

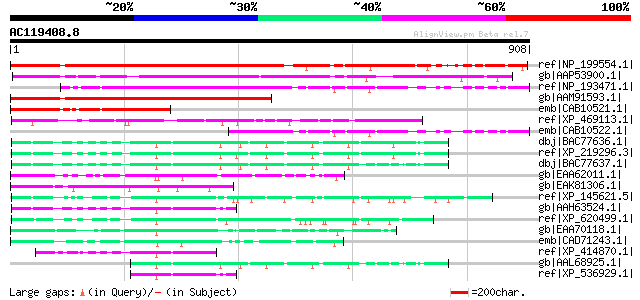
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.8 + phase: 0
(908 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_199554.1| expressed protein [Arabidopsis thaliana] 826 0.0
gb|AAP53900.1| putative DNA-binding protein [Oryza sativa (japon... 717 0.0
ref|NP_193471.1| expressed protein [Arabidopsis thaliana] 645 0.0
gb|AAM91593.1| DNA-binding protein-like [Arabidopsis thaliana] 607 e-172
emb|CAB10521.1| hypothetical protein [Arabidopsis thaliana] gi|7... 372 e-101
ref|XP_469113.1| putative DNA-binding protein [Oryza sativa (jap... 246 2e-63
emb|CAB10522.1| DNA-binding protein homolog [Arabidopsis thalian... 239 2e-61
dbj|BAC77636.1| retinoblastoma binding protein 6 isoform 1 [Homo... 183 3e-44
ref|XP_219296.3| PREDICTED: similar to PACT [Rattus norvegicus] 179 3e-43
dbj|BAC77637.1| retinoblastoma binding protein 6 isoform 2 [Homo... 172 3e-41
gb|EAA62011.1| hypothetical protein AN7431.2 [Aspergillus nidula... 169 3e-40
gb|EAK81306.1| hypothetical protein UM00321.1 [Ustilago maydis 5... 169 4e-40
ref|XP_145621.5| PREDICTED: similar to retinoblastoma-binding pr... 167 1e-39
gb|AAH63524.1| RBBP6 protein [Homo sapiens] 160 2e-37
ref|XP_620499.1| PREDICTED: similar to retinoblastoma-binding pr... 157 1e-36
gb|EAA70118.1| hypothetical protein FG09892.1 [Gibberella zeae P... 153 3e-35
emb|CAD71243.1| conserved hypothetical protein [Neurospora crass... 143 2e-32
ref|XP_414870.1| PREDICTED: similar to retinoblastoma-binding pr... 140 2e-31
gb|AAL68925.1| p53-associated cellular protein PACT [Homo sapiens] 118 1e-24
ref|XP_536929.1| PREDICTED: similar to retinoblastoma-binding pr... 105 5e-21
>ref|NP_199554.1| expressed protein [Arabidopsis thaliana]
Length = 879
Score = 826 bits (2134), Expect = 0.0
Identities = 486/947 (51%), Positives = 582/947 (61%), Gaps = 111/947 (11%)
Query: 1 MAVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDE 60
MA+YYKFKSARDYD+I+MDGPFISVG LK+KIFE+KHLG G D D+VV+NAQTNEEYLDE
Sbjct: 1 MAIYYKFKSARDYDTIAMDGPFISVGILKDKIFETKHLGTGKDLDIVVSNAQTNEEYLDE 60
Query: 61 EMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSA 120
MLIPKNTSVLIRRVPGRPR+ ++T QE +++NKV D + ++ P D SA
Sbjct: 61 AMLIPKNTSVLIRRVPGRPRITVITTQE--------PRIQNKVEDVQAETTNFPVADPSA 112
Query: 121 MKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQE-APPTSTVDEESKIKALIDTPALDWQH 179
++ ED ++DEFG DLYSIPD Q I A VDEESKI+ALIDTPALDWQ
Sbjct: 113 AEFPED-EYDEFGTDLYSIPDTQDAQHIIPRPHLATADDKVDEESKIQALIDTPALDWQQ 171
Query: 180 -QGSD-FGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPN 237
QG D FGAGRG+GRGM GRM G RGFG+ERKTPP GYVCHRC +PGHFIQHCPTNGDPN
Sbjct: 172 RQGQDTFGAGRGYGRGMPGRMNG-RGFGMERKTPPPGYVCHRCNIPGHFIQHCPTNGDPN 230
Query: 238 YDVKRVKQPTGIPRSMLMVNPQGSYALPNGSVAVLKPNEAAFEKEMEGMPSTTRSVGDLP 297
YDVKRVK PTGIP+SMLM P GSY+LP+G+VAVLKPNE AFEKEMEG+PSTTRSVG+LP
Sbjct: 231 YDVKRVKPPTGIPKSMLMATPDGSYSLPSGAVAVLKPNEDAFEKEMEGLPSTTRSVGELP 290
Query: 298 PELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCVCGAMNVLADDLLPNKTLR 357
PEL CPLC VMKDA LTSKCC+KSFCDKCIRD+IISKSMCVCG +VLADDLLPNKTLR
Sbjct: 291 PELKCPLCKEVMKDAALTSKCCYKSFCDKCIRDHIISKSMCVCGRSDVLADDLLPNKTLR 350
Query: 358 DTINRILESGNSSTENAGSTYQVQDMESARCPQPKIPSPTSSAASKGGLKISPVYDGTTN 417
DTINRILE+GN STEN GS + D+ESARCP PK SPT+S ASKG K + +
Sbjct: 351 DTINRILEAGNDSTENVGSVGHIPDLESARCPPPKALSPTTSVASKGEKKPVLSNNNDAS 410
Query: 418 IQDTAVETKVVSAPPQTSEHVKIPRAGDVSEATHESKSVKE-PVSQGSAQVVEEEVQQKL 476
+E +++ P+ S V + + D E+T S VKE VS+ + Q +EE+QQ++
Sbjct: 411 TLKAPMEVAEITSAPRASAEVNVEKPVDACESTQGSVIVKEATVSKLNTQAPKEEMQQQV 470
Query: 477 VPTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGPPP------------------ 518
E D QW P DL +YMMQMGP P
Sbjct: 471 AAGEP--------------DMQWNPVPDLAGPDYMMQMGPGPQYFNGMQPGFNGVQPGFN 516
Query: 519 GYNPYWNGMQPCMDGFMAPYAGPM-HMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ 577
G P +NG P +GF P+ G M M YG P DM F GM N M DPF Q
Sbjct: 517 GVQPGFNGFHPGFNGFGGPFPGAMPPFMGYGLNPMDMGFGGGM------NMMHPDPFMAQ 570
Query: 578 GYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVE-----RDF 632
G+ P I PPPHRDLAE MN+ M R+E EAR A+ RKRENERR E RD
Sbjct: 571 GFGFPNI-PPPHRDLAEMGNRMNLQRAMMGRDEAEARNAEMLRKRENERRPEGGKMFRDG 629
Query: 633 SKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPD--SSHREVEPPRP 690
R +S S +K++ PP DY + R +R SP+ + + + P R
Sbjct: 630 ENSRMMMNNGTSASASSINPNKSRQAPPPPIHDYDRRRRPEKRLSPEHPPTRKNISPSRD 689
Query: 691 TKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRR--------HHHRTEASSKKST 742
+KRKS+ ER DR RD + RHQD D HDR + RR HHR E
Sbjct: 690 SKRKSERYPDER-DRQRDRE-RSRHQDVDREHDRTRDRRDEDRSRDHRHHRGETER---- 743
Query: 743 DPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFS 802
++ RK SEP SSEP P T AE + K+SVF+
Sbjct: 744 ---SQHHHRKRSEP-------PSSEP---------PVPATKAE------IENNLKSSVFA 778
Query: 803 RISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYE 862
RISFP EE ++ K+RK+ +SS+T T +AS SA A T + + + + DYE
Sbjct: 779 RISFPEEE--TSSGKRRKVPSSSSTSV-TDPSASASAAAAVGTSVHRHSSRKEIEVADYE 835
Query: 863 SSDDDEDDERHFKRRPSRYEPSPPPPVDDWVEE----GRHSRGTRDR 905
SSD+D RHFKR+PSRY SPP V D E+ + +G R R
Sbjct: 836 SSDED----RHFKRKPSRYARSPPVVVSDVSEDKLRYSKRGKGERSR 878
>gb|AAP53900.1| putative DNA-binding protein [Oryza sativa (japonica cultivar-group)]
gi|37534622|ref|NP_921613.1| putative DNA-binding protein
[Oryza sativa (japonica cultivar-group)]
Length = 1066
Score = 717 bits (1852), Expect = 0.0
Identities = 444/924 (48%), Positives = 549/924 (59%), Gaps = 129/924 (13%)
Query: 6 KFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEMLIP 65
KFKSARDYDSI ++G FISV LKE+IFESKHLGRGTDFDL+++NAQT+EEY DE +IP
Sbjct: 178 KFKSARDYDSIPIEGQFISVANLKERIFESKHLGRGTDFDLMISNAQTDEEYADEATMIP 237
Query: 66 KNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSAMKYVE 125
KNTSVLIRR+PGRPR PIVTE E ++ +E +V +T P+ S+ AD ++MKY E
Sbjct: 238 KNTSVLIRRIPGRPRKPIVTEPEE------TKAMEGRVEETMPSGSAFLAD--ASMKYPE 289
Query: 126 DSDWD-EFGNDLY---SIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQG 181
+S+WD EFGNDLY S+P QL Q+++ + + VDE+SKIKALIDT ALD+
Sbjct: 290 ESEWDDEFGNDLYVSDSVPSQLGSQAVDASE-----NKVDEDSKIKALIDTSALDYSQIP 344
Query: 182 SDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDVK 241
+G GRG+GRGMGGRM GGRGFG HFIQHCPTNGD +D+K
Sbjct: 345 DGYGGGRGYGRGMGGRMMGGRGFG-------------------HFIQHCPTNGDARFDMK 385
Query: 242 RVKQPTGIPRSMLMVNPQGSYALPNGSVAVLKPNEAAFEKEMEGMPSTTRSVGDLPPELH 301
R+K PTGIP+SMLM P GSYALP+G+ AVLKPNEAAFEKE+EG+P TTRS+GDLPPEL
Sbjct: 386 RMKPPTGIPKSMLMATPDGSYALPSGAGAVLKPNEAAFEKEIEGLP-TTRSLGDLPPELR 444
Query: 302 CPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCVCGAMNVLADDLLPNKTLRDTIN 361
CPLC VMKDAVLTSKCCF+SFCDKCIRDYII+KSMCVCGA ++LADDLLPNKTLR+TI+
Sbjct: 445 CPLCKEVMKDAVLTSKCCFRSFCDKCIRDYIINKSMCVCGATSILADDLLPNKTLRETIS 504
Query: 362 RILES-GNSSTENAGSTYQVQDMESARCPQPKIPSPTSSAASKGGLKISPVYDGTTNIQD 420
RILE+ SSTEN GS QVQDMESA QPK+ SP SAASK K +P + +
Sbjct: 505 RILEAPPTSSTENVGSMVQVQDMESAIPVQPKVRSPAVSAASKEEPKRTPAPVEESPDVE 564
Query: 421 TAVETKVVSAPPQTSEHVKIPRAGDVSEATHESKSVKEPVSQGSAQVVEEEVQQKLVPTE 480
+ E K + +S+ K+P DV E T ESK +KE + + V E + + +V +
Sbjct: 565 SHSEVKTTNVDMSSSDK-KVPALPDVVEGTMESKILKEKTPEATPVVKESQEKMPVVGEQ 623
Query: 481 AGKKKKKKKVRMPTNDFQWKPPHDLGAENYM-MQMGPPPGYNPYWNGMQPC-MDGFMAPY 538
KKKKKKKVR P N WKP D GAEN+ M MGP G+NPYW G P MD AP+
Sbjct: 624 VVKKKKKKKVRAPGNAEDWKPYQDFGAENFAGMPMGPAGGFNPYWGGGMPLPMDYMGAPF 683
Query: 539 AGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQGYMMPPIPPPPHRDLAEF--- 595
GPM M Y GP+D PF G+ +P DPF GYMMP +P RDL+E
Sbjct: 684 PGPMPYMGYPPGPFD-PFGGGV--------LPQDPFMPPGYMMPAVP----RDLSELAVN 730
Query: 596 SMGMNVPPPAMSREEFEARKADARRKRE----NERRVERDFSKDRDFGREVSSVGDVSSI 651
SMGMN+ PP +SR+EFE RK D RR+RE NER ER S++R+ RE
Sbjct: 731 SMGMNMGPPVVSRDEFELRKPDNRRRREMERFNERERERGHSRERERERE---------- 780
Query: 652 KSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDY 711
ER RE E R R D + DR+RD +
Sbjct: 781 -------------------RERERERERERERERERDRDQNRDRDRDQERDRDRERDRER 821
Query: 712 HDRHQDRDYX---HDRHQHRRHHHRTEAS--SKKSTDPVTKST---SRKSSEPDIKSKSR 763
R + R+ +D R R+ +S + +S P S SR+S S +
Sbjct: 822 ESRREARESSGANNDSTTSMRPKARSRSSQPADRSAPPPPASPDRHSRRSPHRSSGSGKK 881
Query: 764 KSSEPVTKSLSLSKTAPTTSAEAAA--------AAAADRKQKA--SVFSRISFPSEEEAA 813
+SS L L P S AA AAAAD + KA SVFSRISFP +
Sbjct: 882 RSSSDRYDDLPLPPPPPPASRHEAAEHAHAKNTAAAADARSKAKGSVFSRISFPGGDGNP 941
Query: 814 AAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGR---------------KSKAVM 858
+ AK+ + S+S AS ++S S KA + N GR + +
Sbjct: 942 SDAKRSRRSSSDKPPAS---SSSSSKKARAAVAEDNDGRHHRRHHREAAAAAEERRRPAA 998
Query: 859 DDY---ESSDDDEDDERHFKRRPS 879
DY E DD+ ++E+HFKRRPS
Sbjct: 999 GDYYGEEEEDDESEEEQHFKRRPS 1022
>ref|NP_193471.1| expressed protein [Arabidopsis thaliana]
Length = 744
Score = 645 bits (1663), Expect = 0.0
Identities = 398/838 (47%), Positives = 494/838 (58%), Gaps = 117/838 (13%)
Query: 90 YLKPLISQKVENKVVDTEPANSSLPADDMSAMKYVEDSDWDEFGNDLYSIPDQLPVQSIN 149
+ +P + KVEN D N+ + AD VED ++DEFGNDLYSIPD V S N
Sbjct: 4 FCRPRVEDKVENVQADM---NNVITADASP----VED-EFDEFGNDLYSIPDAPAVHSNN 55
Query: 150 MIQEAPPTSTVDEESKIKALIDTPALDWQHQGSD-FGAGRGFGRGMGGRMGGGRGFGLER 208
+ ++ P DEE+K+KALIDTPALDW QG+D FG GRG+GRGM GRMGG RGFG+ER
Sbjct: 56 LCHDSAPAD--DEETKLKALIDTPALDWHQQGADSFGPGRGYGRGMAGRMGG-RGFGMER 112
Query: 209 KTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDVKRVKQPTGIPRSMLMVNPQGSYALPNGS 268
TPP GYVCHRC V GHFIQHC TNG+PN+DVKRVK PTGIP+SMLM P GSY+LP+G+
Sbjct: 113 TTPPPGYVCHRCNVSGHFIQHCSTNGNPNFDVKRVKPPTGIPKSMLMATPNGSYSLPSGA 172
Query: 269 VAVLKPNEAAFEKEMEGMPSTTRSVGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCI 328
VAVLKPNE AFEKEMEG+ STTRSVG+ PPEL CPLC VM+DA L SKCC KS+CDKCI
Sbjct: 173 VAVLKPNEDAFEKEMEGLTSTTRSVGEFPPELKCPLCKEVMRDAALASKCCLKSYCDKCI 232
Query: 329 RDYIISKSMCVCGAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARC 388
RD+II+KSMCVCGA +VLADDLLPNKTLRDTINRILESGNSS ENAGS QVQDMES RC
Sbjct: 233 RDHIIAKSMCVCGATHVLADDLLPNKTLRDTINRILESGNSSAENAGSMCQVQDMESVRC 292
Query: 389 PQPKIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSE 448
P PK SPT+SAAS G K +P + T+ ++E +++ ++E VK+ + D S
Sbjct: 293 PPPKALSPTTSAASGGEKKPAPSNNNETSTLKPSIEIAEITSAWASAEIVKVEKPVDASA 352
Query: 449 ATHESKSVKEP-VSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGA 507
S + KE VSQ + Q +EE+ Q++ E GK+KKKK T DL
Sbjct: 353 NIQGSSNGKEAAVSQLNTQPPKEEMPQQVASGEQGKRKKKKPRMSGT---------DLAG 403
Query: 508 ENYMMQMGPPPGYNPYWNGMQPCMDGFMAPYAGPM---HMMDYGHGPYDMPFPNGMPHDP 564
+YMM MGP PG N Y+NG QP +G + G + +G + PFP MP P
Sbjct: 404 PDYMMPMGPGPG-NQYFNGFQPGFNGVQHGFNGVQPGFNGFHHGFNGFPGPFPGAMP--P 460
Query: 565 FAN----GMPH-DPFGMQGYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADAR 619
F G+ H DPF QG+ P IPPP +RDLAE MN+ P M REEFEA+K + +
Sbjct: 461 FVGYGFGGVIHPDPFAAQGFGFPNIPPP-YRDLAEMGNRMNLQHPIMGREEFEAKKTEMK 519
Query: 620 RKRENERR------VERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRS 673
RKRENE R V RD K R +S S +K K++ PP S + R RS
Sbjct: 520 RKRENEIRRSEGGNVVRDSEKSRIMN---NSAVTSSPVKPKSRQGPPPPISSDYDRRRRS 576
Query: 674 ERPSPD--SSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHH 731
+R SP+ SS R PPR + RKS+ DRH D D HDR + R
Sbjct: 577 DRSSPERQSSRRFTSPPRSSSRKSER---------------DRHHDLDSEHDRRRDR--- 618
Query: 732 HRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAA 791
+ T RK + +S KSS T +
Sbjct: 619 --------------PRETDRKH-----RKRSEKSSSDPTVEI------------------ 641
Query: 792 ADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGG 851
D K++VF+RISFP E ++ K+RK S SS + SV+ + H S
Sbjct: 642 -DDNNKSNVFTRISFPEE----SSGKQRKTSKSSPAPPES----SVAPVSSGRRHHSRRE 692
Query: 852 RKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPP-PPVDDWVEEGRHSRGTRDRKHR 908
R+ M +Y+SSDD++ RHFKR+PSRY+ SP P D E RHS+ ++ + R
Sbjct: 693 RE----MVEYDSSDDED---RHFKRKPSRYKRSPSVAPSDAGDEHFRHSKRSKGERAR 743
>gb|AAM91593.1| DNA-binding protein-like [Arabidopsis thaliana]
Length = 462
Score = 607 bits (1565), Expect = e-172
Identities = 307/461 (66%), Positives = 354/461 (76%), Gaps = 13/461 (2%)
Query: 1 MAVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDE 60
MA+YYKFKSARDYD+I+MDGPFISVG LK+KIFE+KHLG G D D+VV+NAQTNEEYLDE
Sbjct: 1 MAIYYKFKSARDYDTIAMDGPFISVGILKDKIFETKHLGTGKDLDIVVSNAQTNEEYLDE 60
Query: 61 EMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSA 120
MLIPKNTSVLIRRVPGRPR+ ++T QE +++NKV D + ++ P D SA
Sbjct: 61 AMLIPKNTSVLIRRVPGRPRITVITTQE--------PRIQNKVEDVQAETTNFPVADPSA 112
Query: 121 MKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQE-APPTSTVDEESKIKALIDTPALDWQH 179
++ ED ++DEFG DLYSIPD Q I A VDEESKI+ALIDTPALDWQ
Sbjct: 113 AEFPED-EYDEFGTDLYSIPDTQDAQHIIPRPHLATADDKVDEESKIQALIDTPALDWQQ 171
Query: 180 -QGSD-FGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPN 237
QG D FGAGRG+GRGM GRM G RGFG+ERKTPP GYVCHRC +PGHFIQHCPTNGDPN
Sbjct: 172 RQGQDTFGAGRGYGRGMPGRMNG-RGFGMERKTPPPGYVCHRCNIPGHFIQHCPTNGDPN 230
Query: 238 YDVKRVKQPTGIPRSMLMVNPQGSYALPNGSVAVLKPNEAAFEKEMEGMPSTTRSVGDLP 297
YDVKRVK PTGIP+SMLM P GSY+LP+G+VAVLKPNE AFEKEMEG+PSTTRSVG+LP
Sbjct: 231 YDVKRVKPPTGIPKSMLMATPDGSYSLPSGAVAVLKPNEDAFEKEMEGLPSTTRSVGELP 290
Query: 298 PELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCVCGAMNVLADDLLPNKTLR 357
PEL CPLC VMKDA LTSKCC+KSFCDKCIRD+IISKSMCVCG +VLADDLLPNKTLR
Sbjct: 291 PELKCPLCKEVMKDAALTSKCCYKSFCDKCIRDHIISKSMCVCGRSDVLADDLLPNKTLR 350
Query: 358 DTINRILESGNSSTENAGSTYQVQDMESARCPQPKIPSPTSSAASKGGLKISPVYDGTTN 417
DTINRILE+GN STEN GS + D+ESARCP PK SPT+S ASKG K + +
Sbjct: 351 DTINRILEAGNDSTENVGSVGHIPDLESARCPPPKALSPTTSVASKGEKKPVLSNNNDAS 410
Query: 418 IQDTAVETKVVSAPPQTSEHVKIPRAGDVSEATHESKSVKE 458
+E +++ P+ S V + + D E+T S VKE
Sbjct: 411 TLKAPMEVAEITSAPRASAEVNVEKPVDACESTQGSVIVKE 451
>emb|CAB10521.1| hypothetical protein [Arabidopsis thaliana]
gi|7268492|emb|CAB78743.1| hypothetical protein
[Arabidopsis thaliana] gi|7485110|pir||D71443
hypothetical protein - Arabidopsis thaliana
Length = 274
Score = 372 bits (955), Expect = e-101
Identities = 190/282 (67%), Positives = 220/282 (77%), Gaps = 16/282 (5%)
Query: 1 MAVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDE 60
MA+YYKFKSARDYD+ISMDGPFI+VG LKEKI+E+KHLG G D D+V++NAQTNEEYLDE
Sbjct: 1 MAIYYKFKSARDYDTISMDGPFITVGLLKEKIYETKHLGSGKDLDIVISNAQTNEEYLDE 60
Query: 61 EMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSA 120
MLIPKNTSVLIRRVPGRPR+ I+T +E P + KVEN D N+ + AD
Sbjct: 61 AMLIPKNTSVLIRRVPGRPRIRIITREE----PRVEDKVENVQAD---MNNVITAD---- 109
Query: 121 MKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQ 180
VED ++DEFGNDLYSIPD V S N+ ++ P DEE+K+KALIDTPALDW Q
Sbjct: 110 ASPVED-EFDEFGNDLYSIPDAPAVHSNNLCHDSAPAD--DEETKLKALIDTPALDWHQQ 166
Query: 181 GSD-FGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYD 239
G+D FG GRG+GRGM GRM GGRGFG+ER TPP GYVCHRC V GHFIQHC TNG+PN+D
Sbjct: 167 GADSFGPGRGYGRGMAGRM-GGRGFGMERTTPPPGYVCHRCNVSGHFIQHCSTNGNPNFD 225
Query: 240 VKRVKQPTGIPRSMLMVNPQGSYALPNGSVAVLKPNEAAFEK 281
VKRVK PTGIP+SMLM P GSY+LP+G+VAVLKPNE K
Sbjct: 226 VKRVKPPTGIPKSMLMATPNGSYSLPSGAVAVLKPNEYVVTK 267
>ref|XP_469113.1| putative DNA-binding protein [Oryza sativa (japonica
cultivar-group)] gi|27764684|gb|AAO23109.1| putative
DNA-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 697
Score = 246 bits (628), Expect = 2e-63
Identities = 228/767 (29%), Positives = 332/767 (42%), Gaps = 145/767 (18%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFES-KH-----LGRGTDFDLVVTNAQTNEE 56
VYYK+KS+++ S+ + F+SV LK+ I S KH GRG D+V++NAQT EE
Sbjct: 4 VYYKYKSSKETISVPVPHSFVSVSELKQLILTSDKHGRGRTRGRGPREDIVLSNAQTGEE 63
Query: 57 YLDEEMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPAD 116
+LDE +IP+N++VL+RR G+ IV
Sbjct: 64 FLDENAMIPQNSTVLVRRTSGQQSEKIVL------------------------------- 92
Query: 117 DMSAMKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALD 176
S+ + +ED +I + + + T DE++ I ++ID L
Sbjct: 93 -FSSREVIEDG----------AIASNKSEITESTSKSCSSTEVQDEDAAIASIIDAAELK 141
Query: 177 WQHQGSDFGAGRGFGRGMGGRMGGG-------------RGFGL----------ERKTPPQ 213
W+ + S G+ GR GR G G R G+ ERKTPP
Sbjct: 142 WEDKPSK--RGQTGGRFTSGRYGHGPVEGETPPPGYVCRSCGVPGHFIQHCQQERKTPPS 199
Query: 214 GYVCHRCKVPGHFIQHCPTNGDPNYDVKRVKQPTGIPR-SMLMVNPQGSYALPNGSVAVL 272
GYVC+RC++PGHFI HCPT GDP +D + K T +P S ++ S P SV+V
Sbjct: 200 GYVCYRCRIPGHFIHHCPTIGDPKFDDYK-KPHTLVPEVSACPIDGIPSALAPAASVSV- 257
Query: 273 KPNEAAFEKEMEGMPSTTRSVGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYI 332
V DLP ELHC LC+ VM DAVLTSKCCF SFCDKCIRDYI
Sbjct: 258 --------------------VDDLPAELHCRLCNKVMADAVLTSKCCFDSFCDKCIRDYI 297
Query: 333 ISKSMCVCGAMNVLADDLLPNKTLRDTINRILESGNSS------TENAGSTYQVQDMESA 386
I++S C+CG + VLADDL+PN+TLR TI+ +L + SS + S V ++
Sbjct: 298 ITQSKCICG-VKVLADDLIPNQTLRSTISNMLATRASSITSGTGKHRSSSGSNVDPNSAS 356
Query: 387 RCPQPKIPSPT-----SSAASKGGLKISPVY-DGTTNIQDTAVETKVVSAPPQTSEHVKI 440
+ P + + SSAA GL+++ + + T V+ +V S + +
Sbjct: 357 QTPSAALEKQSKDHHISSAAPDAGLQVATEHVSHLEHKLTTGVDLEV--KDEGNSAGILV 414
Query: 441 PRAGDVSEATHESKSVKEPVSQGSAQVVEEEVQQKLVPTEAGKKKKKK----KVRMPTND 496
+ V A K V + S+ + + ++ K T+ KKK+KK K+ P N
Sbjct: 415 EKI--VPTADARLKDVTDSTSKPA--TISGTMEPKASKTDQLKKKRKKADSTKIVHPNN- 469
Query: 497 FQWKPPHDLGAENYMMQMGPPPGY-NPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMP 555
NY + P Y NPY +G D +M AG M Y GPY +
Sbjct: 470 -----------ANYGYSIPFDPAYCNPYVSGYPWLTDPYMYGPAG-MPYGGYPMGPYGVN 517
Query: 556 FPNGMPHDPFANGMPHDPFGMQGYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARK 615
MP F + M + + + + D A P ++ + E
Sbjct: 518 SIGNMPLQ-FPSAMQGNLANTHCWETQSMIHWANEDAAR---------PRLAAKPKEPEP 567
Query: 616 ADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSER 675
A+ R E +R+ D + SS D + + +SDY ++ HRS +
Sbjct: 568 ANQSRSSERNQRLSSSHGTDPSYKTSRSS-SDRRDHRRSIDYVEDHRSSDYAED-HRSSK 625
Query: 676 PSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXH 722
SS R ++ +S +S R+ D DRH H
Sbjct: 626 RMRASSPSPTGGDRHSRARSRYSSRKLTYEDSSPTGGDRHSRARSSH 672
>emb|CAB10522.1| DNA-binding protein homolog [Arabidopsis thaliana]
gi|7268493|emb|CAB78744.1| DNA-binding protein homolog
[Arabidopsis thaliana] gi|7488095|pir||E71443 probable
DNA-binding protein - Arabidopsis thaliana
Length = 459
Score = 239 bits (611), Expect = 2e-61
Identities = 193/544 (35%), Positives = 261/544 (47%), Gaps = 104/544 (19%)
Query: 383 MESARCPQPKIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPR 442
MES RCP PK SPT+SAAS G K +P + T+ ++E +++ ++E VK+ +
Sbjct: 1 MESVRCPPPKALSPTTSAASGGEKKPAPSNNNETSTLKPSIEIAEITSAWASAEIVKVEK 60
Query: 443 AGDVSEATHESKSVKEP-VSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKP 501
D S S + KE VSQ + Q +EE+ Q++ E K+KKKK RM D
Sbjct: 61 PVDASANIQGSSNGKEAAVSQLNTQPPKEEMPQQVASGEQAGKRKKKKPRMSGTD----- 115
Query: 502 PHDLGAENYMMQMGPPPGYNPYWNGMQPCMDGFMAPYAGPM---HMMDYGHGPYDMPFPN 558
L +YMM MGP PG N Y+NG QP +G + G + +G + PFP
Sbjct: 116 ---LAGPDYMMPMGPGPG-NQYFNGFQPGFNGVQHGFNGVQPGFNGFHHGFNGFPGPFPG 171
Query: 559 GMPHDPFAN----GMPH-DPFGMQGYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEA 613
MP PF G+ H DPF QG+ P IPPP +RDLAE MN+ P M REEFEA
Sbjct: 172 AMP--PFVGYGFGGVIHPDPFAAQGFGFPNIPPP-YRDLAEMGNRMNLQHPIMGREEFEA 228
Query: 614 RKADARRKRENERR------VERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYH 667
+K + +RKRENE R V RD K R +S S +K K++ PP S +
Sbjct: 229 KKTEMKRKRENEIRRSEGGNVVRDSEKSRIMN---NSAVTSSPVKPKSRQGPPPPISSDY 285
Query: 668 QNRHRSERPSPD--SSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRH 725
R RS+R SP+ SS R PPR + RKS+ DRH D D HDR
Sbjct: 286 DRRRRSDRSSPERQSSRRFTSPPRSSSRKSER---------------DRHHDLDSEHDRR 330
Query: 726 QHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAE 785
+ R + T RK + +S KSS T +
Sbjct: 331 RDR-----------------PRETDRKH-----RKRSEKSSSDPTVEI------------ 356
Query: 786 AAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTT 845
D K++VF+RISFP E ++ K+RK S SS + SV+ +
Sbjct: 357 -------DDNNKSNVFTRISFPEE----SSGKQRKTSKSSPAPPES----SVAPVSSGRR 401
Query: 846 HLSNGGRKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPP-PPVDDWVEEGRHSRGTRD 904
H S R+ M +Y+SSDD++ RHFKR+PSRY+ SP P D E RHS+ ++
Sbjct: 402 HHSRRERE----MVEYDSSDDED---RHFKRKPSRYKRSPSVAPSDAGDEHFRHSKRSKG 454
Query: 905 RKHR 908
+ R
Sbjct: 455 ERAR 458
>dbj|BAC77636.1| retinoblastoma binding protein 6 isoform 1 [Homo sapiens]
gi|33620769|ref|NP_008841.2| retinoblastoma-binding
protein 6 isoform 1 [Homo sapiens]
Length = 1792
Score = 183 bits (464), Expect = 3e-44
Identities = 218/879 (24%), Positives = 345/879 (38%), Gaps = 190/879 (21%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEM 62
V+YKF S +YD+++ DG IS+ LK++I + L + D DL +TNAQT EEY D+
Sbjct: 4 VHYKFSSKLNYDTVTFDGLHISLCDLKKQIMGREKL-KAADCDLQITNAQTKEEYTDDNA 62
Query: 63 LIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSAMK 122
LIPKN+SV++RR+P + + S + TEPA M+ K
Sbjct: 63 LIPKNSSVIVRRIP--------------IGGVKSTSKTYVISRTEPA--------MATTK 100
Query: 123 YVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQGS 182
++DS SI ++ N+ + + EE KIKA++ G
Sbjct: 101 AIDDSS--------ASISLAQLTKTANLAE-----ANASEEDKIKAMMS-------QSGH 140
Query: 183 DFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDV-K 241
++ + +G PP Y C RC PGH+I++CPTNGD N++
Sbjct: 141 EYDPINYMKKPLG--------------PPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGP 186
Query: 242 RVKQPTGIPRSML-----------MVNPQGSYALPN---GSVAVLKPNEAAFEKEMEGMP 287
R+K+ TGIPRS + M+ G YA+P + A+ K + F E P
Sbjct: 187 RIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPE---EP 243
Query: 288 STTRSVGD-LPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISK---SMCVCGAM 343
S++ D +P EL C +C ++M DAV+ CC S+CD+CIR ++ + C
Sbjct: 244 SSSSEEDDPIPDELLCLICKDIMTDAVVI-PCCGNSYCDECIRTALLESDEHTCPTCHQN 302
Query: 344 NVLADDLLPNKTLRDTINRIL-ESG-------------------------------NSST 371
+V D L+ NK LR +N E+G S
Sbjct: 303 DVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPI 362
Query: 372 ENAGSTYQVQDMESARCPQPKIPSPTSSAAS-------KGGLKISPVYDGTTNIQDTAVE 424
+ S+ P P I S TS+ +S +PV D T + +V
Sbjct: 363 SRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATV-SISVH 421
Query: 425 TKVVSAPPQTSEHVKIPRAGDVSE-----------ATHESKSVKEPVSQGSAQVVEEEVQ 473
++ P + S++ +P A SE A E K + PV + + + +
Sbjct: 422 SEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLH 481
Query: 474 QKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLG---AENYMMQMGPPPGYNPYWNGMQ-- 528
+L+PT + R W+ + LG + ++ G Y G
Sbjct: 482 GQLIPTTG--PVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHS 539
Query: 529 -----------PCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ 577
P F+ P+ Y P+ +P P G+P F+ P
Sbjct: 540 ERSQRTQGPSLPATPVFVPVPPPPL----YPPPPHTLPLPPGVPPPQFSPQFPPGQPPPA 595
Query: 578 GYMMPP--IPPPPHR----------DLAEFSMGMNVPPPAMSREEFEARKADARRKRENE 625
GY +PP PP P A + P +SREEF + RR +E E
Sbjct: 596 GYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEF---YREQRRLKEEE 652
Query: 626 RRVERDFSKDRDFGREVSSVGDVSS--IKSKTKPIPPSSASDYHQN------------RH 671
++ + DF +E+ + +S ++ P S S Y ++ R
Sbjct: 653 KKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRS 712
Query: 672 RSERPSPDSSH-REVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRH 730
RS S SH R P R+ R R R + YH R + R +RR+
Sbjct: 713 RSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYH-RSRSRS-----PPYRRY 766
Query: 731 HHRTEA-SSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEP 768
H R+ + + + P ++ + +E + ++ R+ P
Sbjct: 767 HSRSRSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPP 805
Score = 37.0 bits (84), Expect = 3.0
Identities = 50/242 (20%), Positives = 82/242 (33%), Gaps = 31/242 (12%)
Query: 609 EEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ 668
E +A + E ++R ERD K + G + D S++ K + P+
Sbjct: 989 EPMDAESITFKSVSEKDKR-ERDKPKAK--GDKTKRKNDGSAVSKKENIVKPAKGPQEKV 1045
Query: 669 N--RHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQ 726
+ R RS R P + E P+ KS S ++ E
Sbjct: 1046 DGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGT------------------ 1087
Query: 727 HRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKS-----KSRKSSEPVTKSLSLSKTAPT 781
R H A + T PV + +K D+KS K K+ +P K+ L
Sbjct: 1088 -PRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEK 1146
Query: 782 TSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKA 841
+ + +S+ +IS E + KRK+ + T +S A
Sbjct: 1147 RKRKTEEKGVDKDFESSSM--KISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSA 1204
Query: 842 PS 843
P+
Sbjct: 1205 PA 1206
>ref|XP_219296.3| PREDICTED: similar to PACT [Rattus norvegicus]
Length = 1789
Score = 179 bits (455), Expect = 3e-43
Identities = 219/879 (24%), Positives = 343/879 (38%), Gaps = 189/879 (21%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEM 62
V+YKF S +YD+++ DG IS+ LK++I + L + D DL +TNAQT EEY D+
Sbjct: 4 VHYKFSSKLNYDTVTFDGLHISLCDLKKQIMGREKL-KAADSDLQITNAQTKEEYTDDNA 62
Query: 63 LIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSAMK 122
LIPKN+SV++RR+P + + S + TEP M K
Sbjct: 63 LIPKNSSVIVRRIP--------------IGGVKSTSKTYVISRTEPV--------MGTTK 100
Query: 123 YVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQGS 182
++DS SI ++ N+ + + EE KIKA++ G
Sbjct: 101 AIDDSS--------ASISLAQLTKTANLAE-----ANASEEDKIKAMMS-------QSGH 140
Query: 183 DFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDV-K 241
++ + + G PP Y C RC PGH+I++CPTNGD N++
Sbjct: 141 EYDPINYMKKTLVG-------------PPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGP 187
Query: 242 RVKQPTGIPRSML-----------MVNPQGSYALPN---GSVAVLKPNEAAFEKEMEGMP 287
R+K+ TGIPRS + M+ G YA+P + A+ K + F E P
Sbjct: 188 RIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPE---EP 244
Query: 288 STTRSVGD-LPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISK---SMCVCGAM 343
S++ D +P EL C +C ++M DAV+ CC S+CD+CIR ++ + C
Sbjct: 245 SSSSEEDDPIPDELLCLICKDIMTDAVVI-PCCGNSYCDECIRTALLESDEHTCPTCHQN 303
Query: 344 NVLADDLLPNKTLRDTINRIL-ESG-------------------------------NSST 371
+V D L+ NK LR +N E+G S
Sbjct: 304 DVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPISAPRPLIQRNLQPLMRSPI 363
Query: 372 ENAGSTYQVQDMESARCPQPKIPS----PTSSAASKGGLKIS---PVYDGTTNIQDTAVE 424
+ S+ P I S P+S A S G S PV D T + +V
Sbjct: 364 SRQQDPLMIPVTSSSAHSTPSISSLTSNPSSLAPSVPGNPSSAPAPVPDITATV-SISVH 422
Query: 425 TKVVSAPPQTSEHVKIPRAGDVSE-----------ATHESKSVKEPVSQGSAQVVEEEVQ 473
++ P + S++ +P A SE A E K + PV + + + +
Sbjct: 423 SEKSDGPFRDSDNKLLPAAALASEHSKGASSIAITALMEEKGYQVPVLGTPSLLGQSLLH 482
Query: 474 QKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLG---AENYMMQMGPPPGYNPYWNGMQ-- 528
+L+PT + R W+ + LG + ++ G Y G
Sbjct: 483 GQLIPTTG--PVRINAARPGGGRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHS 540
Query: 529 -----------PCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ 577
P F+ P+ Y P+ +P P G+P F+ P
Sbjct: 541 ERSQRTQGPSLPATPVFVPVPPPPL----YPPPPHTLPLPPGVPPPQFSPQFPPGQPPPA 596
Query: 578 GYMMPP--IPPPPHR----------DLAEFSMGMNVPPPAMSREEFEARKADARRKRENE 625
GY +PP PP P A + P +SREEF + RR +E E
Sbjct: 597 GYSVPPPGFPPAPANISTPWVSSGVQTAHSNTIPTTQAPPLSREEF---YREQRRLKEEE 653
Query: 626 RRVERDFSKDRDFGREVSSVGDVSS--IKSKTKPIPPSSASDYHQN------------RH 671
++ + DF +E+ + +S ++ P S S Y ++ R
Sbjct: 654 KKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRS 713
Query: 672 RSERPSPDSSH-REVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRH 730
RS S SH R P R+ R R R + YH R + R +RR+
Sbjct: 714 RSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYH-RSRSRS-----PPYRRY 767
Query: 731 HHRTEA-SSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEP 768
H R+ + + + P ++ + +E + ++ R+ P
Sbjct: 768 HSRSRSPQAFRGQSPTKRNVPQGETEREYFNRYREVPPP 806
Score = 39.7 bits (91), Expect = 0.46
Identities = 56/277 (20%), Positives = 103/277 (36%), Gaps = 33/277 (11%)
Query: 600 NVPPPAMSREEFEARKADARRKRENERRVERDFSKDRD-FGREVSSVGDVSS-IKSKTKP 657
N PP + + +R E + V+++ K+R+ E S G SS K + P
Sbjct: 1531 NSSPPRDKKPHDHKASYETKRSCEETKSVDKNAGKEREKHAAEASRNGKESSGSKLPSIP 1590
Query: 658 IPPSSASDYH----QNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDR--DRDYDY 711
PP + Q + +P P SH + R S RE ++ + DY+
Sbjct: 1591 NPPDPPMEKELAAGQIEKSTIKPKPQLSH--------SSRLSSDLTRETDEAAFEPDYNE 1642
Query: 712 HDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTK 771
D + + E +++K+T+ + +T +S+P +SS V+
Sbjct: 1643 SDSESNVSVKEEEAVASVSKDLKEKATEKATESLNVAT---ASQPGADRSQSQSSPSVSP 1699
Query: 772 SLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEAST 831
S S S + T + +++A++A+ + +++ KK K
Sbjct: 1700 SRSHSPSGSQTRSHSSSASSAESQDS----------KKKKKKKDKKKHKKHKKHKKHKKH 1749
Query: 832 AATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDE 868
A K+ H +K+K D E DD+
Sbjct: 1750 AGVDGDVEKSQKHKHKKKKAKKNK----DKEKEKDDQ 1782
>dbj|BAC77637.1| retinoblastoma binding protein 6 isoform 2 [Homo sapiens]
gi|33620716|ref|NP_061173.1| retinoblastoma-binding
protein 6 isoform 2 [Homo sapiens]
Length = 1758
Score = 172 bits (437), Expect = 3e-41
Identities = 218/864 (25%), Positives = 339/864 (39%), Gaps = 194/864 (22%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEM 62
V+YKF S +YD+++ DG IS+ LK++I + L + D DL +TNAQT EEY D+
Sbjct: 4 VHYKFSSKLNYDTVTFDGLHISLCDLKKQIMGREKL-KAADCDLQITNAQTKEEYTDDNA 62
Query: 63 LIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSAMK 122
LIPKN+SV++RR+P + + S + TEPA M+ K
Sbjct: 63 LIPKNSSVIVRRIP--------------IGGVKSTSKTYVISRTEPA--------MATTK 100
Query: 123 YVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQGS 182
++DS SI ++ N+ + + EE KIKA++ G
Sbjct: 101 AIDDSS--------ASISLAQLTKTANLAE-----ANASEEDKIKAMMS-------QSGH 140
Query: 183 DFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDV-K 241
++ + +G PP Y C RC PGH+I++CPTNGD N++
Sbjct: 141 EYDPINYMKKPLG--------------PPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGP 186
Query: 242 RVKQPTGIPRSML-----------MVNPQGSYALPN---GSVAVLKPNEAAFEKEMEGMP 287
R+K+ TGIPRS + M+ G YA+P + A+ K + F E P
Sbjct: 187 RIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPE---EP 243
Query: 288 STTRSVGD-LPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISK---SMCVCGAM 343
S++ D +P EL C +C ++M DAV+ CC S+CD+CIR ++ + C
Sbjct: 244 SSSSEEDDPIPDELLCLICKDIMTDAVVI-PCCGNSYCDECIRTALLESDEHTCPTCHQN 302
Query: 344 NVLADDLLPNKTLRDTINRIL-ESG-------------------------------NSST 371
+V D L+ NK LR +N E+G S
Sbjct: 303 DVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPI 362
Query: 372 ENAGSTYQVQDMESARCPQPKIPSPTSSAAS-------KGGLKISPVYDGTTNIQDTAVE 424
+ S+ P P I S TS+ +S +PV D T + +V
Sbjct: 363 SRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATV-SISVH 421
Query: 425 TKVVSAPPQTSEHVKIPRAGDVSE-----------ATHESKSVKEPVSQGSAQVVEEEVQ 473
++ P + S++ +P A SE A E K + PV + + + +
Sbjct: 422 SEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLH 481
Query: 474 QKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLG---AENYMMQMGPPPGYNPYWNGMQ-- 528
+L+PT + R W+ + LG + ++ G Y G
Sbjct: 482 GQLIPTTG--PVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHS 539
Query: 529 -----------PCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ 577
P F+ P+ Y P+ +P P G+P F+ P
Sbjct: 540 ERSQRTQGPSLPATPVFVPVPPPPL----YPPPPHTLPLPPGVPPPQFSPQFPPGQPPPA 595
Query: 578 GYMMPP--IPPPPHR----------DLAEFSMGMNVPPPAMSREEFEARKADARRKRENE 625
GY +PP PP P A + P +SREEF E
Sbjct: 596 GYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEF-----------YRE 644
Query: 626 RRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREV 685
+R ++ SK G SS S SK++ S S Y ++ RS S S
Sbjct: 645 QRRLKEESKSPYSG---SSYSRSSYTYSKSRSGSTRSRS-YSRSFSRSHSRSYSRS---- 696
Query: 686 EPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEA-SSKKSTDP 744
PP P R+ R R R + YH R + R +RR+H R+ + + + P
Sbjct: 697 -PPYP--RRGRGKSRNYRSRSRSHGYH-RSRSRS-----PPYRRYHSRSRSPQAFRGQSP 747
Query: 745 VTKSTSRKSSEPDIKSKSRKSSEP 768
++ + +E + ++ R+ P
Sbjct: 748 NKRNVPQGETEREYFNRYREVPPP 771
Score = 37.0 bits (84), Expect = 3.0
Identities = 50/242 (20%), Positives = 82/242 (33%), Gaps = 31/242 (12%)
Query: 609 EEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ 668
E +A + E ++R ERD K + G + D S++ K + P+
Sbjct: 955 EPMDAESITFKSVSEKDKR-ERDKPKAK--GDKTKRKNDGSAVSKKENIVKPAKGPQEKV 1011
Query: 669 N--RHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQ 726
+ R RS R P + E P+ KS S ++ E
Sbjct: 1012 DGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGT------------------ 1053
Query: 727 HRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKS-----KSRKSSEPVTKSLSLSKTAPT 781
R H A + T PV + +K D+KS K K+ +P K+ L
Sbjct: 1054 -PRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEK 1112
Query: 782 TSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKA 841
+ + +S+ +IS E + KRK+ + T +S A
Sbjct: 1113 RKRKTEEKGVDKDFESSSM--KISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSA 1170
Query: 842 PS 843
P+
Sbjct: 1171 PA 1172
>gb|EAA62011.1| hypothetical protein AN7431.2 [Aspergillus nidulans FGSC A4]
gi|67900888|ref|XP_680700.1| hypothetical protein
AN7431_2 [Aspergillus nidulans FGSC A4]
gi|49107797|ref|XP_411568.1| hypothetical protein
AN7431.2 [Aspergillus nidulans FGSC A4]
Length = 628
Score = 169 bits (429), Expect = 3e-40
Identities = 177/627 (28%), Positives = 262/627 (41%), Gaps = 109/627 (17%)
Query: 2 AVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEE 61
+V++KFKS ++ ++ DG ISV LK +I LG GTDF+L + N T EEY D+
Sbjct: 4 SVHFKFKSQKEPSRVTFDGTGISVFELKREIINQSRLGDGTDFELSIYNEDTGEEYDDDT 63
Query: 62 MLIPKNTSVLIRRVP-GRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANS-SLPADDMS 119
+IP++TSV+ RR+P RP K ++ V K+ P N+ S+P ++ S
Sbjct: 64 SVIPRSTSVIARRLPASRPG-----------KGGAARYVSGKM----PVNARSVPRNEQS 108
Query: 120 AMKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQH 179
N S P Q PV N +QE T EE KI AL + A W+
Sbjct: 109 T------------SNRTVSNPTQ-PVS--NGVQELHNAQT--EEEKINALFNLQANQWRE 151
Query: 180 QGSDFGAGR--GFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPN 237
Q + FGRG G+ + PP GY+C+RC+ GH+IQ CPTN DP
Sbjct: 152 QQQEMANATPVPFGRGR------GKPVNVPDHPPPPGYLCYRCREKGHWIQACPTNNDPK 205
Query: 238 YDVK-RVKQPTGIPRSM---------LMVNPQ-------GSYALPNGSVAVLKPNEAAFE 280
+D K RVK+ TGIPRS+ L+++ G +G + KP++A++E
Sbjct: 206 FDGKYRVKRSTGIPRSLQTKVEKPESLLIDGSTEDLRNTGVMVNADGDFIITKPDQASWE 265
Query: 281 KEMEGMPSTTRSVGDLPPELH----------CPLCSNVMKDAVLTSKCCFKSFCDKCIRD 330
E + +T + + H CP+ + + T CC K++C+ CI +
Sbjct: 266 LYQEKVKATAAAAAEAAAAEHSKELQARGLECPIDKRMFLEPTQT-PCCQKTYCNDCITN 324
Query: 331 YIISKSMCV--CGAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARC 388
+I CG VL D+L N + I TE A S + + +
Sbjct: 325 ALIESDFVCPGCGTEGVLLDNLSANDEMLSKIKAY------ETEKADSKKEKEKQLTPTE 378
Query: 389 PQPKIPSP---TSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGD 445
QP +P + + KG + +PV +D V S P + K R+ D
Sbjct: 379 VQPDNNTPVHISDTTERKGDSRSAPVDSKKRPAEDDPVTG--TSEEPGSGSSNKKQRSQD 436
Query: 446 VSEATHESKSVKEPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDL 505
+T S+ E S + Q + Q G +P F P
Sbjct: 437 AQSSTETSEPRTENTST-TFQPLPFNPQMSF-----GMPGFMPGPGLPGMPF---PDAAF 487
Query: 506 GAE--NYMMQMGPPPGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHD 563
G E +M MG P G G P MD P++M+++ P N P+
Sbjct: 488 GGEGMGFMNPMGLPAG-----PGFPPNMDHNW----NPLNMLNF--NPLSNGMYNNRPNG 536
Query: 564 PFANGM----PHDPFGMQGYMMPPIPP 586
PF NG ++ G Q M +PP
Sbjct: 537 PFLNGFGAPNAYNGTGDQSMNMLAMPP 563
>gb|EAK81306.1| hypothetical protein UM00321.1 [Ustilago maydis 521]
gi|49067292|ref|XP_397936.1| hypothetical protein
UM00321.1 [Ustilago maydis 521]
Length = 622
Score = 169 bits (428), Expect = 4e-40
Identities = 129/428 (30%), Positives = 187/428 (43%), Gaps = 47/428 (10%)
Query: 2 AVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEE 61
+VYYKFKS ++ I+ DG ISV LK +I +GRGTDFDL V N+ TN+EY D+
Sbjct: 5 SVYYKFKSQKEPSRITFDGTGISVWDLKREIILQNKMGRGTDFDLGVYNSDTNDEYKDDN 64
Query: 62 MLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPAN--------SSL 113
L+P++T V++RR+P P P + Y+ + N DT A+ S +
Sbjct: 65 FLVPRSTQVIVRRLP--PSKPGRGTAQNYVADV------NGAADTTGASRFSGTANASGI 116
Query: 114 PADDMSAMKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPP--TSTVDEESKIKALID 171
D + Y G S+ P S P +T DE +I A+
Sbjct: 117 KPVDARSQAYRGPMTMRFDGGKDPSMSSTQPSSSTGPSSGGGPAAAATGDEADRIAAMFQ 176
Query: 172 TPALDWQHQGSDFGAGRGFGRGMGGRMGG-GRGFGLER-------KTPPQGYVCHRCKVP 223
W R R GG R +R + PP GYVC RC P
Sbjct: 177 ATTEQWDETQERMSHATYRDRSGQARRGGPPRPMATQRAQQQLPDRPPPIGYVCFRCGKP 236
Query: 224 GHFIQHCPTNGDPNYDVK-RVKQPTGIPRSMLMVNPQ--------GSYALPNGSVAVLKP 274
GH+IQ CPTN D YD + R K+ TGIP+SML Q G +G+ V +
Sbjct: 237 GHWIQDCPTNDDKEYDNRPRFKRTTGIPKSMLKTVEQPTDEDHRAGVMLTADGTYVVARV 296
Query: 275 NEAAFEKEMEGMPSTTRS----VGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRD 330
++ ++ K T+S P L CPLCS +++DAV+T CC +C++CI+
Sbjct: 297 DQESWRKNRVRTKPLTQSDVYQSAPTDPSLACPLCSKLLRDAVVT-PCCQTKYCEECIQT 355
Query: 331 YIISKSMCVCGAMNVLAD------DLLPNKTLRDTINRILE-SGNSSTENAGSTYQVQDM 383
+++ +AD DL K +++ + +E S E A Q D
Sbjct: 356 HLLEHEFLCAECEKRIADLEQLQPDLETRKRVKEYVKETIEQSEREIAEEASKVEQTDDA 415
Query: 384 ESARCPQP 391
++ P
Sbjct: 416 GDSKVGSP 423
>ref|XP_145621.5| PREDICTED: similar to retinoblastoma-binding protein 6 isoform 2
[Mus musculus]
Length = 1049
Score = 167 bits (424), Expect = 1e-39
Identities = 241/987 (24%), Positives = 383/987 (38%), Gaps = 241/987 (24%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEM 62
V+YKF S Y++I+ DG IS+ LK++I + L + + DL +TNA+T EEY D+
Sbjct: 4 VHYKFSSKLSYNTITFDGLHISLFYLKKQIMGREKL-KTANSDLQITNAETEEEYTDDNA 62
Query: 63 LIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSAMK 122
LIPKN+SV++RR+P ++ K + K T + + P M +
Sbjct: 63 LIPKNSSVIVRRIP-----------------VVGVKSKGK---TYQISHTKPV--MGTTR 100
Query: 123 YVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAP-PTSTVDEESKIKALIDTPALDWQHQG 181
V DS P+ +I+ A + EE KIKA++ + H
Sbjct: 101 AVNDSS--------------APMSLAQLIETANLAEANASEEDKIKAMM----IQSGHLY 142
Query: 182 SDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDVK 241
+ + + G+ PP Y C C PGH+I++CPTN + N++ +
Sbjct: 143 NPINSMKKTPVGL----------------PPPSYTCFCCGKPGHYIKNCPTNANKNFESR 186
Query: 242 -RVKQPTGIPRSML-----------MVNPQGSYALPN---GSVAVLKPNEAAFEKEMEGM 286
R+++ TGIPRS + M+ G YA+P + A+ K + F E
Sbjct: 187 PRIRKSTGIPRSFMMEVKDPNMKGAMLTKTGQYAIPTLNAEAYAIGKKEKPPFLPEEPSS 246
Query: 287 PSTTRSVGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISK---SMCVCGAM 343
S++ V +P EL C +C + M DA + CC S+CD+CIR ++ + C
Sbjct: 247 SSSSEEVDPVPDELLCLICKDTMTDAAII-PCCGNSYCDECIRTALLESDEHTCPTCHQN 305
Query: 344 NVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARCP---------QPKIP 394
+V D L+ NK LR +N ++ ST++ G QV P QP +
Sbjct: 306 DVSPDALVANKVLRQAVNN-FKNQTDSTKSLGK--QVTPSPPPLPPPSALIQQNLQPPMK 362
Query: 395 SPT------------------------------SSAASKGGLKIS---PVYDGTTNIQDT 421
SPT SSA S G S PV D T + +
Sbjct: 363 SPTSRQQDPLKVPVTSFSAHPTPSVTSLASNLSSSAPSVPGNPSSAPAPVPDTTARVSIS 422
Query: 422 -----------AVETKVVSAPPQTSEHVKIPRAGDVSEATHESKSVKEPVSQGSAQVVEE 470
+ K++SA TSEH K + V+ A + K + PV + S + +
Sbjct: 423 DHSEKSDGPFRDSDNKLLSAAALTSEHSKEASSIAVA-ALMQEKCSQVPVLETSPLLEQS 481
Query: 471 EVQQKLVPT--------------EAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGP 516
+ ++ +PT + G + K ++ + PP + +E + G
Sbjct: 482 LLHEQFIPTTGPLRINAAHPSGGQPGWEHPNKLGKLVS------PPQQIRSEERLCYRGI 535
Query: 517 PPG--YNPYWNGMQ----PCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMP 570
G Y+ Q P F+ P P+ + P+ + P G+P F P
Sbjct: 536 KRGQDYSEQSQSSQGPPRPATPVFV-PIPPPLMQLS---PPHMLLLPPGVPAPRFPPWFP 591
Query: 571 HDPFGMQGYMMPP---IPPPPHRDLAEFSMGMN---------VPPPAMSREEFEARKADA 618
GY +PP P P + A S G++ P +SREEF +
Sbjct: 592 PSQPPPAGYSVPPPGFPPAPANISTAWVSSGVHTAHSNTIPTTQAPLLSREEF--YREQK 649
Query: 619 RRKRENE--------RRVERDFSKDRD---FGREVSSVGDVSSIKSKTKPIPPSS----- 662
R K+E++ R SK R R S S +S ++ P S
Sbjct: 650 RLKKESKFPYSGSSYSRSSYTDSKSRSGSTHSRSYSQSFSCSHSRSSSRSSPSSRRGRGK 709
Query: 663 -ASDYHQNR----HRSERPSPDSSH---REVEPP-----RPTKRKSDHSEREREDRDRDY 709
+D Q+R HRS SP R PP PTK E+ER
Sbjct: 710 ICNDCSQSRSHGYHRSRSRSPPYRRYHSRSRSPPAFRGQSPTKLDVPQEEKERY------ 763
Query: 710 DYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTD-----PVTKSTSRKSSEPDIKSKSRK 764
+ R+++ S ++ T+ P + + S+ +I +
Sbjct: 764 --------------QEWERKYYQWVPISLQQKTEAVPGTPFPQEYCKVISKQNILQVQQW 809
Query: 765 SS-----EPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKR 819
P+ S+S TA TS + + K SVF + PS A ++ K
Sbjct: 810 EKWSYLLNPLVLLPSVSHTAENTSGKNTVENTSGNK---SVFIHHTLPSIIRTAMSSAKD 866
Query: 820 KLSASSTTEASTAATASVS-AKAPSTT 845
+SA+ +TE + T ++ + PST+
Sbjct: 867 PMSAAPSTELRSGVTLGIAREQCPSTS 893
>gb|AAH63524.1| RBBP6 protein [Homo sapiens]
Length = 919
Score = 160 bits (405), Expect = 2e-37
Identities = 128/413 (30%), Positives = 191/413 (45%), Gaps = 89/413 (21%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEM 62
V+YKF S +YD+++ DG IS+ LK++I + L + D DL +TNAQT EEY D+
Sbjct: 4 VHYKFSSKLNYDTVTFDGLHISLCDLKKQIMGREKL-KAADCDLQITNAQTKEEYTDDNA 62
Query: 63 LIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSAMK 122
LIPKN+SV++RR+P + + S + TEPA M+ K
Sbjct: 63 LIPKNSSVIVRRIP--------------IGGVKSTSKTYVISRTEPA--------MATTK 100
Query: 123 YVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQGS 182
++DS SI ++ N+ + + EE KIKA++ G
Sbjct: 101 AIDDSS--------ASISLAQLTKTANLAE-----ANASEEDKIKAMMS-------QSGH 140
Query: 183 DFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDV-K 241
++ + +G PP Y C RC PGH+I++CPTNGD N++
Sbjct: 141 EYDPINYMKKPLG--------------PPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGP 186
Query: 242 RVKQPTGIPRSML-----------MVNPQGSYALPN---GSVAVLKPNEAAFEKEMEGMP 287
R+K+ TGIPRS + M+ G YA+P + A+ K + F E P
Sbjct: 187 RIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPE---EP 243
Query: 288 STTRSVGD-LPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISK---SMCVCGAM 343
S++ D +P EL C +C ++M DAV+ CC S+CD+CIR ++ + C
Sbjct: 244 SSSSEEDDPIPDELLCLICKDIMTDAVVI-PCCGNSYCDECIRTALLESDEHTCPTCHQN 302
Query: 344 NVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARCPQPKIPSP 396
+V D L+ NK LR +N N E Y + + P P IP P
Sbjct: 303 DVSPDALIANKFLRQAVN------NFKNETG---YTKRLRKQLPPPPPPIPPP 346
>ref|XP_620499.1| PREDICTED: similar to retinoblastoma-binding protein 6 isoform 2
[Mus musculus]
Length = 788
Score = 157 bits (398), Expect = 1e-36
Identities = 206/859 (23%), Positives = 326/859 (36%), Gaps = 195/859 (22%)
Query: 3 VYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEEM 62
V+YKF S Y++I+ DG IS+ LK++I + L G DL + NA+T EEY D+
Sbjct: 4 VHYKFSSKLSYNTITFDGLHISLFYLKKQIMGREKLKTGNS-DLQIINAETEEEYTDDNA 62
Query: 63 LIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENK---VVDTEPANSSLPADDMS 119
LIPKN+SV++RR+P ++ K ++K + T+P M
Sbjct: 63 LIPKNSSVIVRRIP-----------------VVGVKSKSKTYGISHTKPV--------MG 97
Query: 120 AMKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAP-PTSTVDEESKIKALIDTPALDWQ 178
K V DS P+ +I+ A + EE KIKA++ +
Sbjct: 98 TTKAVNDSS--------------APMSLAQLIETANLAEANASEEDKIKAMM----IQSG 139
Query: 179 HQGSDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNY 238
H + + G+ PP Y C RC PGH+I++CPTN D N+
Sbjct: 140 HLYNPINYMKKTPVGL----------------PPPSYTCFRCGKPGHYIKNCPTNADKNF 183
Query: 239 DVK-RVKQPTGIPRSML-----------MVNPQGSYALPN---GSVAVLKPNEAAFEKEM 283
+ + R+++ TGIPRS + M+ G Y +P + A+ K + F E
Sbjct: 184 ESRPRIRKSTGIPRSFMMEVKDPNMKGAMLTKTGQYVIPTINAEAYAIGKKEKPPFLPEE 243
Query: 284 EGMPSTTRSVGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISK---SMCVC 340
S++ V +P E C +C + M DA + CC S+CD+CIR ++ + C
Sbjct: 244 PSSSSSSEEVDPVPDEFLCLICKDTMTDAAII-PCCGNSYCDECIRTALLESDEHTCPTC 302
Query: 341 GAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTY--------------------QV 380
+V D L+ NK LR +N ++ T+ G +
Sbjct: 303 HQNDVSPDALVANKVLRQAVNN-FKNQTDYTKRLGKQVTPSPPPLPPPSALIQQNLQPPM 361
Query: 381 QDMESARCPQPKIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKI 440
+ + S + KIP +SSA++ ++ + +++ +V AP E
Sbjct: 362 KSLTSRQQDPLKIPMTSSSASAHPTPSVTSLASNSSS-SAPSVPGNPSPAPAPVPETTAR 420
Query: 441 PRAGDVSEATHES-KSVKEPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQW 499
D+SE + S + + + +A E + A ++K+ +V +
Sbjct: 421 VSVSDLSEKSDGSFRESENQLLPAAALTSEHSKGASSIAVTAPMEEKRGQVAV------L 474
Query: 500 KPPHDLGA----ENYMMQMGP-------PPGYNPYW----------NGMQPCMDGFMAPY 538
+ P LG E + GP P G P W + Q + Y
Sbjct: 475 ETPPSLGQSLLHEQSIPTTGPVRINAAHPSGGQPGWEHSNKIGKLVSPTQQIRSEERSCY 534
Query: 539 AGPMHMMDY------GHGP---------------------YDMPFPNGMPHDPFANGMPH 571
G DY GP + +P P G+P F +P
Sbjct: 535 RGIKRGRDYSEPSQRSQGPSRPATPVFVPIPPPLLPPSPPHMLPLPPGVPAPRFPPRVPP 594
Query: 572 DPFGMQGYMMPP---IPPPPHRDLAEFSMGM-----NVPP----PAMSREEFEARKADAR 619
GY +PP P P + A FS G+ N P P +SREEF + R
Sbjct: 595 GQPPTAGYSVPPPGFPPAPANISTACFSPGVPTAHSNTMPTTQAPLLSREEF--YREQKR 652
Query: 620 RKRENE--------RRVERDFSKDRD---FGREVSSVGDVSSIKSKTKPIPPSS------ 662
K E++ R SK R R S S +S ++ P S
Sbjct: 653 LKEESKFPYSGSSYSRSSYTDSKSRSGSAHSRSYSQSFSCSHSRSSSRSSPSSRRGRGKI 712
Query: 663 ASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXH 722
+D Q+R RPS S P R + +S R D ++R
Sbjct: 713 CNDCSQSRSHGYRPSRSRS----PPYRRYRSRSRSPPAFRGQSPTKLDVPREEKERYQEW 768
Query: 723 DRHQHRRHHHRTEASSKKS 741
+R ++ + +R ASS+ +
Sbjct: 769 ERKYYQWYENRQRASSRNT 787
>gb|EAA70118.1| hypothetical protein FG09892.1 [Gibberella zeae PH-1]
gi|46136753|ref|XP_390068.1| hypothetical protein
FG09892.1 [Gibberella zeae PH-1]
Length = 617
Score = 153 bits (386), Expect = 3e-35
Identities = 185/715 (25%), Positives = 277/715 (37%), Gaps = 145/715 (20%)
Query: 2 AVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDEE 61
+V++KFKS R+ + DG ISV LK +I LG GTDFDL + +EEY D+
Sbjct: 4 SVFFKFKSQREPTRVEFDGTGISVFELKREIITKSGLGDGTDFDLHIYTDDNSEEYDDDT 63
Query: 62 MLIPKNTSVLIRRVPGRPRLPIVTEQEYYL--KPLISQKVENKVVDTEPANSSLPADDMS 119
+IP++T+V+ RR P P Y+ K ++ K + T A+ S P+ D
Sbjct: 64 TIIPRSTTVIARRQPALK--PGAGRAARYVSGKMPVTAKNAGRKEQTAKASVSKPSSD-- 119
Query: 120 AMKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQH 179
+I+ + A + EE K+ A+ W
Sbjct: 120 ---------------------------AISQMNNA-----MTEEEKMAAMFAAQTEQWSA 147
Query: 180 QGSDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYD 239
Q + + G R + PP GY+C+RC GH+IQ CPTN DP +D
Sbjct: 148 QQEEMSHQTPVFKA-----GAKRPANVPDHDPPNGYICYRCGNKGHWIQLCPTNDDPEFD 202
Query: 240 VK-RVKQPTGIPRSMLM----------------VNPQGSYALPNGSVAVLKPNEAA---F 279
+ RVK+ TGIPRS L P G G + +P++A+ F
Sbjct: 203 NRPRVKRTTGIPRSFLQKVDKSVVLAQTDGDETKRPSGIMVNAEGDFVIAEPDKASWEQF 262
Query: 280 EKEMEGMPSTTRSVGDLPPE---LHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYII-SK 335
+ + + +T GD + L C + + + + T CC K+FC+ CI + +I S
Sbjct: 263 QAKAKSSATTNAPAGDKEIQEQGLECSIDKKMFIEPMKT-PCCQKTFCNDCITNALIESD 321
Query: 336 SMC-VCGAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARCPQPKIP 394
+C C + VL DDL P++ I L ++ E A+ P P P
Sbjct: 322 FVCPACQSEGVLIDDLQPDEEASKKIQEYL----------------KEKEIAKSPPP--P 363
Query: 395 SPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSEATHESK 454
SP AS+G D N + E KV + ++ + + T +S
Sbjct: 364 SP---KASEGAKADGESQDKPQNEDIASTEQKVEN---ESHDKTMASAKEKSNSPTSQSA 417
Query: 455 SVKEPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQM 514
++K P + V VP +A K+ K ++D K P D +N +
Sbjct: 418 TIKTPPTGPKGLV--------SVPHDANKQVDGAKT---SDDNSKKRPADDILDNPRIPK 466
Query: 515 GPPPGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPH--- 571
GP N Q M+G M+ M G +M F NGMP P GMP+
Sbjct: 467 GPKAMQNQ--QNQQNMMNGM------GMNGMGMNMGMNNMMF-NGMPMMPNMMGMPNMGN 517
Query: 572 --------DPFGMQGYM-MPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKR 622
+ GM G M MP P GMN M+ + +
Sbjct: 518 MNMGMPNMNMMGMPGMMGMPGFP------------GMNGGFGGMNGWDMGMNGMNGNMGN 565
Query: 623 ENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPS 677
+ ++ +FG+ V D KP+ P HQN+ R RPS
Sbjct: 566 MGNMNGGMNGFQNGNFGQGVGPDEDAYF----RKPVNPHR----HQNKQRRVRPS 612
>emb|CAD71243.1| conserved hypothetical protein [Neurospora crassa]
gi|32413048|ref|XP_327004.1| hypothetical protein
[Neurospora crassa] gi|28922429|gb|EAA31662.1|
hypothetical protein [Neurospora crassa]
Length = 673
Score = 143 bits (361), Expect = 2e-32
Identities = 151/620 (24%), Positives = 246/620 (39%), Gaps = 117/620 (18%)
Query: 2 AVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEE-YLDE 60
+V++KFKS ++ + DG ISV LK +I LG GTDFDL++ + +E Y D+
Sbjct: 4 SVFFKFKSNKEPTRVEFDGTGISVFELKREIILKSALGDGTDFDLIIAADEGMKEVYDDD 63
Query: 61 EMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSA 120
+IP++T+V+ RR+P KV+ + + +P ++
Sbjct: 64 TTIIPRSTTVIARRMPA--------------------KVQGRGGAARYVSGKMPVHAKNS 103
Query: 121 MKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQ 180
+ + + P PV +N S + EE K+ A+ ++ +
Sbjct: 104 SRKEQPIV------KALAKPVANPVVQLN--------SAMTEEEKMAAVFQAQTENFTAR 149
Query: 181 GSDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDV 240
+ + + G + + PPQGY+C+RC GH+IQ CPTN +P+YD
Sbjct: 150 EEEMATQQYVAKS-----GPKKPANVPDHDPPQGYICYRCGEKGHWIQLCPTNDNPDYDN 204
Query: 241 K-RVKQPTGIPRSMLMV-----------------NPQGSYALPNGSVAVLKPNEAAFEKE 282
+ RVK+ TGIP+S L P G +G + +P++A++E +
Sbjct: 205 RPRVKRTTGIPKSFLKTVDKATALGQTGDGDETKTPSGIMVNADGEFVIAEPDKASWE-Q 263
Query: 283 MEGMPSTTRSVGDLPPE---------LHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYII 333
+ + + PE L CP+ + D + T CC K++C+ CI + +I
Sbjct: 264 FQAKAKSNSAAQKATPEGDKEIQERGLECPIDHKLFIDPMKT-PCCEKTYCNDCITNALI 322
Query: 334 SKSMCV--CGAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARCPQP 391
C + +L DDL ++ D I L NS + E ++ P
Sbjct: 323 ESDFICPGCKSDGILIDDLKADEEAVDKIKAFLAEKNSKAK-----------EGSQSP-- 369
Query: 392 KIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSEATH 451
SP S A+K SP +Q T ETK K P D + +
Sbjct: 370 --GSPNSPTAAK-----SPTSTDAPAVQTTTEETK---------PKCKSPTPTDTAPSAS 413
Query: 452 ESKSVKEPVSQGSAQVVEE-EVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENY 510
+ S ++ SA ++ Q PT+A ++++ ++V K P + EN
Sbjct: 414 QQASQQQTSQASSAASMQSGSTGQTTPPTQANREERSQQVSK-------KRPAEHALENP 466
Query: 511 MMQMGPPPGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMP-FPNGMPHDPFAN-- 567
+ P Q M M G M M +G M P P P
Sbjct: 467 KIPKAPKAMQQKTQQTQQQAM---MEQMMGGMPGMPGMNGMNAMNCMPQMFPAMPMMGNF 523
Query: 568 ---GMPHDPFGMQGYMMPPI 584
GMP+ GM +MM P+
Sbjct: 524 GGMGMPNMNMGMMNHMMNPM 543
>ref|XP_414870.1| PREDICTED: similar to retinoblastoma-binding protein 6 isoform 2;
proliferation potential-related protein; RB-binding
Q-protein 1; retinoblastoma-binding protein 6 [Gallus
gallus]
Length = 2028
Score = 140 bits (352), Expect = 2e-31
Identities = 108/338 (31%), Positives = 163/338 (47%), Gaps = 68/338 (20%)
Query: 45 DLVVTNAQTNEEYLDEEMLIPKNTSVLIRRVP-GRPRLPIVTEQEYYLKPLISQKVENKV 103
DL +TNAQT EEY D+ LIPKN+SV++RR+P G + T + Y + P +K+
Sbjct: 691 DLQITNAQTKEEYTDDNALIPKNSSVIVRRIPIGGVK---ATSKTYVMTPK-----SHKI 742
Query: 104 VDTEPANSSLPADDMSAMKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEE 163
V+T + + P S K ++DS SI ++ N+ + + EE
Sbjct: 743 VETPFRSRTEPVSGTS--KAIDDSS--------ASISLAQLTKTANLAE-----ANASEE 787
Query: 164 SKIKALIDTPALDWQHQGSDFGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVP 223
KIKA++ G ++ + +G PP Y C RC P
Sbjct: 788 DKIKAMMS-------QSGHEYDPINYMKKPLG--------------PPPPSYTCFRCGKP 826
Query: 224 GHFIQHCPTNGDPNYD-VKRVKQPTGIPRSML-----------MVNPQGSYALPN---GS 268
GH+I++CPTNGD N++ V R+K+ TGIPRS + M+ G YA+P +
Sbjct: 827 GHYIKNCPTNGDKNFESVPRIKKSTGIPRSFMMEVKDPNTKGAMLTNTGKYAIPTIDAEA 886
Query: 269 VAVLKPNEAAFEKEMEGMPSTTRSVGD-LPPELHCPLCSNVMKDAVLTSKCCFKSFCDKC 327
A+ K + F E PS++ D +P EL C +C ++M DAV+ CC S+CD+C
Sbjct: 887 YAIGKKEKPPFLPE---EPSSSSEEDDPIPDELLCLICKDIMTDAVVI-PCCGNSYCDEC 942
Query: 328 IRDYIISK---SMCVCGAMNVLADDLLPNKTLRDTINR 362
IR ++ + C +V D L+ NK LR + R
Sbjct: 943 IRTALLESEEHTCPTCHQTDVSPDALIANKFLRQPLMR 980
Score = 46.6 bits (109), Expect = 0.004
Identities = 75/312 (24%), Positives = 111/312 (35%), Gaps = 33/312 (10%)
Query: 591 DLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSS 650
D+ N+ PA + A+ + + E E K SS DV
Sbjct: 1712 DIKSTQPSSNIGKPASVVKNVSAKAPNPIKHNEKEMEPLEKIQKTTKEVSYESSQHDVKG 1771
Query: 651 IKS-------KTKPIPPS-SASDYHQNRHRSERPSPDSSHREVEPPRP---TKRKSDHSE 699
KS KTK S S D + R +P D S R E ++ D
Sbjct: 1772 SKSSVLNEKGKTKERDHSLSDKDTSEKRKSGVQPEKDHSERATEQGNGKNISQSSKDSRS 1831
Query: 700 REREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEP-DI 758
E+ D R D +RD D R H SS K D ++ RK S +
Sbjct: 1832 SEKHDTGRGSTAKDFTPNRDKKSDHDSSREH------SSSKRRDEKSELARRKDSPSRNR 1885
Query: 759 KSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKK 818
+S S + S+P + SK S ++ + + DRKQ + S S EE K
Sbjct: 1886 ESTSGQKSKPRDERAEPSKKGAGDSKRSSYSPSRDRKQSDHKTAHDSKRSLEEHKPLDKN 1945
Query: 819 RKLSASSTTEAS-------TAATASVSAKAPSTTHLSNGGRKSKA---VMDDYESSDDDE 868
S T EA+ + + ++VSAK T + +K KA V D +
Sbjct: 1946 -----SETDEAAFVPDYNESDSESNVSAKDEETVGKNCKEQKEKAIDKVKQDTAAPATKS 2000
Query: 869 DDERHFKRRPSR 880
+H K++ +
Sbjct: 2001 QKHKHKKKKSKK 2012
Score = 38.5 bits (88), Expect = 1.0
Identities = 60/277 (21%), Positives = 103/277 (36%), Gaps = 43/277 (15%)
Query: 584 IPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVS 643
+ PP +D +V P + +E K DA RE R+E K ++ +
Sbjct: 1407 VAAPPKKD--------SVTKPTKASQE----KVDA--DREKSPRMEPLVKKAKEELPKTD 1452
Query: 644 SVGDVSSIKS-KTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDH----- 697
SV SS K KT P R P H E P + K K DH
Sbjct: 1453 SVKASSSQKDEKTLGTP------------RKVHPKVAKDHPEARPAKEEKAKKDHPKELK 1500
Query: 698 SEREREDRDRDYDYHDRHQDRDYXHDRHQHR---RHHHRTEASSKKSTDPVT---KSTSR 751
SE+ D+ ++ + D ++ + + + EA+S K++ P T K++ +
Sbjct: 1501 SEKPSSKEDKSKKAVEKSKSSDAKAEKRKRKADEKADKEHEAASVKASKPETAESKTSPK 1560
Query: 752 KSSEPDIKSKSR-----KSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISF 806
+EPD + R KS +T + + T + K+ A S
Sbjct: 1561 GKTEPDGEKGERTPEKDKSGFLITPAKKIKLNRETGKKIVSGENVPPVKEPAEKVEPSSS 1620
Query: 807 PSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPS 843
++E +RK++A+ + ++ S S+ S
Sbjct: 1621 KVKQEKVKGKVRRKVTAADGSSSTLVDYTSTSSTGGS 1657
>gb|AAL68925.1| p53-associated cellular protein PACT [Homo sapiens]
Length = 1625
Score = 118 bits (295), Expect = 1e-24
Identities = 161/656 (24%), Positives = 249/656 (37%), Gaps = 137/656 (20%)
Query: 211 PPQGYVCHRCKVPGHFIQHCPTNGDPNYDV-KRVKQPTGIPRSML-----------MVNP 258
PP Y C RC PGH+I++CPTNGD N++ R+K+ TGIPRS + M+
Sbjct: 22 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 81
Query: 259 QGSYALPN---GSVAVLKPNEAAFEKEMEGMPSTTRSVGD-LPPELHCPLCSNVMKDAVL 314
G YA+P + A+ K + F E PS++ D +P EL C +C ++M DAV+
Sbjct: 82 TGKYAIPTTDAEAYAIGKKEKPPFLPE---EPSSSSEEDDPIPDELLCLICKDIMTDAVV 138
Query: 315 TSKCCFKSFCDKCIRDYIISK---SMCVCGAMNVLADDLLPNKTLRDTINRIL-ESG--- 367
CC S+CD+CIR ++ + C +V D L+ NK LR +N E+G
Sbjct: 139 I-PCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYTK 197
Query: 368 ----------------------------NSSTENAGSTYQVQDMESARCPQPKIPSPTSS 399
S + S+ P P I S TS+
Sbjct: 198 RLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIPVTSSSTHPAPSISSLTSN 257
Query: 400 AAS-------KGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSE---- 448
+S +PV D T + +V ++ P + S++ +P A SE
Sbjct: 258 QSSLAPPVSGNPSSAPAPVPDITATV-SISVHSEKSDGPFRDSDNKILPAAALASEHSKG 316
Query: 449 -------ATHESKSVKEPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKP 501
A E K + PV + + + + +L+PT + R W+
Sbjct: 317 TSSIAITALMEEKGYQVPVLGTPSLLGQSLLHGQLIPTTG--PVRINTARPGGGRPGWEH 374
Query: 502 PHDLG---AENYMMQMGPPPGYNPYWNGMQ-------------PCMDGFMAPYAGPMHMM 545
+ LG + ++ G Y G P F+ P+
Sbjct: 375 SNKLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPATPVFVPVPPPPL--- 431
Query: 546 DYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQGYMMPP--IPPPPHR----------DLA 593
Y P+ +P P G+P F+ P GY +PP PP P A
Sbjct: 432 -YPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSVPPPGFPPAPANLSTPWVSSGVQTA 490
Query: 594 EFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKS 653
+ P +SREEF E+R ++ SK G SS S S
Sbjct: 491 HSNTIPTTQAPPLSREEF-----------YREQRRLKEESKSPYSG---SSYSRSSYTYS 536
Query: 654 KTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHD 713
K++ S S Y ++ RS S S PP P R+ R R R + YH
Sbjct: 537 KSRSGSTRSRS-YSRSFSRSHSRSYSRS-----PPYP--RRGRGKSRNYRSRSRSHGYH- 587
Query: 714 RHQDRDYXHDRHQHRRHHHRTEA-SSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEP 768
R + R +RR+H R+ + + + P ++ + +E + ++ R+ P
Sbjct: 588 RSRSRS-----PPYRRYHSRSRSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPP 638
Score = 37.0 bits (84), Expect = 3.0
Identities = 50/242 (20%), Positives = 82/242 (33%), Gaps = 31/242 (12%)
Query: 609 EEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ 668
E +A + E ++R ERD K + G + D S++ K + P+
Sbjct: 822 EPMDAESITFKSVSEKDKR-ERDKPKAK--GDKTKRKNDGSAVSKKENIVKPAKGPQEKV 878
Query: 669 N--RHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQ 726
+ R RS R P + E P+ KS S ++ E
Sbjct: 879 DGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGT------------------ 920
Query: 727 HRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKS-----KSRKSSEPVTKSLSLSKTAPT 781
R H A + T PV + +K D+KS K K+ +P K+ L
Sbjct: 921 -PRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEK 979
Query: 782 TSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKA 841
+ + +S+ +IS E + KRK+ + T +S A
Sbjct: 980 RKRKTEEKGVDKDFESSSM--KISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSA 1037
Query: 842 PS 843
P+
Sbjct: 1038 PA 1039
>ref|XP_536929.1| PREDICTED: similar to retinoblastoma-binding protein 6 isoform 2
[Canis familiaris]
Length = 1632
Score = 105 bits (263), Expect = 5e-21
Identities = 71/205 (34%), Positives = 101/205 (48%), Gaps = 32/205 (15%)
Query: 211 PPQGYVCHRCKVPGHFIQHCPTNGDPNYDV-KRVKQPTGIPRSML-----------MVNP 258
PP Y C RC PGH+I++CPTNGD N++ R+K+ TGIPRS + M+
Sbjct: 21 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 80
Query: 259 QGSYALPN---GSVAVLKPNEAAFEKEMEGMPSTTRSVGD-LPPELHCPLCSNVMKDAVL 314
G YA+P + A+ K + F E PS++ D +P EL C +C ++M DAV+
Sbjct: 81 TGKYAIPTIDAEAYAIGKKEKPPFLPE---EPSSSSEEDDPIPDELLCLICKDIMTDAVV 137
Query: 315 TSKCCFKSFCDKCIRDYIISK---SMCVCGAMNVLADDLLPNKTLRDTINRILESGNSST 371
CC S+CD+CIR ++ + C +V D L+ NK LR +N N
Sbjct: 138 I-PCCGNSYCDECIRTALLESDDHTCPTCHQNDVSPDALIANKFLRQAVN------NFKN 190
Query: 372 ENAGSTYQVQDMESARCPQPKIPSP 396
E Y + + P P IP P
Sbjct: 191 ETG---YTKRLRKQLPPPPPPIPPP 212
Score = 40.8 bits (94), Expect = 0.21
Identities = 63/293 (21%), Positives = 109/293 (36%), Gaps = 36/293 (12%)
Query: 621 KRENERRVERDFSKDR-----------DFGREVSSVGDVSSIK-SKTKPIPPSSAS---- 664
KR+N + E+D + DR +E + S++ S TK P+
Sbjct: 1258 KRKNSAQPEKDSNLDRLSEQGNFKSLSQSSKETRTSDKHDSVRGSSTKDFTPNRDKKTDY 1317
Query: 665 ---DYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYX 721
+Y ++ R ER + + R+ PPR S + RE+RD +
Sbjct: 1318 DNREYSSSKRRDERN--ELTRRKDSPPRSKDSASGQKTKPREERDLPKKGTGDSKKSSSS 1375
Query: 722 HDRHQHRRHHHRTEASSKKSTD---PVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKT 778
R + + H H+ +K+ ++ PV K+ + + +++++ K S L+
Sbjct: 1376 PSRDK-KPHDHKATYDTKRPSEEIKPVDKNPCKDREKHMLEARNNKESSGNKLLYVLNPP 1434
Query: 779 APTTSAEAAAAA----AADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAAT 834
P E A KQ+ S SR+S E AA + + S +
Sbjct: 1435 DPQIEKEQVPGQTDKNAVKPKQQLSHSSRLSSDLTRETDEAAFEPDYNESD-------SE 1487
Query: 835 ASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPPP 887
++VS K T+ + K K V +S D + R S+ PS P
Sbjct: 1488 SNVSVKEEETSGKVSKELKDKVVEKTKDSLDTAASVQIGTARSQSQSSPSVSP 1540
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.130 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,713,171,971
Number of Sequences: 2540612
Number of extensions: 84060982
Number of successful extensions: 474999
Number of sequences better than 10.0: 7728
Number of HSP's better than 10.0 without gapping: 1146
Number of HSP's successfully gapped in prelim test: 6977
Number of HSP's that attempted gapping in prelim test: 408817
Number of HSP's gapped (non-prelim): 39650
length of query: 908
length of database: 863,360,394
effective HSP length: 137
effective length of query: 771
effective length of database: 515,296,550
effective search space: 397293640050
effective search space used: 397293640050
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC119408.8


