

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.8 + phase: 0
(393 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
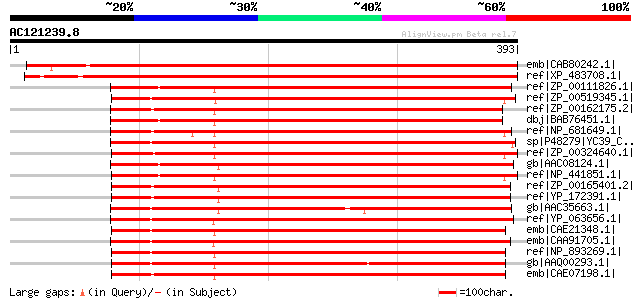
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB80242.1| putative protein [Arabidopsis thaliana] gi|30804... 636 0.0
ref|XP_483708.1| putative NADPH-dependent reductase [Oryza sativ... 624 e-177
ref|ZP_00111826.1| COG0702: Predicted nucleoside-diphosphate-sug... 340 4e-92
ref|ZP_00519345.1| Isoflavone reductase [Crocosphaera watsonii W... 333 4e-90
ref|ZP_00162175.2| COG0702: Predicted nucleoside-diphosphate-sug... 328 1e-88
dbj|BAB76451.1| all4752 [Nostoc sp. PCC 7120] gi|25534778|pir||A... 328 2e-88
ref|NP_681649.1| chaperon-like protein for quinone binding in ph... 324 3e-87
sp|P48279|YC39_CYAPA Hypothetical 36.4 kDa protein ycf39 gi|1146... 323 6e-87
ref|ZP_00324640.1| COG0702: Predicted nucleoside-diphosphate-sug... 320 5e-86
gb|AAC08124.1| hypothetical chloroplast ORF 39. [Porphyra purpur... 319 8e-86
ref|NP_441851.1| ycf39 gene product [Synechocystis sp. PCC 6803]... 310 5e-83
ref|ZP_00165401.2| COG0702: Predicted nucleoside-diphosphate-sug... 301 2e-80
ref|YP_172391.1| chaperon-like protein for quinone binding in ph... 300 5e-80
gb|AAC35663.1| hypothetical chloroplast RF39 [Guillardia theta] ... 294 4e-78
ref|YP_063656.1| conserved hypothetical plastid protein [Gracila... 283 6e-75
emb|CAE21348.1| putative chaperon-like protein for quinone bindi... 275 2e-72
emb|CAA91705.1| ORF319 [Odontella sinensis] gi|11467527|ref|NP_0... 272 1e-71
ref|NP_893269.1| putative chaperon-like protein for quinone bind... 254 4e-66
gb|AAQ00293.1| NADPH-dependent reductase [Prochlorococcus marinu... 253 5e-66
emb|CAE07198.1| putative chaperon-like protein for quinone bindi... 252 2e-65
>emb|CAB80242.1| putative protein [Arabidopsis thaliana] gi|3080425|emb|CAA18744.1|
putative protein [Arabidopsis thaliana]
gi|22137054|gb|AAM91372.1| At4g35250/F23E12_190
[Arabidopsis thaliana] gi|18087583|gb|AAL58922.1|
AT4g35250/F23E12_190 [Arabidopsis thaliana]
gi|15236930|ref|NP_195251.1| vestitone reductase-related
[Arabidopsis thaliana] gi|7486019|pir||T06132
hypothetical protein F23E12.190 - Arabidopsis thaliana
Length = 395
Score = 636 bits (1640), Expect = 0.0
Identities = 324/392 (82%), Positives = 339/392 (85%), Gaps = 15/392 (3%)
Query: 14 PGRVVTRTREAFHALSPS------------PSPSPPHFVEFCKGGRRGRRIIRVGVKCNS 61
P ++VTR H S S P + P F R I V V C
Sbjct: 7 PAQLVTRGNLIHHNSSSSSSGRLSWRRSLTPENTIPLFPSSSSSSLNRERSIVVPVTC-- 64
Query: 62 NSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW 121
S AVNL PGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW
Sbjct: 65 -SAAAVNLAPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW 123
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
GATVVNADLSKPETIPATLVG+HTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY
Sbjct: 124 GATVVNADLSKPETIPATLVGIHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 183
Query: 182 VFYSIHNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSV 241
VFYSIHNCDKHPEVPLMEIKYCTE FL++SGLNHI IRLCGFMQGLIGQYAVPILEEKSV
Sbjct: 184 VFYSIHNCDKHPEVPLMEIKYCTEKFLQESGLNHITIRLCGFMQGLIGQYAVPILEEKSV 243
Query: 242 WGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDA 301
WGTDAPTR+AYMDTQDIARLT IALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDA
Sbjct: 244 WGTDAPTRVAYMDTQDIARLTLIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDA 303
Query: 302 NVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDI 361
NVTTVPVSVLR+TRQLTRFF+WTNDVADRLAFSEVL+SDTVFS PM ET +LLGVD KD+
Sbjct: 304 NVTTVPVSVLRVTRQLTRFFQWTNDVADRLAFSEVLSSDTVFSAPMTETNSLLGVDQKDM 363
Query: 362 ITLEKYLQDYFTNILKKLKDLKAQSKQSDIFF 393
+TLEKYLQDYF+NILKKLKDLKAQSKQSDI+F
Sbjct: 364 VTLEKYLQDYFSNILKKLKDLKAQSKQSDIYF 395
>ref|XP_483708.1| putative NADPH-dependent reductase [Oryza sativa (japonica
cultivar-group)] gi|51979719|ref|XP_507606.1| PREDICTED
OSJNBb0011H15.39-1 gene product [Oryza sativa (japonica
cultivar-group)] gi|51979717|ref|XP_507605.1| PREDICTED
OSJNBb0011H15.39-1 gene product [Oryza sativa (japonica
cultivar-group)] gi|51965082|ref|XP_507325.1| PREDICTED
OSJNBb0011H15.39-1 gene product [Oryza sativa (japonica
cultivar-group)] gi|42408967|dbj|BAD10223.1| putative
NADPH-dependent reductase [Oryza sativa (japonica
cultivar-group)] gi|50725537|dbj|BAD33006.1| putative
NADPH-dependent reductase [Oryza sativa (japonica
cultivar-group)]
Length = 386
Score = 624 bits (1608), Expect = e-177
Identities = 309/383 (80%), Positives = 343/383 (88%), Gaps = 6/383 (1%)
Query: 12 ATPGRVVTRTREAFHALSPSPSPSPPHFVE-FCKGGRRGRRIIRVGVKCNSNSERAVNLG 70
A P ++ + R A ALS S + P H + +G R++ V CN+ + ++
Sbjct: 9 ALPSQLASPARRA--ALSRSATARPRHHHHPLLRAPPKGCRLV---VTCNAQTAVPTSIA 63
Query: 71 PGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADL 130
GTPVRPTSILVVGATGTLGRQ+VRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADL
Sbjct: 64 QGTPVRPTSILVVGATGTLGRQVVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADL 123
Query: 131 SKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCD 190
SKPETIPATLVG+HTVIDCATGRPEEPI+TVDWEGKVALIQCAKAMGIQKYVFYSIHNCD
Sbjct: 124 SKPETIPATLVGIHTVIDCATGRPEEPIRTVDWEGKVALIQCAKAMGIQKYVFYSIHNCD 183
Query: 191 KHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRI 250
KHPEVPLMEIK+CTE F++D+GL++++IRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRI
Sbjct: 184 KHPEVPLMEIKHCTEKFIQDAGLDYLIIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRI 243
Query: 251 AYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSV 310
AYMDTQD+ARLTFIA+RNEK + KLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPV+V
Sbjct: 244 AYMDTQDVARLTFIAMRNEKASKKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVAV 303
Query: 311 LRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQD 370
LR TRQLTRFF+WTNDVADRLAFSEVL+SDT+FSVPM +TY LLGVD+KDI+TLEKYLQD
Sbjct: 304 LRFTRQLTRFFQWTNDVADRLAFSEVLSSDTIFSVPMNDTYQLLGVDSKDILTLEKYLQD 363
Query: 371 YFTNILKKLKDLKAQSKQSDIFF 393
YFTNILKKLKDLKAQSKQ+DIFF
Sbjct: 364 YFTNILKKLKDLKAQSKQTDIFF 386
>ref|ZP_00111826.1| COG0702: Predicted nucleoside-diphosphate-sugar epimerases [Nostoc
punctiforme PCC 73102]
Length = 332
Score = 340 bits (872), Expect = 4e-92
Identities = 171/313 (54%), Positives = 224/313 (70%), Gaps = 3/313 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
++L+VGATGTLGRQ+ RRA+DEGY VRCLVR A FL++WGA +V +L P+T+ A
Sbjct: 2 TLLIVGATGTLGRQVARRAIDEGYKVRCLVRSSKKAA-FLKEWGAELVPGNLRYPDTLAA 60
Query: 139 TLVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
LVGV VID +T RP + IK VDWEGKVALIQ AKA G+++++F+SI + DK+PEVP
Sbjct: 61 ALVGVTQVIDASTSRPTDSLSIKQVDWEGKVALIQAAKAAGVERFIFFSILDADKYPEVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LMEIK CTE FL +SGLN+ ++RL GFMQGLIGQY +PILE + VW T + IAYMDTQ
Sbjct: 121 LMEIKRCTELFLAESGLNYTILRLAGFMQGLIGQYGIPILEGQPVWVTGNSSPIAYMDTQ 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIA+ AL + + G RAW+ +E+I LCERL+G+DA VT +P+S+LR R
Sbjct: 181 DIAKFAIRALSVPETENQAFPVVGTRAWSAEEIINLCERLSGKDARVTRMPISLLRAVRG 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
L R F+W +VADRLAF+EVL S + M E Y + G+D + TLE YLQ+YF+ I+
Sbjct: 241 LMRSFQWGWNVADRLAFTEVLASGKQLNASMDEVYTVFGLDPQQTTTLESYLQEYFSRIM 300
Query: 377 KKLKDLKAQSKQS 389
KKLK++ Q ++
Sbjct: 301 KKLKEVDYQKNKN 313
>ref|ZP_00519345.1| Isoflavone reductase [Crocosphaera watsonii WH 8501]
gi|67852054|gb|EAM47568.1| Isoflavone reductase
[Crocosphaera watsonii WH 8501]
Length = 325
Score = 333 bits (855), Expect = 4e-90
Identities = 164/320 (51%), Positives = 222/320 (69%), Gaps = 8/320 (2%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+L+VGATGTLGRQIVR ALDEG++VRCLVR A FL++WGA ++ D KPET+P
Sbjct: 3 LLIVGATGTLGRQIVRCALDEGHEVRCLVR-NARKAAFLKEWGAELMMGDFCKPETLPRV 61
Query: 140 LVGVHTVIDCATGRPEEPI--KTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L G+ VID A RP + + K +DW GKV LIQ K G+ +Y+F+S+ N +K+P+VPL
Sbjct: 62 LEGMEAVIDAAAARPTDSLSMKEIDWNGKVNLIQAVKESGVDRYIFFSLLNAEKYPDVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M IK+CTE FL++SGLN+ ++R CGFMQGLIGQYAVP+L+ ++VW + T IAYMDTQD
Sbjct: 122 MNIKHCTEKFLKESGLNYTILRPCGFMQGLIGQYAVPMLDNQAVWISGESTPIAYMDTQD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
+ARLT L + + G +AWT +E+I LCERL+ + + VP+ +LR R+
Sbjct: 182 VARLTIRVLEVPETQKQTYPMIGTKAWTPEEIIDLCERLSDKRVKIARVPLGLLRFLRRF 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
TRFF+WT +++DRLAF+EVL S + M + Y LGVD K+ TLE Y Q+YF+ ILK
Sbjct: 242 TRFFQWTYNISDRLAFAEVLASGKPLNASMDDVYKTLGVDVKETTTLEAYFQEYFSRILK 301
Query: 378 KLKDL-----KAQSKQSDIF 392
KLK++ KA+ K+ F
Sbjct: 302 KLKEIDYEKNKAKKKKKTPF 321
>ref|ZP_00162175.2| COG0702: Predicted nucleoside-diphosphate-sugar epimerases
[Anabaena variabilis ATCC 29413]
Length = 328
Score = 328 bits (842), Expect = 1e-88
Identities = 163/306 (53%), Positives = 218/306 (70%), Gaps = 3/306 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
++L+VGATGTLGRQ+ RRA+DEGY VRCLVR P A FL++WGA +V DL +P+T+
Sbjct: 2 TLLIVGATGTLGRQVARRAIDEGYKVRCLVRS-PKRAAFLKEWGAELVRGDLCQPQTLAE 60
Query: 139 TLVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L GV VID AT R + IK VDWEG++ALIQ AKA +++++F+SI + DK+PEVP
Sbjct: 61 ALEGVTAVIDAATSRATDSLTIKQVDWEGQIALIQAAKAASVERFIFFSIIDADKYPEVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LMEIK CTE FL +SG+N+ V+RL GFMQGLIGQY +PILE + VW T A + +AYMDT
Sbjct: 121 LMEIKRCTELFLAESGINYTVLRLAGFMQGLIGQYGIPILEGQPVWVTGASSPVAYMDTL 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIA+ AL + + G RAW+ +E+I LCERL+G++A V +P+++LR R
Sbjct: 181 DIAKFAVRALSVPETEKQAFPVVGTRAWSAEEIINLCERLSGKEAKVRRMPINLLRAVRG 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
L +FF+W ++ADRLAF+EVL S + M E Y + G++ + TLE YLQ+YF+ IL
Sbjct: 241 LAKFFQWGWNIADRLAFTEVLASGRPLNASMDEVYTVFGLEKEQSTTLESYLQEYFSRIL 300
Query: 377 KKLKDL 382
KKLK L
Sbjct: 301 KKLKQL 306
>dbj|BAB76451.1| all4752 [Nostoc sp. PCC 7120] gi|25534778|pir||AH2399 hypothetical
protein all4752 [imported] - Nostoc sp. (strain PCC
7120) gi|17232244|ref|NP_488792.1| hypothetical protein
all4752 [Nostoc sp. PCC 7120]
Length = 328
Score = 328 bits (840), Expect = 2e-88
Identities = 163/306 (53%), Positives = 218/306 (70%), Gaps = 3/306 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
++L+VGATGTLGRQ+ RRA+DEGY VRCLVR A FL++WGA +V DL +P+T+
Sbjct: 2 TLLIVGATGTLGRQVARRAIDEGYKVRCLVRSAKRAA-FLKEWGAELVRGDLCQPQTLVE 60
Query: 139 TLVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L GV VID AT R + IK VDWEG++ALIQ AKA G+++++F+SI + DK+PEVP
Sbjct: 61 ALEGVTAVIDAATSRATDSLTIKQVDWEGQIALIQAAKAAGVERFIFFSIIDADKYPEVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LMEIK CTE FL +SG+N+ V+RL GFMQGLIGQY +PILE + VW T A + +AYMDT
Sbjct: 121 LMEIKRCTELFLAESGINYTVLRLAGFMQGLIGQYGIPILEGQPVWVTGASSPVAYMDTL 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIA+ AL + + G RAW+ +E+I LCERL+G++A V +P+++LR R
Sbjct: 181 DIAKFAVRALSVPETEKQAFPVLGTRAWSAEEIINLCERLSGKEAKVRRMPINLLRAVRG 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
L +FF+W ++ADRLAF+EVL S + M E Y + G++ + TLE YLQ+YF+ IL
Sbjct: 241 LAKFFQWGWNIADRLAFTEVLASGRPLNASMDEVYTVFGLEKEQSTTLENYLQEYFSRIL 300
Query: 377 KKLKDL 382
KKLK L
Sbjct: 301 KKLKQL 306
>ref|NP_681649.1| chaperon-like protein for quinone binding in photosystem II
[Thermosynechococcus elongatus BP-1]
gi|22294581|dbj|BAC08411.1| ycf39 [Thermosynechococcus
elongatus BP-1]
Length = 330
Score = 324 bits (831), Expect = 3e-87
Identities = 164/318 (51%), Positives = 218/318 (67%), Gaps = 8/318 (2%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
++ +VG TGTLGRQIVRRALDEG+ V C VR PA A FLR+WGAT++ +L ++I
Sbjct: 2 NVFIVGGTGTLGRQIVRRALDEGHHVYCFVRS-PAKATFLREWGATILQGNLCAADSILE 60
Query: 139 TL--VGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPE 194
L VID + RP + I VDW+GKV LIQ A+A I+ +F+SI +P+
Sbjct: 61 ALKYAKAAVVIDASATRPTDTLTIAAVDWQGKVNLIQAAQAADIEHLIFFSIMRAQDYPQ 120
Query: 195 VPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMD 254
VPLM+IK+CTE+FLR+SGLN+ ++R CGF QGLIGQYA+PILE +S+W T IAYMD
Sbjct: 121 VPLMQIKHCTEDFLRESGLNYTILRPCGFFQGLIGQYAIPILENQSIWVLGESTAIAYMD 180
Query: 255 TQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLT 314
TQD+A+ A+ GK AG RAWT E+I LCE L+GQ A +T +P+ VLR
Sbjct: 181 TQDVAKFAVRAIDRPATYGKTFDLAGTRAWTADEIIQLCENLSGQQAKITRLPIGVLRAA 240
Query: 315 RQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTN 374
RQ T++F+WT +++DRLAF+EV+ S VF+ PM + Y +D +TLE+YLQDYFT
Sbjct: 241 RQATQWFQWTWNISDRLAFTEVIASGQVFNAPMEDVYRTFDLDPNATLTLEEYLQDYFTR 300
Query: 375 ILKKLKDL---KAQSKQS 389
IL+KLK+L KA+ K+S
Sbjct: 301 ILRKLKELDYEKAKQKKS 318
>sp|P48279|YC39_CYAPA Hypothetical 36.4 kDa protein ycf39 gi|11467300|ref|NP_043157.1|
hypothetical protein CypaCp020 [Cyanophora paradoxa]
gi|1016101|gb|AAA81188.1| ycf39 gene product
gi|7484252|pir||T06845 hypothetical protein ycf39 -
Cyanophora paradoxa cyanelle
Length = 321
Score = 323 bits (828), Expect = 6e-87
Identities = 164/320 (51%), Positives = 224/320 (69%), Gaps = 7/320 (2%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
SILV+GATGTLGRQIVR ALDEGY VRCLVR A FL++WGA ++ DLS+PE++
Sbjct: 2 SILVIGATGTLGRQIVRSALDEGYQVRCLVR-NLRKAAFLKEWGAKLIWGDLSQPESLLP 60
Query: 139 TLVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L G+ +ID +T RP +P + VD +GK ALI AKAM I+K++F+SI N +K+ +VP
Sbjct: 61 ALTGIRVIIDTSTSRPTDPAGVYQVDLKGKKALIDAAKAMKIEKFIFFSILNSEKYSQVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LM IK TE L++SGLN+ + +LCGF QGLIGQYAVPIL++++VW T T IAYMDT
Sbjct: 121 LMRIKTVTEELLKESGLNYTIFKLCGFFQGLIGQYAVPILDQQTVWITTESTSIAYMDTI 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIAR T +L ++ N ++ G R+W + ++I LCERL+GQ+A VT VP++ L L R
Sbjct: 181 DIARFTLRSLVLKETNNRVFPLVGTRSWNSADIIQLCERLSGQNAKVTRVPIAFLELARN 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
+ FFEW ++ADRLAF+EVL+ F+ M E Y + +++ LE YLQ+YF+ IL
Sbjct: 241 TSCFFEWGWNIADRLAFTEVLSKSQFFNSSMDEVYKIFKIESTSTTILESYLQEYFSRIL 300
Query: 377 KKLKDLKAQSKQ----SDIF 392
K+LK++ +Q Q SD+F
Sbjct: 301 KRLKEINSQQSQKKKTSDLF 320
>ref|ZP_00324640.1| COG0702: Predicted nucleoside-diphosphate-sugar epimerases
[Trichodesmium erythraeum IMS101]
Length = 325
Score = 320 bits (820), Expect = 5e-86
Identities = 160/324 (49%), Positives = 223/324 (68%), Gaps = 11/324 (3%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+L++G+TGTLGRQIVR ALDEGY+VRC++R + A FL++WGA +V +L KP+T+
Sbjct: 3 LLILGSTGTLGRQIVRHALDEGYEVRCVIRSY-SKASFLKEWGAELVGGNLCKPKTLIPA 61
Query: 140 LVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L G+ VID AT R + IK VDWEGKV+LIQ A GI++++F+S N +K+ +VPL
Sbjct: 62 LEGIDAVIDAATARATDALSIKQVDWEGKVSLIQTLVAQGIERFIFFSFLNAEKYSQVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
+EIK CTE F+ +SGL + +++ CGF+QGLIGQYAVPIL++++VW + IAYMDTQD
Sbjct: 122 LEIKRCTELFIAESGLKYTILKPCGFLQGLIGQYAVPILDKQAVWVPGLGSAIAYMDTQD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
IA+ AL + K GPRAW E+I +CERL+G+ A VT +++LR+ RQ
Sbjct: 182 IAKFAVGALSVPETENKSFPVVGPRAWDANEIIRMCERLSGEKAKVTRTNINLLRIFRQF 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
TR F+W +VADRLAF+EVL S PM + Y+L G++ ++I TLE YLQ+Y++ I+K
Sbjct: 242 TRSFQWGWNVADRLAFAEVLASGQPIIAPMDDVYSLFGINPQEITTLESYLQEYYSRIMK 301
Query: 378 KLKDL--------KAQSKQSDIFF 393
KLK++ K SK FF
Sbjct: 302 KLKEIEYEQAKLGKTTSKNKPFFF 325
>gb|AAC08124.1| hypothetical chloroplast ORF 39. [Porphyra purpurea]
gi|11465704|ref|NP_053848.1| ORF39 [Porphyra purpurea]
gi|1723342|sp|P51238|YC39_PORPU Hypothetical 35.7 kDa
protein ycf39 (ORF319) gi|2147564|pir||S73159
hypothetical protein 39 - red alga (Porphyra purpurea)
chloroplast
Length = 319
Score = 319 bits (818), Expect = 8e-86
Identities = 155/314 (49%), Positives = 222/314 (70%), Gaps = 3/314 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
++LV+GATGTLGRQIVRRALDEGY+V+C+VR A FL++WGA +V DL PE+I
Sbjct: 2 TLLVIGATGTLGRQIVRRALDEGYNVKCMVRNLRKSA-FLKEWGAELVYGDLKLPESILQ 60
Query: 139 TLVGVHTVIDCATGRPEEPIKT--VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
+ GV VID +T RP +P T +D +GK+ALI+ AKA +Q+++F+SI N D++P+VP
Sbjct: 61 SFCGVTAVIDASTSRPSDPYNTEQIDLDGKIALIEAAKAAKVQRFIFFSILNADQYPKVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LM +K N+L+ S +++ V L GF QGLI QYA+PIL++KSVW T T IAY+DTQ
Sbjct: 121 LMNLKSQVVNYLQKSSISYTVFSLGGFFQGLISQYAIPILDKKSVWVTGESTPIAYIDTQ 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
D A+L +L ++L G +AWT+ E+ITLCE+L+GQ ++ +P+S+L+ R+
Sbjct: 181 DAAKLVIKSLGVPSTENRILPLVGNKAWTSAEIITLCEKLSGQKTQISQIPLSLLKALRK 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
+T+ +WT +++DRLAF+EVLTS +F PM E Y +L +D ++I+LEKY Q+YF IL
Sbjct: 241 ITKTLQWTWNISDRLAFAEVLTSGEIFMAPMNEVYEILSIDKSEVISLEKYFQEYFGKIL 300
Query: 377 KKLKDLKAQSKQSD 390
K LKD+ + Q D
Sbjct: 301 KVLKDISYEQSQQD 314
>ref|NP_441851.1| ycf39 gene product [Synechocystis sp. PCC 6803]
gi|1653617|dbj|BAA18529.1| ycf39 gene product
[Synechocystis sp. PCC 6803] gi|7469557|pir||S76400
hypothetical protein - Synechocystis sp. (strain PCC
6803)
Length = 326
Score = 310 bits (794), Expect = 5e-83
Identities = 156/325 (48%), Positives = 218/325 (67%), Gaps = 12/325 (3%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+LVVG TGTLGRQIVR+A+D+G+ V CLVR A FL++WGAT+V ++ KPET+
Sbjct: 3 VLVVGGTGTLGRQIVRQAIDQGHTVVCLVRSLRKAA-FLKEWGATIVGGNICKPETLSPA 61
Query: 140 LVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L + VID +T R + I+ VDWEGK+ LI+ + GI+K+VF+SI ++P+VPL
Sbjct: 62 LENIDAVIDASTARATDSLTIRQVDWEGKLNLIRAVQKAGIKKFVFFSILRAAEYPKVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M+IK CTE FL + L++ +++L GFMQGLIGQYA+PIL+ +SVW T T IAYM+TQD
Sbjct: 122 MDIKNCTEKFLAQTNLDYTILQLAGFMQGLIGQYAIPILDNQSVWQTGENTPIAYMNTQD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
+A+ A+ + + K G RAW E+I LCER++G +A ++ VP++VLR R
Sbjct: 182 VAKFAVRAVELDSVARKTYPVVGSRAWGATEIIQLCERMSGNNARISQVPMAVLRFMRSF 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
TRFF+WT + +DRLAFSEVL S + MA Y G+D K+ TLE YLQ+YF I+K
Sbjct: 242 TRFFQWTYNASDRLAFSEVLASGKALTADMAPVYEQFGLDPKETTTLESYLQEYFGRIIK 301
Query: 378 KLKDL---------KAQSKQSDIFF 393
KLK+L + + K+++ FF
Sbjct: 302 KLKELDYEVNPTQTEGKKKKNNFFF 326
>ref|ZP_00165401.2| COG0702: Predicted nucleoside-diphosphate-sugar epimerases
[Synechococcus elongatus PCC 7942]
Length = 320
Score = 301 bits (771), Expect = 2e-80
Identities = 149/311 (47%), Positives = 209/311 (66%), Gaps = 3/311 (0%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+LVVGATGTLGRQI RRALDEG+ VRCLVR P +FLR+WG +V DL++PE++
Sbjct: 3 VLVVGATGTLGRQIARRALDEGHRVRCLVRS-PKRGNFLREWGCDLVRGDLTQPESLTFA 61
Query: 140 LVGVHTVIDCATGRPEEPIKT--VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L G+ VID AT R + + VDW+GKV LI+ A G+Q++VF SI + +KH +VPL
Sbjct: 62 LEGIEAVIDAATTRSTDSLSCYDVDWQGKVNLIKAATEAGVQRFVFCSIIDAEKHRDVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M+IKYCTE FLR SGLN+ ++RL GFMQGLI ++A+P+LE ++ + T IAY+ T D
Sbjct: 122 MDIKYCTEEFLRQSGLNYTILRLAGFMQGLIAEFAIPVLEGRTAFITQDSDPIAYLSTLD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
IAR AL + L GP+AW+ E+ LCERL+G++ + +P++ + ++
Sbjct: 182 IARFAVAALTTPATEKQTLPVVGPKAWSGLEIFRLCERLSGKETKIARLPLATVSAMKRF 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
RFF+W ++ADRLAFSEV+ F+ MA TY G+D +I LE YL DYF+ +L+
Sbjct: 242 FRFFQWGWNIADRLAFSEVMACGRPFTADMAATYTAFGIDPAEITGLESYLNDYFSVMLR 301
Query: 378 KLKDLKAQSKQ 388
+LK+L+ K+
Sbjct: 302 RLKELEFDQKK 312
>ref|YP_172391.1| chaperon-like protein for quinone binding in photosystem II
[Synechococcus elongatus PCC 6301]
gi|56686649|dbj|BAD79871.1| chaperon-like protein for
quinone binding in photosystem II [Synechococcus
elongatus PCC 6301]
Length = 320
Score = 300 bits (768), Expect = 5e-80
Identities = 149/311 (47%), Positives = 208/311 (65%), Gaps = 3/311 (0%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+LVVGATGTLGRQI RRALDEG+ VRCLVR P +FLR+WG +V DL++PE++
Sbjct: 3 VLVVGATGTLGRQIARRALDEGHRVRCLVRS-PKRGNFLREWGCDLVRGDLTQPESLTFA 61
Query: 140 LVGVHTVIDCATGRPEEPIKT--VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L G+ VID AT R + + VDW+GKV LI+ A G+Q++VF SI + +KH +VPL
Sbjct: 62 LEGIEAVIDAATTRSTDSLSCYDVDWQGKVNLIKAATEAGVQRFVFCSIIDAEKHRDVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M+IKYCTE FLR SGLN+ ++RL GFMQGLI ++A+P+LE ++ T IAY+ T D
Sbjct: 122 MDIKYCTEEFLRQSGLNYTILRLAGFMQGLIAEFAIPVLEGRTALITQDSDPIAYLSTLD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
IAR AL + L GP+AW+ E+ LCERL+G++ + +P++ + ++
Sbjct: 182 IARFAVAALTTPATEKQTLPVVGPKAWSGLEIFRLCERLSGKETKIARLPLATVSAMKRF 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
RFF+W ++ADRLAFSEV+ F+ MA TY G+D +I LE YL DYF+ +L+
Sbjct: 242 FRFFQWGWNIADRLAFSEVMACGRPFTADMAATYTAFGIDPAEITGLESYLNDYFSVMLR 301
Query: 378 KLKDLKAQSKQ 388
+LK+L+ K+
Sbjct: 302 RLKELEFDQKK 312
>gb|AAC35663.1| hypothetical chloroplast RF39 [Guillardia theta]
gi|11467677|ref|NP_050729.1| hypothetical chloroplast
RF39 [Guillardia theta] gi|6136619|sp|O78472|YC39_GUITH
Hypothetical 35.9 kDa protein ycf39
Length = 314
Score = 294 bits (752), Expect = 4e-78
Identities = 146/316 (46%), Positives = 217/316 (68%), Gaps = 9/316 (2%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
S+LV+GATGTLGRQIVRRALDEGY+V CLVR A FL++WGA ++ DLS PET+P
Sbjct: 2 SLLVIGATGTLGRQIVRRALDEGYEVSCLVR-NLRKAYFLKEWGAELLYGDLSLPETLPT 60
Query: 139 TLVGVHTVIDCATGRPEEPIKT--VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L + +ID +T RP +P K +D EGK+AL++ AK GI+++VF+S+ N + +P
Sbjct: 61 NLTKITAIIDASTARPSDPYKAEKIDLEGKIALVEAAKVAGIKRFVFFSVLNAQNYRHLP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
L+ +K E +L+ S L + +L GF QGLI QYA+PILE++++W T T+I Y+DT
Sbjct: 121 LVNLKCRMEEYLQTSELEYTTFQLSGFFQGLISQYAIPILEKQTIWITGEYTKINYIDTN 180
Query: 257 DIARLTFIALRNEKING---KLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRL 313
DIA+ A+R+ +NG + + G ++W ++E+I LCERL+GQ AN+T +P+ +L
Sbjct: 181 DIAK---FAVRSLSLNGTIKRTIPLVGLKSWNSEEIIQLCERLSGQKANITKIPLQLLVF 237
Query: 314 TRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFT 373
+ LT FFEWT ++++RLAF+EVL + + MA TY+ G + DI +LE Y+QDYF
Sbjct: 238 FKYLTGFFEWTTNISERLAFAEVLNARQDITSEMASTYSQFGFEPNDITSLESYMQDYFG 297
Query: 374 NILKKLKDLKAQSKQS 389
IL++LK+L + +++
Sbjct: 298 RILRRLKELSDERERT 313
>ref|YP_063656.1| conserved hypothetical plastid protein [Gracilaria tenuistipitata
var. liui] gi|50657746|gb|AAT79731.1| conserved
hypothetical plastid protein [Gracilaria tenuistipitata
var. liui]
Length = 323
Score = 283 bits (724), Expect = 6e-75
Identities = 137/314 (43%), Positives = 213/314 (67%), Gaps = 3/314 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
S+LV+GATGTLGRQIVRRALDEG+ V+CLVR A FL++WGA +V DL PETIP
Sbjct: 2 SLLVIGATGTLGRQIVRRALDEGFQVKCLVR-NFRKAAFLKEWGADLVYGDLKLPETIPP 60
Query: 139 TLVGVHTVIDCATGRPEE--PIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
TL+G+ +ID +T R + +D GK LI+ A+ +++Y+F+SI +++ E+P
Sbjct: 61 TLLGITAIIDASTARTSDFHSATQIDLHGKCILIKSAQKACVRRYIFFSIFASEQYNEIP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LM +K N+LR SG+++ + L GF QGLI QYA+PIL+ ++VW D + + Y+DTQ
Sbjct: 121 LMRMKTIIANYLRQSGIDYTIFNLAGFFQGLIVQYALPILDNQTVWIADDSSSVPYIDTQ 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIA+L +L K+L G ++W + E+I+LCE+L+GQ + ++ +P+ +L R+
Sbjct: 181 DIAKLAIKSLSTIATKNKILPMVGSKSWNSTEIISLCEKLSGQRSQISRIPLVILVFFRR 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
LT F+W+ +++DRLAF++VL S+ F+ MAE Y+LL ++ ++ LE Y Q+YF+ ++
Sbjct: 241 LTSLFQWSWNISDRLAFTKVLLSNNSFNTSMAEVYDLLQINEQETERLEDYFQEYFSKVM 300
Query: 377 KKLKDLKAQSKQSD 390
KKLK++ +S +
Sbjct: 301 KKLKEISYKSTSQE 314
>emb|CAE21348.1| putative chaperon-like protein for quinone binding in photosystem
II [Prochlorococcus marinus str. MIT 9313]
gi|33863443|ref|NP_895003.1| putative chaperon-like
protein for quinone binding in photosystem II
[Prochlorococcus marinus str. MIT 9313]
Length = 320
Score = 275 bits (702), Expect = 2e-72
Identities = 129/307 (42%), Positives = 202/307 (65%), Gaps = 3/307 (0%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+L+VG TGTLGRQI RRA+D G+ VRC+VR +P FL++WG + DL PETI +
Sbjct: 3 VLLVGGTGTLGRQIARRAIDAGHQVRCMVR-KPRKGAFLQEWGCELTCGDLLDPETIDYS 61
Query: 140 LVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L G+ VID AT RP++ + T DW+GK+ L++ + G+ +YVF S+ +KH VPL
Sbjct: 62 LDGIDAVIDAATSRPDDSASVYTTDWDGKLNLLRACEKAGVTRYVFLSLLAAEKHLNVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M+IK+CTE L DS ++ +++ FMQGLIGQ A+P+LE ++VW ++ PT +AYM+TQD
Sbjct: 122 MDIKFCTERLLADSSFDYTILQGVAFMQGLIGQIAIPVLENQTVWVSETPTAVAYMNTQD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
+AR AL + + GP+AWT++E++ CE+ + + A V V ++ L++++
Sbjct: 182 VARFAVAALERPETIRRSFPVVGPKAWTSEEIVQFCEKSSSKTAKVIRVSPFLIGLSQRV 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
FFE + ++A+RLAF+EV PM +TY+ G+D+ + LE Y+ +Y+ ILK
Sbjct: 242 VSFFEQSVNMAERLAFAEVTGGGIALDAPMDDTYSCFGLDSSETTPLESYISEYYDTILK 301
Query: 378 KLKDLKA 384
+L++++A
Sbjct: 302 RLREMEA 308
>emb|CAA91705.1| ORF319 [Odontella sinensis] gi|11467527|ref|NP_043673.1| ORF319
[Odontella sinensis] gi|1351766|sp|P49534|YC39_ODOSI
Hypothetical 36.3 kDa protein ycf39 (ORF319)
gi|7484310|pir||S78332 conserved hypothetical protein
319 - Odontella sinensis chloroplast
Length = 319
Score = 272 bits (696), Expect = 1e-71
Identities = 135/312 (43%), Positives = 200/312 (63%), Gaps = 3/312 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
S+L++G TGTLGRQ+V +AL +GY VRCLVR A+FL++WGA ++ DLS+PETIP
Sbjct: 2 SLLIIGGTGTLGRQVVLQALTKGYQVRCLVR-NFRKANFLKEWGAELIYGDLSRPETIPP 60
Query: 139 TLVGVHTVIDCATGRPEE--PIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L G+ VID +T RP + +K VDW+GK ALI+ A+A ++ +VF S N ++ +P
Sbjct: 61 CLQGITAVIDTSTSRPSDLDTLKQVDWDGKCALIEAAQAANVKHFVFCSSQNVEQFLNIP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LME+K+ E L+ S + + V RL GF QGLI QYA+P+LE + T+ T ++YMDTQ
Sbjct: 121 LMEMKFGIETKLQQSNIPYTVFRLAGFYQGLIEQYAIPVLENLPILVTNENTCVSYMDTQ 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIA+ +L+ + + G + W + E+I LCE+LAGQ A V +P+ +L+L Q
Sbjct: 181 DIAKFCLRSLQLPETKNRTFVLGGQKGWVSSEIINLCEQLAGQSAKVNKIPLFLLKLVSQ 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
+ FFEW +++DRLAF+E+L + FS + Y VD +I L+ Y +YF +L
Sbjct: 241 IFGFFEWGQNISDRLAFAEILNVENDFSKSTFDLYKTFKVDDSEITQLDDYFLEYFIRLL 300
Query: 377 KKLKDLKAQSKQ 388
K+L+D+ + Q
Sbjct: 301 KRLRDINFEDIQ 312
>ref|NP_893269.1| putative chaperon-like protein for quinone binding in photosystem
II [Prochlorococcus marinus subsp. pastoris str.
CCMP1986] gi|33640076|emb|CAE19611.1| putative
chaperon-like protein for quinone binding in photosystem
II [Prochlorococcus marinus subsp. pastoris str.
CCMP1986]
Length = 320
Score = 254 bits (648), Expect = 4e-66
Identities = 123/307 (40%), Positives = 198/307 (64%), Gaps = 3/307 (0%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
IL+VGATGTLGRQI ++A++EG++VRC VR P A FL++WG + +L I
Sbjct: 3 ILLVGATGTLGRQIAKQAIEEGHEVRCFVR-NPRKASFLQEWGCELTKGNLLNSGDIDYA 61
Query: 140 LVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L + VID ATGRPE+ I DW+GK+ L ++ I++ +F SI + +K VPL
Sbjct: 62 LQDIEVVIDSATGRPEDSKSIYETDWDGKLNLFNACESKKIKRVIFLSILSTEKFRNVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M++KYCTE L S ++ + + FMQG+I Q+A+P+L+ ++VW + PT+IAYM+TQD
Sbjct: 122 MDVKYCTEKLLEKSNFDYTIFKCAAFMQGVISQFAIPVLDSQAVWMSGTPTKIAYMNTQD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
+A++ ++ K + L GP+AW + EVI+LCE+ + + A + V ++++T+ +
Sbjct: 182 MAKIIVSSINKPKSYKRSLPLVGPKAWDSDEVISLCEKYSNKKAKIFRVSPFLIKVTQNV 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
FF+ +V++RLAF+EV +S M+ TY LL + +D +LE Y+++Y+ ILK
Sbjct: 242 VSFFQDALNVSERLAFAEVTSSGVPLDDDMSNTYELLELKKEDSTSLESYIKEYYQQILK 301
Query: 378 KLKDLKA 384
+LK+++A
Sbjct: 302 RLKEMEA 308
>gb|AAQ00293.1| NADPH-dependent reductase [Prochlorococcus marinus subsp. marinus
str. CCMP1375] gi|33240698|ref|NP_875640.1|
NADPH-dependent reductase [Prochlorococcus marinus
subsp. marinus str. CCMP1375]
Length = 320
Score = 253 bits (647), Expect = 5e-66
Identities = 125/308 (40%), Positives = 203/308 (65%), Gaps = 5/308 (1%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+L+VG TGTLGRQI ++A+D G+ VRC+VR RP A +L++WG + DL + + + +
Sbjct: 3 VLIVGGTGTLGRQIAKKAIDAGFQVRCMVR-RPRKASYLQEWGCELTQGDLLRQKDLEYS 61
Query: 140 LVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L GV +ID AT RP++P + DWEGK+ L + +++G+++ VF S+ +K VPL
Sbjct: 62 LNGVDALIDAATSRPDDPRSVYETDWEGKLNLYRACESIGVKRVVFLSLLAAEKFRNVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M+IK+CTE +L DS L+ +++ FMQG+IGQ+A+PIL+ + VW + + IAYM+T+D
Sbjct: 122 MDIKFCTERYLLDSSLDFTILQGAAFMQGVIGQFAIPILDSQPVWISGNSSNIAYMNTED 181
Query: 258 IARLTFIAL-RNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
+A AL R++ I G L GPR+W ++EVI +CE + + A V V +++R+ +
Sbjct: 182 MASFAIAALSRSQTIRGVYLV-VGPRSWKSEEVIKICEGFSDKKAKVLRVSPALIRIAKT 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
L FF+ T +VA+RL F++V S + PM +TY G+D +L+ Y+++Y+ IL
Sbjct: 241 LVSFFQATTNVAERLEFADVSGSGIELNAPMEKTYKDFGLDISLTTSLDSYIKEYYEVIL 300
Query: 377 KKLKDLKA 384
K+L++++A
Sbjct: 301 KRLREMEA 308
>emb|CAE07198.1| putative chaperon-like protein for quinone binding in photosystem
II [Synechococcus sp. WH 8102]
gi|33865217|ref|NP_896776.1| putative chaperon-like
protein for quinone binding in photosystem II
[Synechococcus sp. WH 8102]
Length = 320
Score = 252 bits (643), Expect = 2e-65
Identities = 123/307 (40%), Positives = 192/307 (62%), Gaps = 3/307 (0%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPAT 139
+LV+G TGTLGRQI RRALD G+DVRC+VR P A FL++WG + DL +P+++
Sbjct: 3 VLVLGGTGTLGRQIARRALDAGHDVRCMVRT-PRKASFLQEWGCELTRGDLLEPDSLDYA 61
Query: 140 LVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPL 197
L GV VID +T RP +P + DW+GK+ L++ + ++++VF S+ H VPL
Sbjct: 62 LDGVDAVIDASTSRPTDPHSVYETDWDGKLNLLRACERAEVKRFVFVSLLGAHGHRSVPL 121
Query: 198 MEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQD 257
M+IK CTEN L S ++ +++ FMQG+I Q+A+P+LE ++VW + +PT IAYM++QD
Sbjct: 122 MDIKACTENLLESSDFDYTILQGAAFMQGVISQFAIPVLESQTVWVSGSPTAIAYMNSQD 181
Query: 258 IARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQL 317
+AR AL ++ G +AW T E++ LCER +G+ A V V +++L + +
Sbjct: 182 MARFAVAALERQETIRGTYPVVGLKAWNTGELVQLCERCSGKTARVFRVQPVLIKLMQGI 241
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNILK 377
FFE +VA+RLAF+EV M +Y G++ + +E Y+ +Y+ ILK
Sbjct: 242 ASFFEPAVNVAERLAFAEVTGGGQALDAQMENSYAAFGLEPSETTEMESYISEYYDTILK 301
Query: 378 KLKDLKA 384
+L++++A
Sbjct: 302 RLREMEA 308
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 685,901,160
Number of Sequences: 2540612
Number of extensions: 30722793
Number of successful extensions: 106876
Number of sequences better than 10.0: 846
Number of HSP's better than 10.0 without gapping: 189
Number of HSP's successfully gapped in prelim test: 657
Number of HSP's that attempted gapping in prelim test: 106304
Number of HSP's gapped (non-prelim): 904
length of query: 393
length of database: 863,360,394
effective HSP length: 130
effective length of query: 263
effective length of database: 533,080,834
effective search space: 140200259342
effective search space used: 140200259342
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC121239.8


