

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.7 + phase: 1 /pseudo
(576 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
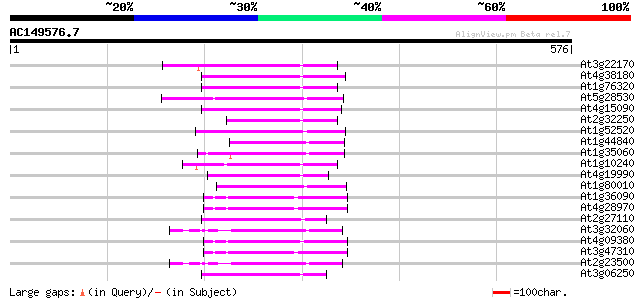
Score E
Sequences producing significant alignments: (bits) Value
At3g22170 far-red impaired response protein, putative 70 2e-12
At4g38180 unknown protein (At4g38180) 69 5e-12
At1g76320 putative phytochrome A signaling protein 65 1e-10
At5g28530 far-red impaired response protein (FAR1) - like 62 7e-10
At4g15090 unknown protein 62 9e-10
At2g32250 Mutator-like transposase 57 3e-08
At1g52520 F6D8.26 56 5e-08
At1g44840 hypothetical protein 55 1e-07
At1g35060 hypothetical protein 55 1e-07
At1g10240 unknown protein 55 1e-07
At4g19990 putative protein 54 2e-07
At1g80010 hypothetical protein 54 2e-07
At1g36090 hypothetical protein 53 4e-07
At4g28970 putative protein 53 5e-07
At2g27110 Mutator-like transposase 52 9e-07
At3g32060 hypothetical protein 52 1e-06
At4g09380 putative protein 51 2e-06
At3g47310 putative protein 50 3e-06
At2g23500 Mutator-like transposase 50 3e-06
At3g06250 unknown protein 50 4e-06
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 70.5 bits (171), Expect = 2e-12
Identities = 47/185 (25%), Positives = 87/185 (46%), Gaps = 8/185 (4%)
Query: 158 ETKKYLTNLKKKKKESITNIKRVYNECHKFKKAKR-----GDLTEMQYLISKLEE-NEYV 211
+T+K + K+ E T I + F+K + GD + +S+++ N
Sbjct: 193 QTRKIYAAMAKQFAEYKTVISLKSDSKSSFEKGRTLSVETGDFKILLDFLSRMQSLNSNF 252
Query: 212 NYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLT 271
Y + + Q V+++FW S + +F V+ +D+TY N Y+MPL VGV Y
Sbjct: 253 FYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQY 312
Query: 272 YSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAIL 331
+G A + + ++W ++ L+ + PKV++T+ D M + V + P++ L
Sbjct: 313 MVLGCALISDESAATYSWLMETWLRAI--GGQAPKVLITELDVVMNSIVPEIFPNTRHCL 370
Query: 332 CYFHV 336
+HV
Sbjct: 371 FLWHV 375
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 69.3 bits (168), Expect = 5e-12
Identities = 38/147 (25%), Positives = 68/147 (45%), Gaps = 2/147 (1%)
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
+ YL +N Y + E Q V ++FW P ++ F F + D+TY++N YR+
Sbjct: 254 LDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRL 313
Query: 258 PLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMM 317
P GV G AF++++ E +F W L + ++ P + TD D +
Sbjct: 314 PFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAM--SAHPPVSITTDHDAVIR 371
Query: 318 NAVANVLPDSSAILCYFHVGKNIRSRI 344
A+ +V P + C +H+ K + ++
Sbjct: 372 AAIMHVFPGARHRFCKWHILKKCQEKL 398
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 65.1 bits (157), Expect = 1e-10
Identities = 33/139 (23%), Positives = 70/139 (49%), Gaps = 2/139 (1%)
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
+++L+ EEN + + E +++++FW ++ + +F V+ +++Y + Y++
Sbjct: 167 LEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKV 226
Query: 258 PLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMM 317
PL VGV +G + + W +Q L + PKV++TD++ ++
Sbjct: 227 PLVLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAM--GGQKPKVMLTDQNNAIK 284
Query: 318 NAVANVLPDSSAILCYFHV 336
A+A VLP++ C +HV
Sbjct: 285 AAIAAVLPETRHCYCLWHV 303
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 62.4 bits (150), Expect = 7e-10
Identities = 42/189 (22%), Positives = 91/189 (47%), Gaps = 6/189 (3%)
Query: 157 GETKKYLTNLKKKKKESITNIKRVYNECHKFKKAKR-GDLTEMQYLISKLEENEYVNYVR 215
G L ++K + + K+ E F KR D E+ L E + +++V
Sbjct: 219 GVVSGQLPFIEKDVRNFVRACKKSVQENDAFMTEKRESDTLELLECCKGLAERD-MDFVY 277
Query: 216 E--KPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYS 273
+ E+Q V++I W + SV+ ++ F V++ D++Y++ Y + L G+ +
Sbjct: 278 DCTSDENQKVENIAWAYGDSVRGYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAML 337
Query: 274 VGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCY 333
+G + + +FTWALQ ++ ++ P+ ++TD D + +A+ +P+++ ++
Sbjct: 338 LGCVLLQDESCRSFTWALQTFVRFMRGRH--PQTILTDIDTGLKDAIGREMPNTNHVVFM 395
Query: 334 FHVGKNIRS 342
H+ + S
Sbjct: 396 SHIVSKLAS 404
>At4g15090 unknown protein
Length = 768
Score = 62.0 bits (149), Expect = 9e-10
Identities = 37/143 (25%), Positives = 71/143 (48%), Gaps = 2/143 (1%)
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
++Y +EN Y + E Q ++++FW S + +F V+ D+TY ++
Sbjct: 171 LEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKL 230
Query: 258 PLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMM 317
PL +GV +G A + + + F W ++ L+ + PKV++TD+D +M
Sbjct: 231 PLALFIGVNHHSQPMLLGCALVADESMETFVWLIKTWLRAM--GGRAPKVILTDQDKFLM 288
Query: 318 NAVANVLPDSSAILCYFHVGKNI 340
+AV+ +LP++ +HV + I
Sbjct: 289 SAVSELLPNTRHCFALWHVLEKI 311
>At2g32250 Mutator-like transposase
Length = 684
Score = 57.0 bits (136), Expect = 3e-08
Identities = 31/114 (27%), Positives = 55/114 (48%), Gaps = 2/114 (1%)
Query: 223 VQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSK 282
V+++FW + + +F V++ D+ Y N YR+P +GV+ +G A +
Sbjct: 199 VRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHHRQYVLLGCALIGEV 258
Query: 283 KEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHV 336
E ++W + LK + P V++TD+D + + V V PD I C + V
Sbjct: 259 SESTYSWLFRTWLKAV--GGQAPGVMITDQDKLLSDIVVEVFPDVRHIFCLWSV 310
>At1g52520 F6D8.26
Length = 703
Score = 56.2 bits (134), Expect = 5e-08
Identities = 35/155 (22%), Positives = 73/155 (46%), Gaps = 4/155 (2%)
Query: 191 KRGDLTEMQYLISKLE-ENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDST 249
KRGD + +++ N Y+ + + ++++FW S + F V+ +DS+
Sbjct: 244 KRGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSS 303
Query: 250 YKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVV 309
Y + + +PL GV T + F+ + +++ W L++ L ++K + P+ +V
Sbjct: 304 YISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVMKRS---PQTIV 360
Query: 310 TDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRI 344
TDR + A++ V P S H+ + I ++
Sbjct: 361 TDRCKPLEAAISQVFPRSHQRFSLTHIMRKIPEKL 395
>At1g44840 hypothetical protein
Length = 926
Score = 54.7 bits (130), Expect = 1e-07
Identities = 28/118 (23%), Positives = 61/118 (50%), Gaps = 2/118 (1%)
Query: 226 IFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKED 285
+F S+ F +L++D T+ Y+ L G + + Y +GFA + S+ ++
Sbjct: 557 MFLAFGASIAGFRHLRRILVVDGTHLKGKYKGVLLTSSGQDANFQVYPLGFAVVDSENDE 616
Query: 286 NFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSR 343
++TW L +++ + + +++DR S++ AV V P ++ C H+ +NI+++
Sbjct: 617 SWTWFFTKLERIIADSKTL--TILSDRHSSILVAVKRVFPQANHGACIIHLCRNIQTK 672
>At1g35060 hypothetical protein
Length = 873
Score = 54.7 bits (130), Expect = 1e-07
Identities = 36/155 (23%), Positives = 74/155 (47%), Gaps = 8/155 (5%)
Query: 194 DLTEMQYLISKLEENEYVNYVREKPESQIVQD-----IFWTHPTSVKLFNTFPTVLIMDS 248
+L E +L+ KL + ++ +P+ + + +F S++ F VL++D
Sbjct: 472 NLAEYLHLL-KLTNPGTITHIETEPDIEDERKERFLYMFLAFGASIQGFKHLRRVLVVDG 530
Query: 249 TYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVV 308
T+ Y+ L G + + Y + FA + S+ +D +TW L +++ N+ + +
Sbjct: 531 THLKGKYKGVLLTASGQDANFQVYPLAFAVVDSENDDAWTWFFTKLERIIADNNTL--TI 588
Query: 309 VTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSR 343
++DR S+ V V P + C H+ +NI++R
Sbjct: 589 LSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQAR 623
>At1g10240 unknown protein
Length = 680
Score = 54.7 bits (130), Expect = 1e-07
Identities = 37/163 (22%), Positives = 75/163 (45%), Gaps = 8/163 (4%)
Query: 178 KRVYNECHKFKKA----KRGDLTEMQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTS 233
K V N FKK + D M I + + N + + + +++I W++ +S
Sbjct: 210 KDVRNLLQSFKKLDPEDENIDFLRMCQSIKEKDPNFKFEFTLDANDK--LENIAWSYASS 267
Query: 234 VKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQM 293
++ + F ++ D+T++ + MPL VGV + + G + + +++WALQ
Sbjct: 268 IQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLRSWSWALQA 327
Query: 294 LLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHV 336
+ N P+ ++TD + + A+A +P + LC + V
Sbjct: 328 FTGFM--NGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWMV 368
>At4g19990 putative protein
Length = 672
Score = 53.9 bits (128), Expect = 2e-07
Identities = 32/125 (25%), Positives = 59/125 (46%), Gaps = 3/125 (2%)
Query: 204 KLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIV 263
KLE+ + + + Q +++IFW + F V+ +D+T+ N Y++PL
Sbjct: 167 KLEDGDVERLLNFFTDMQSLRNIFWVDAKGRFDYTCFSDVVSIDTTFIKNEYKLPLVAFT 226
Query: 264 GVTSTYLTYSVGFAFMMS-KKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVAN 322
GV +GF +++ + + F W + LK + + P+V++T D + AV
Sbjct: 227 GVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAM--HGCRPRVILTKHDQMLKEAVLE 284
Query: 323 VLPDS 327
V P S
Sbjct: 285 VFPSS 289
>At1g80010 hypothetical protein
Length = 696
Score = 53.9 bits (128), Expect = 2e-07
Identities = 30/133 (22%), Positives = 62/133 (46%), Gaps = 2/133 (1%)
Query: 213 YVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 272
Y+ + + ++++FW + ++ F VL+ D+T +N Y +PL VG+ T
Sbjct: 262 YLMDLADDGSLRNVFWIDARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTI 321
Query: 273 SVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILC 332
+G + + + + W + L + P++ +T++ +M AV+ V P + L
Sbjct: 322 LLGCGLLADQSFETYVWLFRAWLTCMLGRP--PQIFITEQCKAMRTAVSEVFPRAHHRLS 379
Query: 333 YFHVGKNIRSRII 345
HV NI ++
Sbjct: 380 LTHVLHNICQSVV 392
>At1g36090 hypothetical protein
Length = 645
Score = 53.1 bits (126), Expect = 4e-07
Identities = 38/149 (25%), Positives = 74/149 (49%), Gaps = 6/149 (4%)
Query: 200 YLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPL 259
Y++ K+ V YV + E + + +F ++ F V+++D+T+ +Y L
Sbjct: 329 YMLEKVNPGT-VTYVELEGEKKF-KYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 386
Query: 260 FEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVV-VTDRDPSMMN 318
+ Y + F + S+K+ ++ W L+ KL SD+P++V ++DR S+
Sbjct: 387 VIATAHDPNHHHYPLAFGIIDSEKDVSWIWFLE---KLKTVYSDVPRLVFISDRHQSIKK 443
Query: 319 AVANVLPDSSAILCYFHVGKNIRSRIITD 347
AV V P++ C +H+ +N+R R+ D
Sbjct: 444 AVKTVYPNALHAACIWHLCQNMRDRVKID 472
>At4g28970 putative protein
Length = 914
Score = 52.8 bits (125), Expect = 5e-07
Identities = 38/149 (25%), Positives = 73/149 (48%), Gaps = 6/149 (4%)
Query: 200 YLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPL 259
Y++ K+ V YV + E + + +F ++ F V+++D+T+ +Y L
Sbjct: 369 YMLEKVNPGT-VTYVELEGEKKF-KYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 426
Query: 260 FEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVV-VTDRDPSMMN 318
+ Y + F + S+K+ ++ W L+ KL SD+P +V ++DR S+
Sbjct: 427 VIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLE---KLKTVYSDVPGLVFISDRHQSIKK 483
Query: 319 AVANVLPDSSAILCYFHVGKNIRSRIITD 347
AV V P++ C +H+ +N+R R+ D
Sbjct: 484 AVKTVYPNALHAACIWHLCQNMRDRVTID 512
>At2g27110 Mutator-like transposase
Length = 851
Score = 52.0 bits (123), Expect = 9e-07
Identities = 31/129 (24%), Positives = 56/129 (43%), Gaps = 4/129 (3%)
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
++Y EN Y + E + ++FW S + F + +D+ Y+ N +R+
Sbjct: 201 LEYFKRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRV 260
Query: 258 PLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKV-VVTDRDPSM 316
P GV G A ++ + + +F W + L ++ D P V +VTD+D ++
Sbjct: 261 PFAPFTGVNHHGQAILFGCALILDESDTSFIWLFKTFLTAMR---DQPPVSLVTDQDRAI 317
Query: 317 MNAVANVLP 325
A V P
Sbjct: 318 QIAAGQVFP 326
>At3g32060 hypothetical protein
Length = 487
Score = 51.6 bits (122), Expect = 1e-06
Identities = 45/177 (25%), Positives = 81/177 (45%), Gaps = 22/177 (12%)
Query: 165 NLKKKKKESITNIKRVYNECHKFKKAKRGDLTEMQYLISKLEENEYVNYVREKPESQIVQ 224
+LK ++ES +I C+ + K D T + YL KL+EN+ YV
Sbjct: 208 DLKGSREESYKDIN-----CYLYMLKKVNDGT-VTYL--KLDENDKFQYV---------- 249
Query: 225 DIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKE 284
F S++ F VLI+D+T+ N Y L Y + FA + + +
Sbjct: 250 --FVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFAVLDGEND 307
Query: 285 DNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIR 341
++ W + L ++ S++ V +TDR+ S++ A+ NV + C +H+ +N++
Sbjct: 308 ASWEWFFEKLKTVVPDTSEL--VFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNVK 362
>At4g09380 putative protein
Length = 960
Score = 50.8 bits (120), Expect = 2e-06
Identities = 37/149 (24%), Positives = 73/149 (48%), Gaps = 6/149 (4%)
Query: 200 YLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPL 259
Y++ K+ V YV + E + + +F ++ F V+++D+T+ +Y L
Sbjct: 369 YMLEKVNPGT-VTYVELEGEKKF-KYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 426
Query: 260 FEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVV-VTDRDPSMMN 318
+ Y + F + S+K+ ++ W L+ L + SD+P +V ++DR S+
Sbjct: 427 VIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLENLKTVY---SDVPGLVFISDRHQSIKK 483
Query: 319 AVANVLPDSSAILCYFHVGKNIRSRIITD 347
AV V P++ C +H+ +N+R R+ D
Sbjct: 484 AVKTVYPNALHAACIWHLCQNMRDRVKID 512
>At3g47310 putative protein
Length = 735
Score = 50.4 bits (119), Expect = 3e-06
Identities = 37/149 (24%), Positives = 72/149 (47%), Gaps = 6/149 (4%)
Query: 200 YLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPL 259
Y++ K+ V YV + E + + +F ++ F T V+++D+T+ +Y L
Sbjct: 318 YMLEKVNPGT-VTYVELEGEKKF-KYLFIALGACIEGFRTMRKVIVVDATHLKTVYGGML 375
Query: 260 FEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVV-VTDRDPSMMN 318
+ Y + F + S+ + ++ W L+ KL SD+P +V ++DR S+
Sbjct: 376 VIATAQDPNHHHYPLAFGIIDSENDVSWIWFLE---KLKTVYSDVPGLVFISDRHQSIKK 432
Query: 319 AVANVLPDSSAILCYFHVGKNIRSRIITD 347
V V P++ C +H+ +N+R R+ D
Sbjct: 433 VVKTVYPNALHAACIWHLCQNMRDRVKID 461
>At2g23500 Mutator-like transposase
Length = 784
Score = 50.4 bits (119), Expect = 3e-06
Identities = 45/177 (25%), Positives = 80/177 (44%), Gaps = 22/177 (12%)
Query: 165 NLKKKKKESITNIKRVYNECHKFKKAKRGDLTEMQYLISKLEENEYVNYVREKPESQIVQ 224
+LK +ES +I C+ + K D T + YL KL+EN+ YV
Sbjct: 338 DLKGSPEESYKDIN-----CYLYMLKKVNDGT-VTYL--KLDENDKFQYV---------- 379
Query: 225 DIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKE 284
F S++ F VLI+D+T+ N Y L Y + FA + + +
Sbjct: 380 --FVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFAVLDGEND 437
Query: 285 DNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIR 341
++ W + L ++ S++ V +TDR+ S++ A+ NV + C +H+ +N++
Sbjct: 438 ASWEWFFEKLKTVVPDTSEL--VFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNVK 492
>At3g06250 unknown protein
Length = 764
Score = 49.7 bits (117), Expect = 4e-06
Identities = 31/128 (24%), Positives = 55/128 (42%), Gaps = 2/128 (1%)
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
+ Y SK E+ Y E + IFW S + F ++ D++Y+ Y +
Sbjct: 330 LDYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYRKGDYSV 389
Query: 258 PLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMM 317
P +G +G A + + ++ F+W Q L+ + + P+ +V D+D +
Sbjct: 390 PFATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAM--SGRRPRSMVADQDLPIQ 447
Query: 318 NAVANVLP 325
AVA V P
Sbjct: 448 QAVAQVFP 455
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.334 0.144 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,105,292
Number of Sequences: 26719
Number of extensions: 497009
Number of successful extensions: 2127
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 2062
Number of HSP's gapped (non-prelim): 94
length of query: 576
length of database: 11,318,596
effective HSP length: 105
effective length of query: 471
effective length of database: 8,513,101
effective search space: 4009670571
effective search space used: 4009670571
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149576.7


