

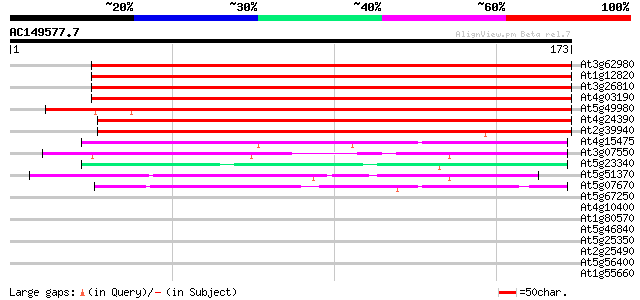
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149577.7 - phase: 0
(173 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g62980 transport inhibitor response 1 (TIR1) 197 2e-51
At1g12820 transport inhibitor response 1, putative 196 7e-51
At3g26810 transport inhibitor response 1-like protein 193 4e-50
At4g03190 putative homolog of transport inhibitor response 1 183 5e-47
At5g49980 transport inhibitor response 1 protein 171 1e-43
At4g24390 transport inhibitor response-like protein 170 4e-43
At2g39940 unknown protein 127 4e-30
At4g15475 unknown protein 60 8e-10
At3g07550 unknown protein 50 5e-07
At5g23340 unknown protein 44 3e-05
At5g51370 putative protein 44 6e-05
At5g07670 unknown protein 41 4e-04
At5g67250 unknown protein 39 0.001
At4g10400 putative protein 39 0.001
At1g80570 unknown protein 38 0.002
At5g46840 unknown protein 38 0.003
At5g25350 leucine-rich repeats containing protein 37 0.004
At2g25490 putative glucose regulated repressor protein 37 0.007
At5g56400 putative protein 34 0.035
At1g55660 hypothetical protein 34 0.035
>At3g62980 transport inhibitor response 1 (TIR1)
Length = 594
Score = 197 bits (502), Expect = 2e-51
Identities = 90/148 (60%), Positives = 110/148 (73%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP+EVLE V ++ KDR+SVSLVCK WY ERW RR VFIGNCYAVSP + RRFP +
Sbjct: 9 FPEEVLEHVFSFIQLDKDRNSVSLVCKSWYEIERWCRRKVFIGNCYAVSPATVIRRFPKV 68
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RSV +KGKP F+DFNLVP WG ++ W+ + Y +LEE+RLKRM V+D+ LE +A S
Sbjct: 69 RSVELKGKPHFADFNLVPDGWGGYVYPWIEAMSSSYTWLEEIRLKRMVVTDDCLELIAKS 128
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F NFK L L SC+GFSTDGLAA+A C+
Sbjct: 129 FKNFKVLVLSSCEGFSTDGLAAIAATCR 156
>At1g12820 transport inhibitor response 1, putative
Length = 577
Score = 196 bits (497), Expect = 7e-51
Identities = 89/148 (60%), Positives = 115/148 (77%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FPDEV+E V + S KDR+S+SLVCK W+ ER+SR+ VFIGNCYA++PE L RRFP +
Sbjct: 4 FPDEVIEHVFDFVASHKDRNSISLVCKSWHKIERFSRKEVFIGNCYAINPERLIRRFPCL 63
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
+S+T+KGKP F+DFNLVP WG +H W+ A LEELRLKRM V+DESL+ L+ S
Sbjct: 64 KSLTLKGKPHFADFNLVPHEWGGFVHPWIEALARSRVGLEELRLKRMVVTDESLDLLSRS 123
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F NFK+L L+SC+GF+TDGLA++A NC+
Sbjct: 124 FANFKSLVLVSCEGFTTDGLASIAANCR 151
>At3g26810 transport inhibitor response 1-like protein
Length = 575
Score = 193 bits (490), Expect = 4e-50
Identities = 89/148 (60%), Positives = 114/148 (76%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FPDEV+E V + S KDR+++SLVCK WY ER+SR+ VFIGNCYA++PE L RRFP +
Sbjct: 4 FPDEVIEHVFDFVTSHKDRNAISLVCKSWYKIERYSRQKVFIGNCYAINPERLLRRFPCL 63
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
+S+T+KGKP F+DFNLVP WG + W+ A LEELRLKRM V+DESLE L+ S
Sbjct: 64 KSLTLKGKPHFADFNLVPHEWGGFVLPWIEALARSRVGLEELRLKRMVVTDESLELLSRS 123
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F NFK+L L+SC+GF+TDGLA++A NC+
Sbjct: 124 FVNFKSLVLVSCEGFTTDGLASIAANCR 151
>At4g03190 putative homolog of transport inhibitor response 1
Length = 585
Score = 183 bits (464), Expect = 5e-47
Identities = 84/148 (56%), Positives = 109/148 (72%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP +VLE +L + S +DR+SVSLVCK W+ ER +R+ VF+GNCYAVSP +TRRFP +
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RS+T+KGKP F+D+NLVP WG W+ A K LEE+R+KRM V+DE LE +A S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRMVVTDECLEKIAAS 124
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F +FK L L SC+GFSTDG+AA+A C+
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIAATCR 152
>At5g49980 transport inhibitor response 1 protein
Length = 619
Score = 171 bits (434), Expect = 1e-43
Identities = 86/167 (51%), Positives = 111/167 (65%), Gaps = 5/167 (2%)
Query: 12 SQGEKNNNMDSNSD-FPDEVLERVLG----MMKSRKDRSSVSLVCKEWYNAERWSRRNVF 66
S K+ N SNS FPD VLE VL + SR DR++ SLVCK W+ E +R VF
Sbjct: 35 SSPNKSRNCISNSQTFPDHVLENVLENVLQFLDSRCDRNAASLVCKSWWRVEALTRSEVF 94
Query: 67 IGNCYAVSPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEE 126
IGNCYA+SP LT+RF +RS+ +KGKPRF+DFNL+P +WGA+ W+ A YP LE+
Sbjct: 95 IGNCYALSPARLTQRFKRVRSLVLKGKPRFADFNLMPPDWGANFAPWVSTMAQAYPCLEK 154
Query: 127 LRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
+ LKRM V+D+ L LA SFP FK L L+ C+GF T G++ VA C+
Sbjct: 155 VDLKRMFVTDDDLALLADSFPGFKELILVCCEGFGTSGISIVANKCR 201
>At4g24390 transport inhibitor response-like protein
Length = 623
Score = 170 bits (430), Expect = 4e-43
Identities = 78/146 (53%), Positives = 102/146 (69%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
+ VLE VL + SR DR++VSLVC+ WY E +R VFIGNCY++SP L RF +RS
Sbjct: 56 ENVLENVLQFLTSRCDRNAVSLVCRSWYRVEAQTRLEVFIGNCYSLSPARLIHRFKRVRS 115
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSFP 147
+ +KGKPRF+DFNL+P NWGA W+ A YP+LE++ LKRM V+D+ L LA SFP
Sbjct: 116 LVLKGKPRFADFNLMPPNWGAQFSPWVAATAKAYPWLEKVHLKRMFVTDDDLALLAESFP 175
Query: 148 NFKALSLLSCDGFSTDGLAAVATNCK 173
FK L+L+ C+GF T G+A VA C+
Sbjct: 176 GFKELTLVCCEGFGTSGIAIVANKCR 201
>At2g39940 unknown protein
Length = 592
Score = 127 bits (318), Expect = 4e-30
Identities = 59/147 (40%), Positives = 94/147 (63%), Gaps = 1/147 (0%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
D+V+E+V+ + KDR S SLVC+ W+ + +R +V + CY +P+ L+RRFPN+RS
Sbjct: 18 DDVIEQVMTYITDPKDRDSASLVCRRWFKIDSETREHVTMALCYTATPDRLSRRFPNLRS 77
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS-F 146
+ +KGKPR + FNL+P NWG + W+ ++ L+ + +RM VSD L+ LA +
Sbjct: 78 LKLKGKPRAAMFNLIPENWGGYVTPWVTEISNNLRQLKSVHFRRMIVSDLDLDRLAKARA 137
Query: 147 PNFKALSLLSCDGFSTDGLAAVATNCK 173
+ + L L C GF+TDGL ++ T+C+
Sbjct: 138 DDLETLKLDKCSGFTTDGLLSIVTHCR 164
>At4g15475 unknown protein
Length = 610
Score = 59.7 bits (143), Expect = 8e-10
Identities = 45/159 (28%), Positives = 79/159 (49%), Gaps = 10/159 (6%)
Query: 23 NSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSP--EILTR 80
N+ P+E++ + ++S+ +R + SLVCK W + ER+SR + IG ++ +L+R
Sbjct: 8 NNCLPEELILEIFRRLESKPNRDACSLVCKRWLSLERFSRTTLRIGASFSPDDFISLLSR 67
Query: 81 RFPNIRSVTMKGKPRFSDFNLVPA-------NWGADIHSWLVVFADKYPFLEELRLKRMA 133
RF I S+ + + S +L P+ + + S DK E ++ +
Sbjct: 68 RFLYITSIHVDERISVSLPSLSPSPKRKRGRDSSSPSSSKRKKLTDKTHSGAE-NVESSS 126
Query: 134 VSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
++D L LA FP + LSL+ C S+ GL ++A C
Sbjct: 127 LTDTGLTALANGFPRIENLSLIWCPNVSSVGLCSLAQKC 165
>At3g07550 unknown protein
Length = 395
Score = 50.4 bits (119), Expect = 5e-07
Identities = 42/165 (25%), Positives = 71/165 (42%), Gaps = 27/165 (16%)
Query: 11 ESQGEKNNNMDSNS-DFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGN 69
E E +NN++++ PD+ L + + S D S L C W N + SRR++
Sbjct: 2 EDVSESDNNVETSIIHLPDDCLSFIFQRLDSVADHDSFGLTCHRWLNIQNISRRSLQFQC 61
Query: 70 CYAV-SPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELR 128
++V +P L++ P++ S +H L ++ +LE L
Sbjct: 62 SFSVLNPSSLSQTNPDVSS--------------------HHLHRLLT----RFQWLEHLS 97
Query: 129 LKRMAV-SDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
L V +D SL+ L + L L C G S DG++ +A+ C
Sbjct: 98 LSGCTVLNDSSLDSLRYPGARLHTLYLDCCFGISDDGISTIASFC 142
>At5g23340 unknown protein
Length = 405
Score = 44.3 bits (103), Expect = 3e-05
Identities = 37/151 (24%), Positives = 57/151 (37%), Gaps = 9/151 (5%)
Query: 23 NSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRF 82
N D+ L VL + S KD+ LVCK W N + R+ + P +L R
Sbjct: 7 NEALTDDELRWVLSRLDSDKDKEVFGLVCKRWLNLQSTDRKKL----AARAGPHMLRRLA 62
Query: 83 PNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKR-MAVSDESLEF 141
+ + + P +D L V ++ + FL L L ++D L
Sbjct: 63 SRFTQIVELDLSQSISRSFYPGVTDSD----LAVISEGFKFLRVLNLHNCKGITDTGLAS 118
Query: 142 LAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
+ + L + C S GL+AVA C
Sbjct: 119 IGRCLSLLQFLDVSYCRKLSDKGLSAVAEGC 149
>At5g51370 putative protein
Length = 446
Score = 43.5 bits (101), Expect = 6e-05
Identities = 44/167 (26%), Positives = 71/167 (42%), Gaps = 14/167 (8%)
Query: 7 KRKKESQGEKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVF 66
K +K K + +DS D +L +++ + + + VSLVCK W N + R++
Sbjct: 13 KNEKPFNSIKPSEIDSTLSLSDSLLLKIIEKLPESQS-NDVSLVCKRWLNLQGQRLRSLK 71
Query: 67 IGNCYAVSPEILTRRFPNIRSVTMKG---KPRFSDFNLVPANWGADIHSWLVVFADKYPF 123
+ + + E LT RFPN+ V + PR + ++ + H L + + F
Sbjct: 72 LLDFDFLLSERLTTRFPNLTHVDLVNACMNPRVNS-GILFCHKSISFH--LSSDSSNWEF 128
Query: 124 LEELRLKRMAV-------SDESLEFLAFSFPNFKALSLLSCDGFSTD 163
LEE L + S ES + L N L LLS G +D
Sbjct: 129 LEENLLHSDVIDRGLRILSRESFDLLNLKVINASELGLLSLAGDCSD 175
Score = 27.3 bits (59), Expect = 4.2
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query: 133 AVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
+VSD L FLA + L L C+G S DG+ A+ C+
Sbjct: 214 SVSDIGLTFLAQGCRSLVKLELSGCEG-SFDGIKAIGQCCE 253
>At5g07670 unknown protein
Length = 476
Score = 40.8 bits (94), Expect = 4e-04
Identities = 41/152 (26%), Positives = 68/152 (43%), Gaps = 16/152 (10%)
Query: 27 PDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIR 86
PD +L RV+ + + + R ++SLVCK W+ R+ + + +S L RFPN+
Sbjct: 66 PDLILIRVIQKIPNSQ-RKNLSLVCKRWFRLHGRLVRSFKVSDWEFLSSGRLISRFPNLE 124
Query: 87 SVTMKGKPRFSDFNLVPANWGADIHSWLVVFA------DKYPFLEELRLKRMAVSDESLE 140
+V + S + P N G ++ +V F + F EE L + + + L+
Sbjct: 125 TVDL-----VSGCLISPPNLGILVNHRIVSFTVGVGSYQSWSFFEE-NLLSVELVERGLK 178
Query: 141 FLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
LA N + L + + S GL VA C
Sbjct: 179 ALAGGCSNLRKLVVTNT---SELGLLNVAEEC 207
>At5g67250 unknown protein
Length = 527
Score = 38.9 bits (89), Expect = 0.001
Identities = 44/154 (28%), Positives = 66/154 (42%), Gaps = 11/154 (7%)
Query: 21 DSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTR 80
D D PDE L V + + DR SLVCK W + SR + + +S LT
Sbjct: 41 DFTGDLPDECLAHVFQFLGAG-DRKRCSLVCKRWLLVDGQSRHRLSLDAKDEIS-SFLTS 98
Query: 81 RFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKR-MAVSDESL 139
F SVT D V + A L + + + L ++L+ ++D +
Sbjct: 99 MFNRFDSVTKLALR--CDRKSVSLSDEA-----LAMISVRCLNLTRVKLRGCREITDLGM 151
Query: 140 EFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
E A + N K LS+ SC+ F G+ A+ +CK
Sbjct: 152 EDFAKNCKNLKKLSVGSCN-FGAKGVNAMLEHCK 184
>At4g10400 putative protein
Length = 326
Score = 38.9 bits (89), Expect = 0.001
Identities = 40/143 (27%), Positives = 62/143 (42%), Gaps = 8/143 (5%)
Query: 20 MDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILT 79
MD S PDEVL ++L + + K S S++ K W W + F Y+ S
Sbjct: 1 MDRISGLPDEVLVKILSFVPT-KVAVSTSILSKRWEFLWMWLTKLKFGSKRYSESEFKRL 59
Query: 80 RRFPNIRSVTMKGKPRFSDFNLVPAN---WGADIHSWLVVFADKYPFLEELRLKRMAVSD 136
+ F + R++ + P F LV ++ DI W+VV +Y + EL++
Sbjct: 60 QCFLD-RNLPLHRAPVIESFRLVLSDSHFKPEDIRMWVVVAVSRY--IRELKIYSSHYG- 115
Query: 137 ESLEFLAFSFPNFKALSLLSCDG 159
E L S K+L +L DG
Sbjct: 116 EKQNILPSSLYTCKSLVILKLDG 138
>At1g80570 unknown protein
Length = 467
Score = 38.1 bits (87), Expect = 0.002
Identities = 38/159 (23%), Positives = 69/159 (42%), Gaps = 23/159 (14%)
Query: 25 DFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPE----ILTR 80
+ PD ++ +L + + DR+S+SL CK +++ + R ++ IG C V L R
Sbjct: 3 ELPDHLVWDILSKLHTTDDRNSLSLSCKRFFSLDNEQRYSLRIG-CGLVPASDALLSLCR 61
Query: 81 RFPNIRSVTMKGKPRFSDFNLVPANWGADI-----HSWLVVFADKYPFLEELRLKRMA-V 134
RFPN+ V ++ + W + + L+V L +L L +
Sbjct: 62 RFPNLSKV-----------EIIYSGWMSKLGKQVDDQGLLVLTTNCHSLTDLTLSFCTFI 110
Query: 135 SDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
+D + L+ S P +L L + G+ ++A CK
Sbjct: 111 TDVGIGHLS-SCPELSSLKLNFAPRITGCGVLSLAVGCK 148
Score = 26.9 bits (58), Expect = 5.5
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 3/48 (6%)
Query: 118 ADKYPFLEELRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGL 165
A K LE + + VSDE L L FP+ L L C G + DG+
Sbjct: 381 AQKLEILELVHCQE--VSDEGL-ILVSQFPSLNVLKLSKCLGVTDDGM 425
>At5g46840 unknown protein
Length = 501
Score = 37.7 bits (86), Expect = 0.003
Identities = 24/92 (26%), Positives = 46/92 (49%), Gaps = 4/92 (4%)
Query: 7 KRKKESQGEKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVF 66
KRKK + E N ++ E+ E+ +G + + D + ++V KE ++ E R VF
Sbjct: 115 KRKKRKRDEIENEYETKKYGSVEMKEKKVGEKRKKADEVADTMVSKEGFDDESKLLRTVF 174
Query: 67 IGNC-YAVSPEILTR---RFPNIRSVTMKGKP 94
+GN V +++ + +F + SV ++ P
Sbjct: 175 VGNLPLKVKKKVILKEFSKFGEVESVRIRSVP 206
>At5g25350 leucine-rich repeats containing protein
Length = 623
Score = 37.4 bits (85), Expect = 0.004
Identities = 26/91 (28%), Positives = 40/91 (43%), Gaps = 14/91 (15%)
Query: 83 PNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRM-AVSDESLEF 141
P++R V++ P SD L A P +E+L L R ++D L
Sbjct: 167 PSLRIVSLWNLPAVSDLGLSE-------------IARSCPMIEKLDLSRCPGITDSGLVA 213
Query: 142 LAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
+A + N L++ SC G +GL A+A C
Sbjct: 214 IAENCVNLSDLTIDSCSGVGNEGLRAIARRC 244
Score = 37.0 bits (84), Expect = 0.005
Identities = 43/173 (24%), Positives = 74/173 (41%), Gaps = 19/173 (10%)
Query: 15 EKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYN---------AERWSRRNV 65
EK ++D P+E L +L + S ++RS+ + V K W N S ++V
Sbjct: 50 EKQTSIDV---LPEECLFEILRRLPSGQERSACACVSKHWLNLLSSISRSEVNESSVQDV 106
Query: 66 FIGNCYAVSPEILTRRFPNIR----SVTMKGKPRFSDFNLVPANWGADIHS-WLVVFADK 120
G + +S + ++ ++R +V + + + + + + L A
Sbjct: 107 EEGEGF-LSRSLEGKKATDLRLAAIAVGTSSRGGLGKLQIRGSGFESKVTDVGLGAVAHG 165
Query: 121 YPFLEELRLKRM-AVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
P L + L + AVSD L +A S P + L L C G + GL A+A NC
Sbjct: 166 CPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENC 218
>At2g25490 putative glucose regulated repressor protein
Length = 628
Score = 36.6 bits (83), Expect = 0.007
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query: 114 LVVFADKYPFLEELRLKRMA-VSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
L+ A+ LE+L L R + ++D+ L +A S PN L+L +C +GL A+A +C
Sbjct: 195 LLEIAEGCAQLEKLELNRCSTITDKGLVAIAKSCPNLTELTLEACSRIGDEGLLAIARSC 254
Score = 30.0 bits (66), Expect = 0.65
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 1/59 (1%)
Query: 114 LVVFADKYPFLEELRLKRMA-VSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATN 171
LV A P L EL L+ + + DE L +A S K++S+ +C G+A++ +N
Sbjct: 221 LVAIAKSCPNLTELTLEACSRIGDEGLLAIARSCSKLKSVSIKNCPLVRDQGIASLLSN 279
>At5g56400 putative protein
Length = 450
Score = 34.3 bits (77), Expect = 0.035
Identities = 36/171 (21%), Positives = 68/171 (39%), Gaps = 32/171 (18%)
Query: 8 RKKESQGEKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSR----- 62
RKK + + N+D SD P+++L +L ++ + D + S V K W + W++
Sbjct: 20 RKKLNCQSRQFNVDKISDLPEDLLVHILSLLPTTNDIVATSGVSKRWESL--WTKVHKLR 77
Query: 63 --RNVFIGNCY-----AVSPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLV 115
++ G Y V ++ + P + S+ + P+ + DI W+
Sbjct: 78 FNDRIYDGKRYDSFLHFVEKSLILHKAPTLVSLRLSVGPKCT---------ADDIGLWIK 128
Query: 116 VFADK---------YPFLEELRLKRMAVSDESLEFLAFSFPNFKALSLLSC 157
+ D+ YP +RL R ++L L A+ L +C
Sbjct: 129 LALDRNICELIIKHYPDHGHIRLSRRLCDSKTLVSLKLKNAILGAIWLPTC 179
>At1g55660 hypothetical protein
Length = 492
Score = 34.3 bits (77), Expect = 0.035
Identities = 33/123 (26%), Positives = 53/123 (42%), Gaps = 17/123 (13%)
Query: 18 NNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVS--- 74
N MD S PDE+L +VL + S KD S S++ W + W + + Y+VS
Sbjct: 50 NLMDKISQLPDELLVKVLSFL-STKDAVSTSILSMRWKSLWMWLPKLEYNFRHYSVSEGQ 108
Query: 75 -------PEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEEL 127
+ + P I S+++K + + + + DI+ W V A + EL
Sbjct: 109 GLARFITSSLRVHKAPAIESLSLKFR-----YGAIGSIKPKDIYLW-VSLAVHVSNVREL 162
Query: 128 RLK 130
LK
Sbjct: 163 SLK 165
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,108,735
Number of Sequences: 26719
Number of extensions: 167979
Number of successful extensions: 633
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 541
Number of HSP's gapped (non-prelim): 120
length of query: 173
length of database: 11,318,596
effective HSP length: 92
effective length of query: 81
effective length of database: 8,860,448
effective search space: 717696288
effective search space used: 717696288
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149577.7


