

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149580.1 + phase: 0 /pseudo
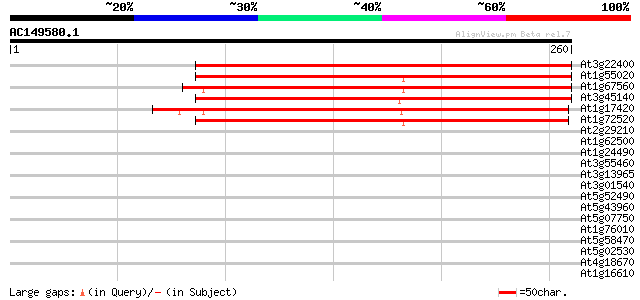
(260 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g22400 putative lipoxygenase 241 4e-64
At1g55020 unknown protein 230 7e-61
At1g67560 putative lipoxygenase 205 2e-53
At3g45140 lipoxygenase (AtLox2) 204 3e-53
At1g17420 lipoxygenase 199 1e-51
At1g72520 putative lipoxygenase 196 1e-50
At2g29210 proline-rich protein like 39 0.003
At1g62500 putative proline-rich cell wall protein (pir|IS52985);... 39 0.003
At1g24490 hypothetical protein 35 0.040
At3g55460 RNA binding protein like 34 0.069
At3g13965 unknown protein 33 0.12
At3g01540 AT3g01540/F4P13_9 33 0.12
At5g52490 fibrillarin-like 33 0.20
At5g43960 unknown protein 33 0.20
At5g07750 putative protein 33 0.20
At1g76010 unknown protein 33 0.20
At5g58470 RNA/ssDNA-binding protein - like 32 0.26
At5g02530 unknown protein 32 0.26
At4g18670 extensin-like protein 32 0.26
At1g16610 arginine/serine-rich protein 32 0.26
>At3g22400 putative lipoxygenase
Length = 864
Score = 241 bits (614), Expect = 4e-64
Identities = 108/174 (62%), Positives = 141/174 (80%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAYA VNDS HQL+SHWL+THAV+EPF+IA+NR LSV+HPIHKLL PH+R TM I
Sbjct: 500 WQLAKAYAAVNDSGYHQLISHWLQTHAVIEPFIIASNRQLSVVHPIHKLLHPHFRDTMNI 559
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R++LIN+ G++E T +Y MEMS+ +YK+WVF E+ LP DL RG+AV+D +S
Sbjct: 560 NALARHVLINSDGVLERTVFPSRYAMEMSSSIYKNWVFTEQALPKDLLKRGVAVEDPNSD 619
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+G++LLIEDYP+A DGLEIW+AIK+WV EY FYY +DK V+ D E++++W EL
Sbjct: 620 NGVKLLIEDYPFAVDGLEIWSAIKTWVTEYCTFYYNNDKTVQTDTEIQSWWTEL 673
>At1g55020 unknown protein
Length = 859
Score = 230 bits (586), Expect = 7e-61
Identities = 106/175 (60%), Positives = 136/175 (77%), Gaps = 1/175 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKA+ VNDS HQL+SHW++THA +EPFVIATNR LSVLHP+ KLL PH+R TM I
Sbjct: 499 WQLAKAFVGVNDSGNHQLISHWMQTHASIEPFVIATNRQLSVLHPVFKLLEPHFRDTMNI 558
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD-WVFAEEGLPTDLKNRGMAVDDDSS 205
NA +R ILIN GGI E T KY MEMS+ +YK+ W F ++ LP +LK RGMAV+D +
Sbjct: 559 NALARQILINGGGIFEITVFPSKYAMEMSSFIYKNHWTFPDQALPAELKKRGMAVEDPEA 618
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
PHGLRL I+DYPYA DGLE+W AI+SWV +Y+ +Y+ ++D++ D EL+A+WKE+
Sbjct: 619 PHGLRLRIKDYPYAVDGLEVWYAIESWVRDYIFLFYKIEEDIQTDTELQAWWKEV 673
>At1g67560 putative lipoxygenase
Length = 917
Score = 205 bits (522), Expect = 2e-53
Identities = 100/183 (54%), Positives = 129/183 (69%), Gaps = 3/183 (1%)
Query: 81 HAGTFNWL--LAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVP 138
H T +W+ LAKA+ ND+ HQLV+HWL+THA +EP++IATNR LS +HP++KLL P
Sbjct: 547 HDATTHWIWKLAKAHVCSNDAGVHQLVNHWLRTHASMEPYIIATNRQLSTMHPVYKLLHP 606
Query: 139 HYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD-WVFAEEGLPTDLKNRG 197
H R T+ INAR+R LIN GGI+ES F KY ME+S+ YK W F EGLP DL RG
Sbjct: 607 HMRYTLEINARARKSLINGGGIIESCFTPGKYAMELSSAAYKSMWRFDMEGLPADLVRRG 666
Query: 198 MAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFW 257
MA +D S+ G+RL+I+DYPYA+DGL IW AIK V+ YV +Y K + D EL+A+W
Sbjct: 667 MAEEDSSAECGVRLVIDDYPYAADGLLIWKAIKDLVESYVKHFYSDSKSITSDLELQAWW 726
Query: 258 KEL 260
E+
Sbjct: 727 DEI 729
>At3g45140 lipoxygenase (AtLox2)
Length = 896
Score = 204 bits (520), Expect = 3e-53
Identities = 93/175 (53%), Positives = 128/175 (73%), Gaps = 1/175 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAK +A+ +D+ HQL+SHWL+THA EP++IA NR LS +HPI++LL PH+R TM I
Sbjct: 534 WNLAKTHAISHDAGYHQLISHWLRTHACTEPYIIAANRQLSAMHPIYRLLHPHFRYTMEI 593
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNRGMAVDDDSS 205
NAR+R L+N GGI+E+ F KY +E+S+ VY K W F +EGLP DL RG+A +D ++
Sbjct: 594 NARARQSLVNGGGIIETCFWPGKYALELSSAVYGKLWRFDQEGLPADLIKRGLAEEDKTA 653
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL I DYP+A+DGL +W AIK WV +YV YY ++ + DEEL+ +W E+
Sbjct: 654 EHGVRLTIPDYPFANDGLILWDAIKEWVTDYVKHYYPDEELITSDEELQGWWSEV 708
>At1g17420 lipoxygenase
Length = 919
Score = 199 bits (506), Expect = 1e-51
Identities = 103/201 (51%), Positives = 133/201 (65%), Gaps = 8/201 (3%)
Query: 67 LPYPPPGPPNS-----TAYHAGTFNWL--LAKAYAVVNDSCCHQLVSHWLKTHAVVEPFV 119
L PP GP + T T NW+ LAKA+ ND+ HQLV+HWL+THA +EPF+
Sbjct: 531 LSLPPHGPKHRSKRVLTPPVDATSNWMWQLAKAHVSSNDAGVHQLVNHWLRTHACLEPFI 590
Query: 120 IATNRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVY 179
+A +R LS +HPI KLL PH R T+ INA +R LI+A G++E F Y MEMSA Y
Sbjct: 591 LAAHRQLSAMHPIFKLLDPHMRYTLEINALARQSLISADGVIEGGFTAGAYGMEMSAAAY 650
Query: 180 K-DWVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVN 238
K W F EGLP DL RGMA+ D + PHGL+LLIEDYPYA+DGL +W+AI++WV YV
Sbjct: 651 KSSWRFDMEGLPADLIRRGMAIPDATQPHGLKLLIEDYPYANDGLLLWSAIQTWVRTYVE 710
Query: 239 FYYESDKDVKDDEELKAFWKE 259
YY + +K D EL++++ E
Sbjct: 711 RYYPNPNLIKTDSELQSWYSE 731
>At1g72520 putative lipoxygenase
Length = 926
Score = 196 bits (497), Expect = 1e-50
Identities = 93/174 (53%), Positives = 126/174 (71%), Gaps = 1/174 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKA+ ND+ HQLV+HWL+THA +EPF++A +R LS +HPI KLL PH R T+ I
Sbjct: 565 WQLAKAHVGSNDAGVHQLVNHWLRTHACLEPFILAAHRQLSAMHPIFKLLDPHMRYTLEI 624
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD-WVFAEEGLPTDLKNRGMAVDDDSS 205
NA +R LI+A G++ES F +Y +E+S+ YK+ W F EGLP DL RGMAV D +
Sbjct: 625 NAVARQTLISADGVIESCFTAGQYGLEISSAAYKNKWRFDMEGLPADLIRRGMAVPDPTQ 684
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
PHGL+LL+EDYPYA+DGL +W+AI++WV YV YY + ++ D EL+A++ E
Sbjct: 685 PHGLKLLVEDYPYANDGLLLWSAIQTWVRTYVERYYANSNLIQTDTELQAWYSE 738
>At2g29210 proline-rich protein like
Length = 878
Score = 38.9 bits (89), Expect = 0.003
Identities = 24/69 (34%), Positives = 29/69 (41%), Gaps = 1/69 (1%)
Query: 7 RSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNPT 66
R R+P+P R PSPP R R PAR PA + AR+H R+ P
Sbjct: 308 RRRRSPSPPARRRRSPSPPARRRRSPSPPARRHRSPTPPARQRRSPSPPARRH-RSPPPA 366
Query: 67 LPYPPPGPP 75
P PP
Sbjct: 367 RRRRSPSPP 375
Score = 33.9 bits (76), Expect = 0.090
Identities = 20/59 (33%), Positives = 26/59 (43%), Gaps = 1/59 (1%)
Query: 7 RSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNP 65
R R+P+P R PSPP R R P +R P +NR S K R+ +P
Sbjct: 367 RRRRSPSPPARRRRSPSPPARRRRSPS-PLYRRNRSPSPLYRRNRSRSPLAKRGRSDSP 424
Score = 32.0 bits (71), Expect = 0.34
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Query: 7 RSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNP 65
R R+P P R S PSPP R R PAR R P +NR S + R+ +P
Sbjct: 358 RRHRSPPPARRRRS-PSPPARRRRSPSPPAR-RRRSPSPLYRRNRSPSPLYRRNRSRSP 414
Score = 29.6 bits (65), Expect = 1.7
Identities = 21/69 (30%), Positives = 25/69 (35%), Gaps = 2/69 (2%)
Query: 7 RSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNPT 66
R R P +GR +S P R R PAR PA + AR+H P
Sbjct: 290 RRHRRPTHEGRRQS--PAPSRRRRSPSPPARRRRSPSPPARRRRSPSPPARRHRSPTPPA 347
Query: 67 LPYPPPGPP 75
P PP
Sbjct: 348 RQRRSPSPP 356
Score = 28.9 bits (63), Expect = 2.9
Identities = 23/74 (31%), Positives = 31/74 (41%), Gaps = 11/74 (14%)
Query: 8 SERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNPTL 67
++R P+P R PSPPP R P+ R G P+ S A H + +P+
Sbjct: 457 AQRLPSPPPRRAGLPSPPPAQR----LPSPPPRRAGLPSP-MRIGGSHAANHLESPSPSS 511
Query: 68 PYPP------PGPP 75
PP P PP
Sbjct: 512 LSPPGRKKVLPSPP 525
>At1g62500 putative proline-rich cell wall protein (pir|IS52985);
similar to ESTs gb|AI239404, gb|R89984, and emb|Z17709
Length = 297
Score = 38.9 bits (89), Expect = 0.003
Identities = 28/76 (36%), Positives = 32/76 (41%), Gaps = 10/76 (13%)
Query: 2 GGQGERSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESR-----A 56
GG G S+ P P G+ G PPP G GGG P G GG GK+ R
Sbjct: 45 GGGGGGSK--PPPHHGGKGGGKPPPHGGKGGGPPHHG---GGGGGGGKSPPVVRPPPVVV 99
Query: 57 RKHTRTHNPTLPYPPP 72
R P + YPPP
Sbjct: 100 RPPPIIRPPPVVYPPP 115
Score = 32.0 bits (71), Expect = 0.34
Identities = 21/64 (32%), Positives = 23/64 (35%), Gaps = 3/64 (4%)
Query: 12 PAPDGRGESGPSPPP---RGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNPTLP 68
P G G G S PP G+GGG P G GG P G P +
Sbjct: 40 PPQHGGGGGGGSKPPPHHGGKGGGKPPPHGGKGGGPPHHGGGGGGGGKSPPVVRPPPVVV 99
Query: 69 YPPP 72
PPP
Sbjct: 100 RPPP 103
>At1g24490 hypothetical protein
Length = 593
Score = 35.0 bits (79), Expect = 0.040
Identities = 22/65 (33%), Positives = 29/65 (43%), Gaps = 13/65 (20%)
Query: 2 GGQGERSERT-------PAPDGRGESGPSP------PPRGRGGGGRPARGDDRGGDPADG 48
GG G + T P+P G S PS PP GGGG P++G G + G
Sbjct: 389 GGSGNGTNSTSTSGGGSPSPGGGSGSPPSTGGGSGSPPSTGGGGGSPSKGGGGGKSDSSG 448
Query: 49 KNRDE 53
K+ +E
Sbjct: 449 KSGEE 453
>At3g55460 RNA binding protein like
Length = 262
Score = 34.3 bits (77), Expect = 0.069
Identities = 17/30 (56%), Positives = 17/30 (56%), Gaps = 3/30 (10%)
Query: 16 GRGESGPSPPPR---GRGGGGRPARGDDRG 42
GRG S P PPPR G GGGG RG G
Sbjct: 18 GRGRSPPPPPPRRGYGGGGGGGGRRGSSHG 47
>At3g13965 unknown protein
Length = 209
Score = 33.5 bits (75), Expect = 0.12
Identities = 22/70 (31%), Positives = 27/70 (38%), Gaps = 15/70 (21%)
Query: 11 TPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNPTLPYP 70
TP+P S P PPPR + P NR+ R + R P P P
Sbjct: 108 TPSPPPPPISKPPPPPRAQAS-------------PPHSNNRESHRNQPPPRRLRP--PSP 152
Query: 71 PPGPPNSTAY 80
PP PP S +
Sbjct: 153 PPPPPPSRVF 162
>At3g01540 AT3g01540/F4P13_9
Length = 619
Score = 33.5 bits (75), Expect = 0.12
Identities = 25/76 (32%), Positives = 31/76 (39%), Gaps = 7/76 (9%)
Query: 2 GGQGERSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTR 61
GG + S P GRG G S G GG G A D R ++G R+ R+R R
Sbjct: 541 GGMNKFSRWGPPSGGRGRGGDS----GYGGRGSFASRDSRS---SNGWGRERERSRSPER 593
Query: 62 THNPTLPYPPPGPPNS 77
+ P PP S
Sbjct: 594 FNRAPPPSSTGSPPRS 609
>At5g52490 fibrillarin-like
Length = 292
Score = 32.7 bits (73), Expect = 0.20
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Query: 24 PPPRGRGGGGRPARGDDRGGDPA----DGKNRDESRAR 57
PP RGRGGG R RG RGG+ + G+ + R R
Sbjct: 3 PPQRGRGGGVRGGRGLARGGEGSAVRGSGRGGESGRGR 40
>At5g43960 unknown protein
Length = 450
Score = 32.7 bits (73), Expect = 0.20
Identities = 22/53 (41%), Positives = 24/53 (44%), Gaps = 10/53 (18%)
Query: 2 GGQGERSERTPAP---------DGRGESG-PSPPPRGRGGGGRPARGDDRGGD 44
G Q ER P P GRG G P+ PRGR GG RG+ GGD
Sbjct: 384 GRQVYIEERRPNPAGVRGARRGGGRGRGGYPTEAPRGRFGGRGSGRGNQDGGD 436
>At5g07750 putative protein
Length = 1289
Score = 32.7 bits (73), Expect = 0.20
Identities = 23/60 (38%), Positives = 25/60 (41%), Gaps = 10/60 (16%)
Query: 2 GGQGERSERTPAPDGRGESGPSPPPRGRGGGGRP-------ARG---DDRGGDPADGKNR 51
GG+G + P P G GP PPP R GG P ARG D RG G R
Sbjct: 1141 GGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGGGPPPPPMLGARGAAVDPRGAGRGRGLPR 1200
Score = 32.3 bits (72), Expect = 0.26
Identities = 16/34 (47%), Positives = 17/34 (49%)
Query: 12 PAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDP 45
P P RG + P PPP RGG P RGG P
Sbjct: 1077 PPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAP 1110
Score = 31.6 bits (70), Expect = 0.45
Identities = 23/68 (33%), Positives = 24/68 (34%), Gaps = 10/68 (14%)
Query: 12 PAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNPTLPYPP 71
P P RG + P PPP GG P RGG P P P PP
Sbjct: 1101 PPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGGAPP---------PPPPPGGRGPGAP-PP 1150
Query: 72 PGPPNSTA 79
P PP A
Sbjct: 1151 PPPPGGRA 1158
Score = 29.3 bits (64), Expect = 2.2
Identities = 15/34 (44%), Positives = 16/34 (46%)
Query: 12 PAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDP 45
P P G + P PPP RGG P RGG P
Sbjct: 1065 PPPPMFGGAQPPPPPPMRGGAPPPPPPPMRGGAP 1098
Score = 28.9 bits (63), Expect = 2.9
Identities = 17/37 (45%), Positives = 17/37 (45%), Gaps = 3/37 (8%)
Query: 12 PAPDGRGESGPSPPPR--GRGGGGRPARGD-DRGGDP 45
P P GRG P PPP GR G P G GG P
Sbjct: 1138 PPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGGGP 1174
Score = 27.3 bits (59), Expect = 8.4
Identities = 14/34 (41%), Positives = 15/34 (43%)
Query: 12 PAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDP 45
P P G + P PPP GG P RGG P
Sbjct: 1053 PPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAP 1086
>At1g76010 unknown protein
Length = 350
Score = 32.7 bits (73), Expect = 0.20
Identities = 27/65 (41%), Positives = 31/65 (47%), Gaps = 12/65 (18%)
Query: 2 GGQGERSERTPAPDGR-GESGPSP-------PPRGRGGGGRPARGDDRGG-DPADGKNRD 52
G QG R P P GR G GPS P +GRGG P++G RGG D G+ R
Sbjct: 261 GPQGRRGYDGP-PQGRGGYDGPSQGRGGYDGPSQGRGGYDGPSQG--RGGYDGPQGRGRG 317
Query: 53 ESRAR 57
R R
Sbjct: 318 RGRGR 322
>At5g58470 RNA/ssDNA-binding protein - like
Length = 422
Score = 32.3 bits (72), Expect = 0.26
Identities = 17/37 (45%), Positives = 19/37 (50%)
Query: 16 GRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRD 52
GRG G RGGGG +G DRGG + G RD
Sbjct: 49 GRGGYGGGGGRGNRGGGGGGYQGGDRGGRGSGGGGRD 85
>At5g02530 unknown protein
Length = 292
Score = 32.3 bits (72), Expect = 0.26
Identities = 21/45 (46%), Positives = 24/45 (52%), Gaps = 8/45 (17%)
Query: 16 GRGESGPSPPPRGRG------GGGRPARGDDRGGDPADGKNRDES 54
GRG G PRG G GGR ARG RGG + G+ RDE+
Sbjct: 229 GRGRGGFMGRPRGGGFGGGNFRGGRGARG--RGGRGSGGRGRDEN 271
>At4g18670 extensin-like protein
Length = 839
Score = 32.3 bits (72), Expect = 0.26
Identities = 22/77 (28%), Positives = 25/77 (31%), Gaps = 1/77 (1%)
Query: 2 GGQGERSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTR 61
GG S TP P G S P+ P G P GG P +
Sbjct: 478 GGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTT-PSPGGSPPSPSISPSPPITVPSP 536
Query: 62 THNPTLPYPPPGPPNST 78
PT P PP P + T
Sbjct: 537 PSTPTSPGSPPSPSSPT 553
Score = 29.6 bits (65), Expect = 1.7
Identities = 19/75 (25%), Positives = 26/75 (34%), Gaps = 1/75 (1%)
Query: 3 GQGERSERTPAPDGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRT 62
G S TP P G S P+ P G P GG P + +
Sbjct: 466 GSPPTSPTTPTPGGSPPSSPTTPTPGGSPPSSPTT-PTPGGSPPSSPTTPSPGGSPPSPS 524
Query: 63 HNPTLPYPPPGPPNS 77
+P+ P P PP++
Sbjct: 525 ISPSPPITVPSPPST 539
>At1g16610 arginine/serine-rich protein
Length = 414
Score = 32.3 bits (72), Expect = 0.26
Identities = 24/79 (30%), Positives = 34/79 (42%), Gaps = 10/79 (12%)
Query: 7 RSERTPAP-DGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGK-NRDESRAR------- 57
R R+P GR S P +GRG GR R P+ + R SR+R
Sbjct: 332 RRSRSPIRRPGRSRSSSISPRKGRGPAGRRGRSSSYSSSPSPRRIPRKISRSRSPKRPLR 391
Query: 58 -KHTRTHNPTLPYPPPGPP 75
K + +++ + PPP PP
Sbjct: 392 GKRSSSNSSSSSSPPPPPP 410
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,434,977
Number of Sequences: 26719
Number of extensions: 379103
Number of successful extensions: 2576
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 2141
Number of HSP's gapped (non-prelim): 352
length of query: 260
length of database: 11,318,596
effective HSP length: 97
effective length of query: 163
effective length of database: 8,726,853
effective search space: 1422477039
effective search space used: 1422477039
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149580.1


