

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149639.1 + phase: 0
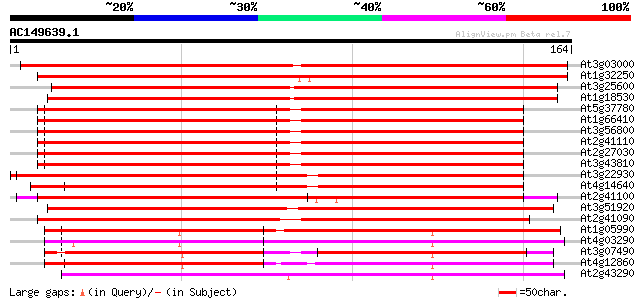
(164 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g03000 putative calmodulin 245 9e-66
At1g32250 calmodulin, putative 235 9e-63
At3g25600 calmodulin, putative 152 8e-38
At1g18530 hypothetical protein 151 2e-37
At5g37780 calmodulin 1 (CAM1) 140 4e-34
At1g66410 calmodulin-4 140 4e-34
At3g56800 calmodulin-3 139 5e-34
At2g41110 calmodulin (cam2) 139 5e-34
At2g27030 calmodulin 139 5e-34
At3g43810 calmodulin 7 138 1e-33
At3g22930 calmodulin, putative 132 6e-32
At4g14640 calmodulin 129 9e-31
At2g41100 calmodulin-like protein 117 3e-27
At3g51920 putative calmodulin 115 1e-26
At2g41090 calcium binding protein (CaBP-22) 109 8e-25
At1g05990 calcium-binding protein, putative 102 7e-23
At4g03290 putative calmodulin 102 9e-23
At3g07490 putative calmodulin 100 6e-22
At4g12860 putative calmodulin 99 8e-22
At2g43290 putative calcium binding protein 98 2e-21
>At3g03000 putative calmodulin
Length = 165
Score = 245 bits (625), Expect = 9e-66
Identities = 123/160 (76%), Positives = 140/160 (86%), Gaps = 2/160 (1%)
Query: 4 KQHQVKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADT 63
K KL DEQ++ELREIFRSFD+N DG+LT+LEL SLLRSLGLKPS +QL+ IQ+AD
Sbjct: 7 KPAPAKLGDEQLAELREIFRSFDQNKDGSLTELELGSLLRSLGLKPSQDQLDTLIQKADR 66
Query: 64 NNNGLIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGH 123
NNNGL+EFSEFVALV P+L+ K PYT++QL+ +FRMFDRDGNG+ITAAELAHSMAKLGH
Sbjct: 67 NNNGLVEFSEFVALVEPDLV--KCPYTDDQLKAIFRMFDRDGNGYITAAELAHSMAKLGH 124
Query: 124 ALTAEELTGMIKEADMDGDGMISFQEFGQAITSAAFDNSW 163
ALTAEELTGMIKEAD DGDG I FQEF QAITSAAFDN+W
Sbjct: 125 ALTAEELTGMIKEADRDGDGCIDFQEFVQAITSAAFDNAW 164
>At1g32250 calmodulin, putative
Length = 166
Score = 235 bits (599), Expect = 9e-63
Identities = 121/158 (76%), Positives = 137/158 (86%), Gaps = 3/158 (1%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
KLD+EQI+ELREIFRSFDRN DG+LTQLEL SLLR+LG+KPS +Q E I +ADT +NGL
Sbjct: 8 KLDEEQINELREIFRSFDRNKDGSLTQLELGSLLRALGVKPSPDQFETLIDKADTKSNGL 67
Query: 69 IEFSEFVALVAPELL-PAK--SPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHAL 125
+EF EFVALV+PELL PAK +PYTEEQL +LFR+FD DGNGFITAAELAHSMAKLGHAL
Sbjct: 68 VEFPEFVALVSPELLSPAKRTTPYTEEQLLRLFRIFDTDGNGFITAAELAHSMAKLGHAL 127
Query: 126 TAEELTGMIKEADMDGDGMISFQEFGQAITSAAFDNSW 163
T ELTGMIKEAD DGDG I+FQEF +AI SAAFD+ W
Sbjct: 128 TVAELTGMIKEADSDGDGRINFQEFAKAINSAAFDDIW 165
>At3g25600 calmodulin, putative
Length = 161
Score = 152 bits (384), Expect = 8e-38
Identities = 77/148 (52%), Positives = 104/148 (70%), Gaps = 1/148 (0%)
Query: 13 EQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFS 72
+QI +L++IF FD + DG+LTQLEL +LLRSLG+KP ++Q+ + + D N NG +EF
Sbjct: 8 DQIKQLKDIFARFDMDKDGSLTQLELAALLRSLGIKPRSDQISLLLNQIDRNGNGSVEFD 67
Query: 73 EFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTG 132
E V + P++ + +EQL ++FR FDRDGNG ITAAELA SMAK+GH LT ELT
Sbjct: 68 ELVVAILPDI-NEEVLINQEQLMEVFRSFDRDGNGSITAAELAGSMAKMGHPLTYRELTE 126
Query: 133 MIKEADMDGDGMISFQEFGQAITSAAFD 160
M+ EAD +GDG+ISF EF + +A D
Sbjct: 127 MMTEADSNGDGVISFNEFSHIMAKSAAD 154
>At1g18530 hypothetical protein
Length = 157
Score = 151 bits (381), Expect = 2e-37
Identities = 78/149 (52%), Positives = 105/149 (70%), Gaps = 1/149 (0%)
Query: 12 DEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEF 71
++QI +L++IF FD + DG+LT LEL +LLRSLGLKPS +Q+ + D+N NG +EF
Sbjct: 2 EDQIRQLKDIFDRFDMDADGSLTILELAALLRSLGLKPSGDQIHVLLASMDSNGNGFVEF 61
Query: 72 SEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELT 131
E V + P+L + EQL ++F+ FDRDGNGFI+AAELA +MAK+G LT +ELT
Sbjct: 62 DELVGTILPDL-NEEVLINSEQLLEIFKSFDRDGNGFISAAELAGAMAKMGQPLTYKELT 120
Query: 132 GMIKEADMDGDGMISFQEFGQAITSAAFD 160
MIKEAD +GDG+ISF EF + +A D
Sbjct: 121 EMIKEADTNGDGVISFGEFASIMAKSAVD 149
>At5g37780 calmodulin 1 (CAM1)
Length = 149
Score = 140 bits (352), Expect = 4e-34
Identities = 69/142 (48%), Positives = 99/142 (69%), Gaps = 3/142 (2%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L DEQISE +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 4 QLTDEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 64 IDFPEFLNLMAKKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 120
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MI+EAD+DGDG I+++EF
Sbjct: 121 EVEEMIREADVDGDGQINYEEF 142
Score = 52.4 bits (124), Expect = 1e-07
Identities = 24/68 (35%), Positives = 43/68 (62%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIE 70
D + EL+E FR FD++ +G ++ EL ++ +LG K + E++E I+ AD + +G I
Sbjct: 79 DTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQIN 138
Query: 71 FSEFVALV 78
+ EFV ++
Sbjct: 139 YEEFVKIM 146
>At1g66410 calmodulin-4
Length = 149
Score = 140 bits (352), Expect = 4e-34
Identities = 69/142 (48%), Positives = 99/142 (69%), Gaps = 3/142 (2%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L DEQISE +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 4 QLTDEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 64 IDFPEFLNLMAKKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 120
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MI+EAD+DGDG I+++EF
Sbjct: 121 EVEEMIREADVDGDGQINYEEF 142
Score = 52.4 bits (124), Expect = 1e-07
Identities = 24/68 (35%), Positives = 43/68 (62%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIE 70
D + EL+E FR FD++ +G ++ EL ++ +LG K + E++E I+ AD + +G I
Sbjct: 79 DTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQIN 138
Query: 71 FSEFVALV 78
+ EFV ++
Sbjct: 139 YEEFVKIM 146
>At3g56800 calmodulin-3
Length = 149
Score = 139 bits (351), Expect = 5e-34
Identities = 69/142 (48%), Positives = 99/142 (69%), Gaps = 3/142 (2%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QISE +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 4 QLTDDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 64 IDFPEFLNLMARKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 120
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MIKEAD+DGDG I+++EF
Sbjct: 121 EVDEMIKEADVDGDGQINYEEF 142
Score = 50.8 bits (120), Expect = 3e-07
Identities = 23/68 (33%), Positives = 43/68 (62%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIE 70
D + EL+E FR FD++ +G ++ EL ++ +LG K + E+++ I+ AD + +G I
Sbjct: 79 DTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQIN 138
Query: 71 FSEFVALV 78
+ EFV ++
Sbjct: 139 YEEFVKVM 146
>At2g41110 calmodulin (cam2)
Length = 149
Score = 139 bits (351), Expect = 5e-34
Identities = 69/142 (48%), Positives = 99/142 (69%), Gaps = 3/142 (2%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QISE +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 4 QLTDDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 64 IDFPEFLNLMARKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 120
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MIKEAD+DGDG I+++EF
Sbjct: 121 EVDEMIKEADVDGDGQINYEEF 142
Score = 50.8 bits (120), Expect = 3e-07
Identities = 23/68 (33%), Positives = 43/68 (62%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIE 70
D + EL+E FR FD++ +G ++ EL ++ +LG K + E+++ I+ AD + +G I
Sbjct: 79 DTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQIN 138
Query: 71 FSEFVALV 78
+ EFV ++
Sbjct: 139 YEEFVKVM 146
>At2g27030 calmodulin
Length = 149
Score = 139 bits (351), Expect = 5e-34
Identities = 69/142 (48%), Positives = 99/142 (69%), Gaps = 3/142 (2%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QISE +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 4 QLTDDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 64 IDFPEFLNLMARKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 120
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MIKEAD+DGDG I+++EF
Sbjct: 121 EVDEMIKEADVDGDGQINYEEF 142
Score = 50.8 bits (120), Expect = 3e-07
Identities = 23/68 (33%), Positives = 43/68 (62%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIE 70
D + EL+E FR FD++ +G ++ EL ++ +LG K + E+++ I+ AD + +G I
Sbjct: 79 DTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQIN 138
Query: 71 FSEFVALV 78
+ EFV ++
Sbjct: 139 YEEFVKVM 146
>At3g43810 calmodulin 7
Length = 149
Score = 138 bits (348), Expect = 1e-33
Identities = 68/142 (47%), Positives = 99/142 (68%), Gaps = 3/142 (2%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QISE +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 4 QLTDDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 64 IDFPEFLNLMARKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 120
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MI+EAD+DGDG I+++EF
Sbjct: 121 EVDEMIREADVDGDGQINYEEF 142
Score = 50.8 bits (120), Expect = 3e-07
Identities = 23/68 (33%), Positives = 43/68 (62%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIE 70
D + EL+E FR FD++ +G ++ EL ++ +LG K + E+++ I+ AD + +G I
Sbjct: 79 DTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQIN 138
Query: 71 FSEFVALV 78
+ EFV ++
Sbjct: 139 YEEFVKVM 146
>At3g22930 calmodulin, putative
Length = 173
Score = 132 bits (333), Expect = 6e-32
Identities = 66/148 (44%), Positives = 101/148 (67%), Gaps = 3/148 (2%)
Query: 3 KKQHQVKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRAD 62
++Q Q +L EQI E +E F FD++ DG +T EL +++RSL P+ ++L+ I D
Sbjct: 21 QQQQQQELTQEQIMEFKEAFCLFDKDGDGCITADELATVIRSLDQNPTEQELQDMITEID 80
Query: 63 TNNNGLIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG 122
++ NG IEFSEF+ L+A +L + +E+L++ F++FD+D NG+I+A+EL H M LG
Sbjct: 81 SDGNGTIEFSEFLNLMANQLQETDA---DEELKEAFKVFDKDQNGYISASELRHVMINLG 137
Query: 123 HALTAEELTGMIKEADMDGDGMISFQEF 150
LT EE+ MIKEAD+DGDG +++ EF
Sbjct: 138 EKLTDEEVDQMIKEADLDGDGQVNYDEF 165
Score = 47.0 bits (110), Expect = 5e-06
Identities = 23/78 (29%), Positives = 48/78 (61%), Gaps = 1/78 (1%)
Query: 1 MNKKQHQVKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQR 60
+N +Q++ D EL+E F+ FD++ +G ++ EL ++ +LG K + E+++ I+
Sbjct: 93 LNLMANQLQETDAD-EELKEAFKVFDKDQNGYISASELRHVMINLGEKLTDEEVDQMIKE 151
Query: 61 ADTNNNGLIEFSEFVALV 78
AD + +G + + EFV ++
Sbjct: 152 ADLDGDGQVNYDEFVRMM 169
>At4g14640 calmodulin
Length = 151
Score = 129 bits (323), Expect = 9e-31
Identities = 63/144 (43%), Positives = 100/144 (68%), Gaps = 3/144 (2%)
Query: 7 QVKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNN 66
+ L +QI+E +E F FD++ DG +T EL +++RSL P+ ++L I D+++N
Sbjct: 3 ETALTKDQITEFKEAFCLFDKDGDGCITVEELATVIRSLDQNPTEQELHDIITEIDSDSN 62
Query: 67 GLIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALT 126
G IEF+EF+ L+A +L + + EE+L++ F++FD+D NG+I+A+EL+H M LG LT
Sbjct: 63 GTIEFAEFLNLMAKKLQESDA---EEELKEAFKVFDKDQNGYISASELSHVMINLGEKLT 119
Query: 127 AEELTGMIKEADMDGDGMISFQEF 150
EE+ MIKEAD+DGDG +++ EF
Sbjct: 120 DEEVEQMIKEADLDGDGQVNYDEF 143
Score = 48.1 bits (113), Expect = 2e-06
Identities = 21/62 (33%), Positives = 42/62 (66%)
Query: 17 ELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFVA 76
EL+E F+ FD++ +G ++ EL+ ++ +LG K + E++E I+ AD + +G + + EFV
Sbjct: 86 ELKEAFKVFDKDQNGYISASELSHVMINLGEKLTDEEVEQMIKEADLDGDGQVNYDEFVK 145
Query: 77 LV 78
++
Sbjct: 146 MM 147
>At2g41100 calmodulin-like protein
Length = 324
Score = 117 bits (293), Expect = 3e-27
Identities = 68/172 (39%), Positives = 98/172 (56%), Gaps = 14/172 (8%)
Query: 3 KKQHQVKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRAD 62
KK KL D+QI+E RE FR FD+N DG++T+ EL +++ SLG + L+ + D
Sbjct: 87 KKTMADKLTDDQITEYRESFRLFDKNGDGSITKKELRTVMFSLGKNRTKADLQDMMNEVD 146
Query: 63 TNNNGLIEFSEFVALVAPELLPAKSP-----------YTEEQL---RQLFRMFDRDGNGF 108
+ +G I+F EF+ L+A ++P T++Q+ R+ FR+FD++G+G+
Sbjct: 147 LDGDGTIDFPEFLYLMAKNQGHDQAPRHTKKTMVDYQLTDDQILEFREAFRVFDKNGDGY 206
Query: 109 ITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEFGQAITSAAFD 160
IT EL +M LG T EL MI EAD DGDG ISF EF +T D
Sbjct: 207 ITVNELRTTMRSLGETQTKAELQDMINEADADGDGTISFSEFVCVMTGKMID 258
Score = 114 bits (286), Expect = 2e-26
Identities = 62/155 (40%), Positives = 94/155 (60%), Gaps = 13/155 (8%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
KL D+QI+E RE FR FD+N DG++T+ EL +++RS+G KP+ L+ + AD + +G
Sbjct: 4 KLTDDQITEYRESFRLFDKNGDGSITKKELGTMMRSIGEKPTKADLQDLMNEADLDGDGT 63
Query: 69 IEFSEFVALVAPELLPAKSP----------YTEEQL---RQLFRMFDRDGNGFITAAELA 115
I+F EF+ ++A ++P T++Q+ R+ FR+FD++G+G IT EL
Sbjct: 64 IDFPEFLCVMAKNQGHDQAPRHTKKTMADKLTDDQITEYRESFRLFDKNGDGSITKKELR 123
Query: 116 HSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
M LG T +L M+ E D+DGDG I F EF
Sbjct: 124 TVMFSLGKNRTKADLQDMMNEVDLDGDGTIDFPEF 158
Score = 67.8 bits (164), Expect = 3e-12
Identities = 33/79 (41%), Positives = 50/79 (62%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QI E RE FR FD+N DG +T EL + +RSLG + +L+ I AD + +G
Sbjct: 183 QLTDDQILEFREAFRVFDKNGDGYITVNELRTTMRSLGETQTKAELQDMINEADADGDGT 242
Query: 69 IEFSEFVALVAPELLPAKS 87
I FSEFV ++ +++ +S
Sbjct: 243 ISFSEFVCVMTGKMIDTQS 261
>At3g51920 putative calmodulin
Length = 151
Score = 115 bits (288), Expect = 1e-26
Identities = 57/148 (38%), Positives = 91/148 (60%), Gaps = 3/148 (2%)
Query: 12 DEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEF 71
DEQI E E F D+++DG +T+ +L +++S+G P AEQL+ + D NG I F
Sbjct: 7 DEQIQEFYEAFCLIDKDSDGFITKEKLTKVMKSMGKNPKAEQLQQMMSDVDIFGNGGITF 66
Query: 72 SEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELT 131
+F+ ++A ++L ++FR+FDRDG+G I+ EL M +G +TAEE
Sbjct: 67 DDFLYIMAQN---TSQESASDELIEVFRVFDRDGDGLISQLELGEGMKDMGMKITAEEAE 123
Query: 132 GMIKEADMDGDGMISFQEFGQAITSAAF 159
M++EAD+DGDG +SF EF + + +A++
Sbjct: 124 HMVREADLDGDGFLSFHEFSKMMIAASY 151
>At2g41090 calcium binding protein (CaBP-22)
Length = 191
Score = 109 bits (272), Expect = 8e-25
Identities = 59/144 (40%), Positives = 90/144 (61%), Gaps = 6/144 (4%)
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
K +QISE RE F +D+N DG +T E +++RSLGL + +L+ I +D + +G
Sbjct: 4 KFTRQQISEFREQFSVYDKNGDGHITTEEFGAVMRSLGLNLTQAELQEEINDSDLDGDGT 63
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I F+EF+ +A K Y+E+ L++ FR+FD D NGFI+AAE+ + L T E
Sbjct: 64 INFTEFLCAMA------KDTYSEKDLKKDFRLFDIDKNGFISAAEMRYVRTILRWKQTDE 117
Query: 129 ELTGMIKEADMDGDGMISFQEFGQ 152
E+ +IK AD+DGDG I+++EF +
Sbjct: 118 EIDEIIKAADVDGDGQINYREFAR 141
>At1g05990 calcium-binding protein, putative
Length = 150
Score = 102 bits (255), Expect = 7e-23
Identities = 52/148 (35%), Positives = 90/148 (60%), Gaps = 4/148 (2%)
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFV 75
+EL+ +F+ FD+N DGT+T EL+ LRSLG+ ++L I++ D N +G ++ EF
Sbjct: 4 TELKRVFQMFDKNGDGTITGKELSETLRSLGIYIPDKELTQMIEKIDVNGDGCVDIDEFG 63
Query: 76 ALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG--HALTAEELTGM 133
L + + + EE +++ F +FD++G+GFIT EL ++ LG T ++ M
Sbjct: 64 ELY--KTIMDEEDEEEEDMKEAFNVFDQNGDGFITVDELKAVLSSLGLKQGKTLDDCKKM 121
Query: 134 IKEADMDGDGMISFQEFGQAITSAAFDN 161
IK+ D+DGDG ++++EF Q + F++
Sbjct: 122 IKKVDVDGDGRVNYKEFRQMMKGGGFNS 149
Score = 48.9 bits (115), Expect = 1e-06
Identities = 23/66 (34%), Positives = 41/66 (61%), Gaps = 2/66 (3%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLK--PSAEQLEGFIQRADTNNNGL 68
+DE+ +++E F FD+N DG +T EL ++L SLGLK + + + I++ D + +G
Sbjct: 73 EDEEEEDMKEAFNVFDQNGDGFITVDELKAVLSSLGLKQGKTLDDCKKMIKKVDVDGDGR 132
Query: 69 IEFSEF 74
+ + EF
Sbjct: 133 VNYKEF 138
>At4g03290 putative calmodulin
Length = 154
Score = 102 bits (254), Expect = 9e-23
Identities = 52/149 (34%), Positives = 85/149 (56%), Gaps = 2/149 (1%)
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFV 75
+EL +F+ FD++ DG +T ELN ++LG+ ++L IQ+ D N +G ++ EF
Sbjct: 4 TELNRVFQMFDKDGDGKITTKELNESFKNLGIIIPEDELTQIIQKIDVNGDGCVDIEEFG 63
Query: 76 ALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG--HALTAEELTGM 133
L ++ + EE +++ F +FDR+G+GFIT EL ++ LG T EE M
Sbjct: 64 ELYKTIMVEDEDEVGEEDMKEAFNVFDRNGDGFITVDELKAVLSSLGLKQGKTLEECRKM 123
Query: 134 IKEADMDGDGMISFQEFGQAITSAAFDNS 162
I + D+DGDG +++ EF Q + F +S
Sbjct: 124 IMQVDVDGDGRVNYMEFRQMMKKGRFFSS 152
Score = 48.1 bits (113), Expect = 2e-06
Identities = 25/68 (36%), Positives = 41/68 (59%), Gaps = 4/68 (5%)
Query: 11 DDEQISE--LREIFRSFDRNNDGTLTQLELNSLLRSLGLK--PSAEQLEGFIQRADTNNN 66
D++++ E ++E F FDRN DG +T EL ++L SLGLK + E+ I + D + +
Sbjct: 73 DEDEVGEEDMKEAFNVFDRNGDGFITVDELKAVLSSLGLKQGKTLEECRKMIMQVDVDGD 132
Query: 67 GLIEFSEF 74
G + + EF
Sbjct: 133 GRVNYMEF 140
>At3g07490 putative calmodulin
Length = 153
Score = 99.8 bits (247), Expect = 6e-22
Identities = 55/146 (37%), Positives = 83/146 (56%), Gaps = 5/146 (3%)
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFV 75
+EL IF+ FDRN DG +T+ ELN L +LG+ + L I++ D N +G ++ EF
Sbjct: 4 AELARIFQMFDRNGDGKITKQELNDSLENLGIYIPDKDLVQMIEKIDLNGDGYVDIEEFG 63
Query: 76 ALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG--HALTAEELTGM 133
L + + EE +R+ F +FD++ +GFIT EL +A LG T E+ M
Sbjct: 64 GLYQTIM---EERDEEEDMREAFNVFDQNRDGFITVEELRSVLASLGLKQGRTLEDCKRM 120
Query: 134 IKEADMDGDGMISFQEFGQAITSAAF 159
I + D+DGDGM++F+EF Q + F
Sbjct: 121 ISKVDVDGDGMVNFKEFKQMMKGGGF 146
Score = 56.2 bits (134), Expect = 8e-09
Identities = 25/61 (40%), Positives = 42/61 (67%)
Query: 91 EEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
+ +L ++F+MFDR+G+G IT EL S+ LG + ++L MI++ D++GDG + +EF
Sbjct: 3 QAELARIFQMFDRNGDGKITKQELNDSLENLGIYIPDKDLVQMIEKIDLNGDGYVDIEEF 62
Query: 151 G 151
G
Sbjct: 63 G 63
Score = 51.2 bits (121), Expect = 2e-07
Identities = 27/66 (40%), Positives = 41/66 (61%), Gaps = 4/66 (6%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKP--SAEQLEGFIQRADTNNNGL 68
D+E+ ++RE F FD+N DG +T EL S+L SLGLK + E + I + D + +G+
Sbjct: 74 DEEE--DMREAFNVFDQNRDGFITVEELRSVLASLGLKQGRTLEDCKRMISKVDVDGDGM 131
Query: 69 IEFSEF 74
+ F EF
Sbjct: 132 VNFKEF 137
>At4g12860 putative calmodulin
Length = 152
Score = 99.4 bits (246), Expect = 8e-22
Identities = 52/145 (35%), Positives = 84/145 (57%), Gaps = 5/145 (3%)
Query: 17 ELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFVA 76
EL +F+ FD+N DG + + EL +S+G+ ++ I + D N +G ++ EF +
Sbjct: 5 ELSRVFQMFDKNGDGKIAKNELKDFFKSVGIMVPENEINEMIAKMDVNGDGAMDIDEFGS 64
Query: 77 LVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG--HALTAEELTGMI 134
L E++ K EE +R+ FR+FD++G+GFIT EL +A +G T E+ MI
Sbjct: 65 LY-QEMVEEKE--EEEDMREAFRVFDQNGDGFITDEELRSVLASMGLKQGRTLEDCKKMI 121
Query: 135 KEADMDGDGMISFQEFGQAITSAAF 159
+ D+DGDGM++F+EF Q + F
Sbjct: 122 SKVDVDGDGMVNFKEFKQMMRGGGF 146
Score = 54.3 bits (129), Expect = 3e-08
Identities = 26/66 (39%), Positives = 41/66 (61%), Gaps = 2/66 (3%)
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKP--SAEQLEGFIQRADTNNNGL 68
+ E+ ++RE FR FD+N DG +T EL S+L S+GLK + E + I + D + +G+
Sbjct: 72 EKEEEEDMREAFRVFDQNGDGFITDEELRSVLASMGLKQGRTLEDCKKMISKVDVDGDGM 131
Query: 69 IEFSEF 74
+ F EF
Sbjct: 132 VNFKEF 137
>At2g43290 putative calcium binding protein
Length = 215
Score = 97.8 bits (242), Expect = 2e-21
Identities = 54/151 (35%), Positives = 83/151 (54%), Gaps = 4/151 (2%)
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFV 75
SEL+ +F+ FD+N DG +T+ ELN L +LG+ + L I + D N +G ++ EF
Sbjct: 64 SELKRVFQMFDKNGDGRITKEELNDSLENLGIYIPDKDLTQMIHKIDANGDGCVDIDEFE 123
Query: 76 ALVAP--ELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG--HALTAEELT 131
+L + + EE ++ F +FD+DG+GFIT EL MA LG T +
Sbjct: 124 SLYSSIVDEHHNDGETEEEDMKDAFNVFDQDGDGFITVEELKSVMASLGLKQGKTLDGCK 183
Query: 132 GMIKEADMDGDGMISFQEFGQAITSAAFDNS 162
MI + D DGDG ++++EF Q + F +S
Sbjct: 184 KMIMQVDADGDGRVNYKEFLQMMKGGGFSSS 214
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,515,757
Number of Sequences: 26719
Number of extensions: 137851
Number of successful extensions: 1101
Number of sequences better than 10.0: 157
Number of HSP's better than 10.0 without gapping: 121
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 459
Number of HSP's gapped (non-prelim): 358
length of query: 164
length of database: 11,318,596
effective HSP length: 92
effective length of query: 72
effective length of database: 8,860,448
effective search space: 637952256
effective search space used: 637952256
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149639.1


