SSY+++ CVY S+ L + WG + A
Sbjct: 267 FVDGLPASSLRSATDDQLVKITGLVACGDISLISSYEKVENCVYTSTLLRKCGRWGSEVA 326
Query: 280 HPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVKAGYGNKVAPFVKPTTVVDVTKENRE 339
+PK+RC W ++E++ ADFYI+D ++G RALVKAG+ ++V P + +V T N E
Sbjct: 327 NPKNRCSKWKLTHAERFAADFYITDAKSGKRALVKAGHDSRVVPLIDENLLV-TTSGNTE 385
Query: 340 LSPNFLGWLADRKLSTDD-RIMRLKEGHIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTG 398
LS WL +R + +++ +++RL+EG+I EG +SV+G++ + + LMI+PP EP+STG
Sbjct: 386 LSSTLRCWLDERNIPSEECQLIRLEEGYIAEGMRLSVIGILSKKDGDLMILPPPEPISTG 445
Query: 399 CQWMRCLLPTGVEGLII 415
C ++ LLPT +G+++
Sbjct: 446 CVFLSFLLPTYFDGIVL 462
>dbj|BAD38005.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 491
Score = 189 bits (479), Expect = 2e-46
Identities = 146/433 (33%), Positives = 207/433 (47%), Gaps = 54/433 (12%)
Query: 8 HQLSSGLYVSG-RPEQPKERQPPTMASRSVP-YTGGDPKKSGELGKMLDIPVLDPKSHPS 65
H++ SG+YVSG P++ KER+ + S + P YTGGD +SGELG+M DI
Sbjct: 101 HKIGSGMYVSGPAPDRGKERRQLSSGSVATPPYTGGDVSRSGELGRMFDI---------- 150
Query: 66 SSSSQLSTGPARSRPNSGQVGKNIT-----GSGTLSRKSTGS---GPIALQPTGLITSGP 117
PA SR +SG + + + SG LS+ S GP P P
Sbjct: 151 ---GGAGVSPASSRRSSGPLPRPLPLLPSPASGPLSQLSHSGLLVGPSPPPPPPQTQQSP 207
Query: 118 VGSGPVGASRRSGQLEQSGSMGKAVYGSAVTSLGEEVKVGFRVSRSVVWVFMVVVAMCLL 177
GS + RR E++ + +A G A LG V V V+ SV L
Sbjct: 208 AGSWRKSSRRR----EEAAAAPEAARGRA--RLG--VSVACYVAASVA------ATAGLG 253
Query: 178 VGVFLMVAVKKNVILFALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDA--ELRGAIDG 235
G F +VA + +L A GG + V WN F +R PD + G
Sbjct: 254 AGAFFLVAWHRWEVLSAAGGAVAAVAAAFAWNVRRRDAEAERFFRRLPDTVFDQSDMPIG 313
Query: 236 QYVKVTGVVTCGSIPLESSYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEK 295
+ VK+TG VTCG PL + + RC++ S +LYE +G CF W +SE
Sbjct: 314 ELVKITGQVTCGHQPLGARFHDAARCIFTSVQLYERRGC----------CFRWQQTHSET 363
Query: 296 YIADFYISDFQTGLRALVKAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLST 355
A+FYISD TG R V+AG G K+ +K T + E + S N W+A LS
Sbjct: 364 RTANFYISDRNTGKRFYVRAGEGGKITWMIKQKTD-SLDGERKGASRNLKSWMASNDLSC 422
Query: 356 DDRIMRLKEGHIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLII 415
D + +KEG EG T SV+GV+++H ++ P+ V+TGCQ+ RC+ P VEGLI+
Sbjct: 423 DGTV-HVKEG---EGDTASVIGVLKKHHAYDIVDAPSGVVTTGCQFTRCMFPVHVEGLIL 478
Query: 416 TCEDNQNADVIAV 428
+++ + +V V
Sbjct: 479 VGDEDPDDEVYMV 491
>gb|AAB86002.1| coenzyme F390 synthetase I [Methanothermobacter thermautotrophicus
str. Delta H] gi|15679524|ref|NP_276641.1| coenzyme F390
synthetase I [Methanothermobacter thermautotrophicus
str. Delta H] gi|7482061|pir||G69070 coenzyme F390
synthetase I - Methanobacterium thermoautotrophicum
(strain Delta H)
Length = 453
Score = 43.9 bits (102), Expect = 0.010
Identities = 24/57 (42%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query: 267 ELYEYKGWGGKSAHPKHRCFTWGS--RYSEKYIADFYISDFQTGLRALVKAGYGNKV 321
E+Y G S PK TWG RY+EKY F F+TG R +V A YG V
Sbjct: 94 EIYTIHETSGTSGRPKSFFLTWGDWQRYAEKYARSFVSQGFETGDRVVVCASYGMNV 150
>gb|AAC48255.2| Prion-like-(q/n-rich)-domain-bearing protein protein 75, isoform a
[Caenorhabditis elegans] gi|25152832|ref|NP_741365.1|
prion-like Q/N-rich domain protein PQN-75, Prion-like
Q/N-rich domain protein (pqn-75) [Caenorhabditis
elegans]
Length = 524
Score = 42.0 bits (97), Expect = 0.037
Identities = 43/138 (31%), Positives = 50/138 (36%), Gaps = 33/138 (23%)
Query: 22 QPKERQPPTMASRSV------PYTGGDPKKSGELGKMLDIPVLDPKSHP----------S 65
Q ER PPT + + P GG K S E + + P P+ P S
Sbjct: 344 QAPERSPPTGSPPTGSPPTGRPPRGGPGKSSEESSESREGPRGGPRGGPRGGPRKSSEES 403
Query: 66 SSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLIT----------- 114
S S + GP RS P G TGS R GS P PTGL +
Sbjct: 404 SESREEPRGPRRSPPT----GSPPTGSPPTGRPPRGSPPTGSPPTGLPSRQKRQAPEDRP 459
Query: 115 --SGPVGSGPVGASRRSG 130
S P GS P G R G
Sbjct: 460 TGSPPTGSPPTGRPHRGG 477
Score = 34.7 bits (78), Expect = 5.8
Identities = 32/114 (28%), Positives = 42/114 (36%), Gaps = 10/114 (8%)
Query: 20 PEQPKERQPPTMASRSVPYTGGDPKKSGELGKM-LDIPVLDPKSHPSS--SSSQLSTGPA 76
P P+ P P TG P+ S G +P + P + S + P
Sbjct: 410 PRGPRRSPPTGSPPTGSPPTGRPPRGSPPTGSPPTGLPSRQKRQAPEDRPTGSPPTGSPP 469
Query: 77 RSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSG 130
RP+ G GK S + + GP PTG S P GS P GA + G
Sbjct: 470 TGRPHRGGPGK----SESSESREGPRGPRRSPPTG---SPPTGSPPTGAPPKKG 516
>ref|YP_117847.1| hypothetical protein nfa16370 [Nocardia farcinica IFM 10152]
gi|54015113|dbj|BAD56483.1| hypothetical protein
[Nocardia farcinica IFM 10152]
Length = 461
Score = 42.0 bits (97), Expect = 0.037
Identities = 41/159 (25%), Positives = 58/159 (35%), Gaps = 28/159 (17%)
Query: 12 SGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQL 71
+G SG P P + P +SR+ P G DP D P DP+ SS
Sbjct: 279 AGTNPSGNPNTPAAQDP---SSRNDPTAGDDPAD--------DSPYEDPQGTNPQSSEPQ 327
Query: 72 STGPARSRPNSGQVGKNIT------GSGTLSRKSTGSGPIALQPTGLI-----------T 114
ST P + P++ G +++ S T S S G G L G T
Sbjct: 328 STVPQSTVPSAAPPGSSLSDPSGRPNSSTPSLGSPGLGAPGLGSPGATPSPGSSVPGAKT 387
Query: 115 SGPVGSGPVGASRRSGQLEQSGSMGKAVYGSAVTSLGEE 153
+ P G G +G ++G G G+ G+E
Sbjct: 388 AQPTGLGTAATRAATGAAGRAGMPGMGAMGAGAGRRGDE 426
>gb|AAM97961.1| Prion-like-(q/n-rich)-domain-bearing protein protein 75, isoform c
[Caenorhabditis elegans] gi|25152837|ref|NP_741366.1|
prion-like Q/N-rich domain protein PQN-75, Prion-like
Q/N-rich domain protein (pqn-75) [Caenorhabditis
elegans] gi|7508879|pir||T28770 hypothetical protein
W03D2.1 - Caenorhabditis elegans
Length = 539
Score = 42.0 bits (97), Expect = 0.037
Identities = 43/138 (31%), Positives = 50/138 (36%), Gaps = 33/138 (23%)
Query: 22 QPKERQPPTMASRSV------PYTGGDPKKSGELGKMLDIPVLDPKSHP----------S 65
Q ER PPT + + P GG K S E + + P P+ P S
Sbjct: 359 QAPERSPPTGSPPTGSPPTGRPPRGGPGKSSEESSESREGPRGGPRGGPRGGPRKSSEES 418
Query: 66 SSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLIT----------- 114
S S + GP RS P G TGS R GS P PTGL +
Sbjct: 419 SESREEPRGPRRSPPT----GSPPTGSPPTGRPPRGSPPTGSPPTGLPSRQKRQAPEDRP 474
Query: 115 --SGPVGSGPVGASRRSG 130
S P GS P G R G
Sbjct: 475 TGSPPTGSPPTGRPHRGG 492
Score = 34.7 bits (78), Expect = 5.8
Identities = 32/114 (28%), Positives = 42/114 (36%), Gaps = 10/114 (8%)
Query: 20 PEQPKERQPPTMASRSVPYTGGDPKKSGELGKM-LDIPVLDPKSHPSS--SSSQLSTGPA 76
P P+ P P TG P+ S G +P + P + S + P
Sbjct: 425 PRGPRRSPPTGSPPTGSPPTGRPPRGSPPTGSPPTGLPSRQKRQAPEDRPTGSPPTGSPP 484
Query: 77 RSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSG 130
RP+ G GK S + + GP PTG S P GS P GA + G
Sbjct: 485 TGRPHRGGPGK----SESSESREGPRGPRRSPPTG---SPPTGSPPTGAPPKKG 531
>gb|AAM97960.1| Prion-like-(q/n-rich)-domain-bearing protein protein 75, isoform b
[Caenorhabditis elegans] gi|25152834|ref|NP_741367.1|
prion-like Q/N-rich domain protein PQN-75, Prion-like
Q/N-rich domain protein (50.4 kD) (pqn-75)
[Caenorhabditis elegans]
Length = 518
Score = 42.0 bits (97), Expect = 0.037
Identities = 43/138 (31%), Positives = 50/138 (36%), Gaps = 33/138 (23%)
Query: 22 QPKERQPPTMASRSV------PYTGGDPKKSGELGKMLDIPVLDPKSHP----------S 65
Q ER PPT + + P GG K S E + + P P+ P S
Sbjct: 338 QAPERSPPTGSPPTGSPPTGRPPRGGPGKSSEESSESREGPRGGPRGGPRGGPRKSSEES 397
Query: 66 SSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLIT----------- 114
S S + GP RS P G TGS R GS P PTGL +
Sbjct: 398 SESREEPRGPRRSPPT----GSPPTGSPPTGRPPRGSPPTGSPPTGLPSRQKRQAPEDRP 453
Query: 115 --SGPVGSGPVGASRRSG 130
S P GS P G R G
Sbjct: 454 TGSPPTGSPPTGRPHRGG 471
Score = 34.7 bits (78), Expect = 5.8
Identities = 32/114 (28%), Positives = 42/114 (36%), Gaps = 10/114 (8%)
Query: 20 PEQPKERQPPTMASRSVPYTGGDPKKSGELGKM-LDIPVLDPKSHPSS--SSSQLSTGPA 76
P P+ P P TG P+ S G +P + P + S + P
Sbjct: 404 PRGPRRSPPTGSPPTGSPPTGRPPRGSPPTGSPPTGLPSRQKRQAPEDRPTGSPPTGSPP 463
Query: 77 RSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSG 130
RP+ G GK S + + GP PTG S P GS P GA + G
Sbjct: 464 TGRPHRGGPGK----SESSESREGPRGPRRSPPTG---SPPTGSPPTGAPPKKG 510
>ref|NP_036631.1| adenomatosis polyposis coli [Rattus norvegicus]
gi|10719873|sp|P70478|APC_RAT Adenomatous polyposis coli
protein (APC protein) gi|1580725|dbj|BAA07609.1| APC
protein [Rattus norvegicus]
Length = 2842
Score = 41.6 bits (96), Expect = 0.048
Identities = 33/127 (25%), Positives = 48/127 (36%), Gaps = 3/127 (2%)
Query: 19 RPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPAR- 77
+P E P T + + + P +SG P P S P S + S P R
Sbjct: 2275 KPAVKSELSPITRQTSHISGSNKGPSRSGSRDSTPSRPTQQPLSRPMQSPGRNSISPGRN 2334
Query: 78 --SRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSGQLEQS 135
S PN + T S KS+GSG ++ G S S G S+ + + +S
Sbjct: 2335 GISTPNKLSQLPRTSSPSTASTKSSGSGKMSYTSPGRQLSQQNLSKQTGLSKNASSIPRS 2394
Query: 136 GSMGKAV 142
S K +
Sbjct: 2395 ESASKGL 2401
>gb|AAH49829.1| Col1a1-prov protein [Xenopus laevis]
Length = 1449
Score = 41.2 bits (95), Expect = 0.062
Identities = 39/131 (29%), Positives = 51/131 (38%), Gaps = 8/131 (6%)
Query: 17 SGRPEQPKERQPP-TMASRSVPYTGGDPKKSGELG------KMLDIPVLDPKSHPSSSSS 69
+G+P +P ER PP +R +P T G P G G D PK P S
Sbjct: 221 AGKPGRPGERGPPGPQGARGLPGTAGLPGMKGHRGFNGLDGAKGDSGPAGPKGEPGSPGE 280
Query: 70 QLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVG-SGPVGASRR 128
+ G R SG+ G+ +R + G+ A P SGP G G VG
Sbjct: 281 NGAPGQVGPRGLSGERGRPGPSGPAGARGNDGAPGAAGPPGSTGPSGPPGFPGGVGPKGD 340
Query: 129 SGQLEQSGSMG 139
+G GS G
Sbjct: 341 AGPQGSRGSDG 351
>dbj|BAA94972.1| type I collagen alpha 1 [Xenopus laevis]
Length = 1447
Score = 41.2 bits (95), Expect = 0.062
Identities = 39/131 (29%), Positives = 51/131 (38%), Gaps = 8/131 (6%)
Query: 17 SGRPEQPKERQPP-TMASRSVPYTGGDPKKSGELG------KMLDIPVLDPKSHPSSSSS 69
+G+P +P ER PP +R +P T G P G G D PK P S
Sbjct: 221 AGKPGRPGERGPPGPQGARGLPGTAGLPGMKGHRGFNGLDGAKGDSGPAGPKGEPGSPGE 280
Query: 70 QLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVG-SGPVGASRR 128
+ G R SG+ G+ +R + G+ A P SGP G G VG
Sbjct: 281 NGAPGQVGPRGLSGERGRPGPSGPAGARGNDGAPGAAGPPGSTGPSGPPGFPGGVGPKGD 340
Query: 129 SGQLEQSGSMG 139
+G GS G
Sbjct: 341 AGPQGSRGSDG 351
>emb|CAG07468.1| unnamed protein product [Tetraodon nigroviridis]
Length = 764
Score = 40.8 bits (94), Expect = 0.081
Identities = 37/125 (29%), Positives = 50/125 (39%), Gaps = 13/125 (10%)
Query: 19 RPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTG--PA 76
RPE P P A R+V GG P S + P P++ PSS+ + ST P
Sbjct: 575 RPEAPA----PKAAPRTVGPVGGSPAPSTQ-------PSSRPQAAPSSTPAPASTPAPPT 623
Query: 77 RSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSGQLEQSG 136
+ PN GQ + ++ + R P P SGP P+ R + E S
Sbjct: 624 ATLPNPGQEPRKLSYAEVCQRPPKDPPPATPAPAPSGPSGPSSGQPLRELRVNKPEEPSS 683
Query: 137 SMGKA 141
S G A
Sbjct: 684 SSGPA 688
>emb|CAA63105.1| coenzyme F390 synthetase [Methanothermobacter thermautotrophicus]
Length = 377
Score = 40.4 bits (93), Expect = 0.11
Identities = 22/57 (38%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Query: 267 ELYEYKGWGGKSAHPKHRCFTWGS--RYSEKYIADFYISDFQTGLRALVKAGYGNKV 321
++Y G S PK TWG RY+EKY F F+ G R +V A YG V
Sbjct: 90 DIYTIHETSGTSGRPKSFFLTWGDWQRYAEKYARSFVSQGFERGDRVVVCASYGMNV 146
>ref|XP_537254.1| PREDICTED: similar to Thrombospondin 3 precursor [Canis familiaris]
Length = 1702
Score = 40.4 bits (93), Expect = 0.11
Identities = 38/137 (27%), Positives = 54/137 (38%), Gaps = 19/137 (13%)
Query: 19 RPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARS 78
R KE + P S P GG+ + S P+SSSS + S
Sbjct: 1216 RETNRKEEEGPVSGSIITPAQGGNSSLT---------------SSPASSSSSTLRKHSSS 1260
Query: 79 RPNSGQVGKNITGSGTLSRKSTGSGPIA---LQPTGLITSGPVGSGPVGASRRSGQLEQS 135
S + T G LS S S P++ + P TS P SGP +S + + S
Sbjct: 1261 SLTSSPASSSTTSLGKLSSSSLTSSPVSGSIITPAQHGTSSPT-SGPASSSSSTLRKHSS 1319
Query: 136 GSMGKAVYGSAVTSLGE 152
S+ + S+ TSLG+
Sbjct: 1320 SSLTSSPASSSTTSLGK 1336
>ref|XP_615675.1| PREDICTED: similar to KIAA0774 protein, partial [Bos taurus]
Length = 877
Score = 40.4 bits (93), Expect = 0.11
Identities = 46/153 (30%), Positives = 63/153 (41%), Gaps = 28/153 (18%)
Query: 13 GLYVSGRPEQPKERQPPTMAS------RSVPYTGG------DPKKSGELGKMLDIPVLDP 60
G+ GR E E QP A+ VP+ G +P+K GE G + P
Sbjct: 341 GVSPLGRKEPAGEVQPGASAAGLSSLAHGVPHPPGAGRDRGEPEKGGEDGSLR----TTP 396
Query: 61 KSHPSSSSSQLSTGPARSRPNSG-QVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVG 119
K P SS + +RS P S ++G+ + S S G G +A P
Sbjct: 397 KPSPGSSRAADQQPTSRSLPGSARKLGEMVPNS-----VSPGDGHVAFVP-----GNSTD 446
Query: 120 SGPVGASRRSGQLEQSGSMGKAVYGSAVTSLGE 152
S P GAS G L S+ V+ SA +S+GE
Sbjct: 447 SKPSGASDEKGVLGDGNSVNTMVFDSA-SSVGE 478
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 771,330,424
Number of Sequences: 2540612
Number of extensions: 34694808
Number of successful extensions: 114506
Number of sequences better than 10.0: 327
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 336
Number of HSP's that attempted gapping in prelim test: 113136
Number of HSP's gapped (non-prelim): 1281
length of query: 428
length of database: 863,360,394
effective HSP length: 131
effective length of query: 297
effective length of database: 530,540,222
effective search space: 157570445934
effective search space used: 157570445934
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122163.2


