

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.9 + phase: 2 /pseudo
(88 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
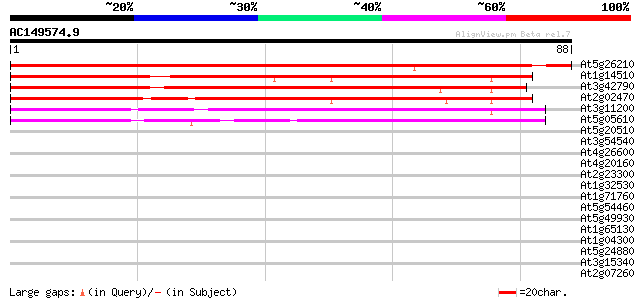
Score E
Sequences producing significant alignments: (bits) Value
At5g26210 nucleic acid binding protein - like 97 1e-21
At1g14510 unknown protein 82 6e-17
At3g42790 nucleic acid binding protein-like 80 1e-16
At2g02470 putative PHD-type zinc finger protein 77 1e-15
At3g11200 putative nucleic acid binding protein 61 9e-11
At5g05610 nucleic acid binding protein-like 57 1e-09
At5g20510 zinc finger protein - like 33 0.019
At3g54540 ABC transporter - like protein 33 0.025
At4g26600 unknown protein 32 0.072
At4g20160 Glu-rich protein 32 0.072
At2g23300 putative receptor-like protein kinase 32 0.072
At1g32530 unknown protein 31 0.12
At1g71760 hypothetical protein 30 0.16
At5g54460 unknown protein 30 0.21
At5g49930 putative protein 30 0.21
At1g65130 hypothetical protein 30 0.21
At1g04300 unknown protein 30 0.21
At5g24880 glutamic acid-rich protein 30 0.27
At3g15340 hypothetical protein 30 0.27
At2g07260 putative protein on FARE2.1 (CDS2) 30 0.27
>At5g26210 nucleic acid binding protein - like
Length = 255
Score = 97.1 bits (240), Expect = 1e-21
Identities = 51/95 (53%), Positives = 68/95 (70%), Gaps = 9/95 (9%)
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE 60
KRLF +NDLPTIFEVV G+AKKQ K+K SVS+++SN+SKS SK AK+SKP K+
Sbjct: 121 KRLFNMVNDLPTIFEVVAGTAKKQGKDKSSVSNNSSNRSKSSSKRGSESRAKFSKPEPKD 180
Query: 61 DD-------EEVDDEEEYQGECTACGENYVSASDE 88
D+ EE D++E+ + +C ACGE+Y A+DE
Sbjct: 181 DEEEEEEGVEEEDEDEQGETQCGACGESY--AADE 213
>At1g14510 unknown protein
Length = 252
Score = 81.6 bits (200), Expect = 6e-17
Identities = 48/88 (54%), Positives = 60/88 (67%), Gaps = 9/88 (10%)
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSK-SGSKARGSE----LAKYSK 55
KRLF INDLPTIFEVVTG+AK + K ++HNS++SK SG K R SE +K S
Sbjct: 121 KRLFQMINDLPTIFEVVTGNAK---QSKDQSANHNSSRSKSSGGKPRHSESHTKASKMSP 177
Query: 56 PPAKEDDEEVDDEEEYQGE-CTACGENY 82
PP KED+ +DE++ QG C ACG+NY
Sbjct: 178 PPRKEDESGDEDEDDEQGAVCGACGDNY 205
>At3g42790 nucleic acid binding protein-like
Length = 250
Score = 80.5 bits (197), Expect = 1e-16
Identities = 46/84 (54%), Positives = 56/84 (65%), Gaps = 5/84 (5%)
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE 60
KRLF IND+PTIFEVVTG AK K+K S ++ N NKSKS SK R SE KE
Sbjct: 122 KRLFNMINDVPTIFEVVTGMAK--AKDKSSAANQNGNKSKSNSKVRTSEGKSSKTKQPKE 179
Query: 61 DDEEVD-DEEEYQGE--CTACGEN 81
+DEE+D D+E+ GE C ACG++
Sbjct: 180 EDEEIDEDDEDDHGETLCGACGDS 203
>At2g02470 putative PHD-type zinc finger protein
Length = 256
Score = 77.4 bits (189), Expect = 1e-15
Identities = 48/92 (52%), Positives = 64/92 (69%), Gaps = 12/92 (13%)
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSE----LAKYSKP 56
KRLF IN+LPTIFEVV+G+A KQ+K+ SV+++NS SG K+R SE +AK S P
Sbjct: 121 KRLFQMINELPTIFEVVSGNA-KQSKDL-SVNNNNSKSKPSGVKSRQSESLSKVAKMSSP 178
Query: 57 PAKEDDEEVDD-----EEEYQGE-CTACGENY 82
P KE++EE D+ E++ QG C ACG+NY
Sbjct: 179 PPKEEEEEEDESEDESEDDEQGAVCGACGDNY 210
>At3g11200 putative nucleic acid binding protein
Length = 246
Score = 61.2 bits (147), Expect = 9e-11
Identities = 36/86 (41%), Positives = 52/86 (59%), Gaps = 5/86 (5%)
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE 60
KRLF+ INDLPT+F+VVTG K KP SS + +KS++G+K K S P E
Sbjct: 120 KRLFSLINDLPTLFDVVTG-RKAMKDNKP--SSDSGSKSRNGTKRSIDGQTKSSTPKLME 176
Query: 61 DDEEVDDEEEYQGE--CTACGENYVS 84
+ E ++EE+ G+ C +CG +Y +
Sbjct: 177 ESYEEEEEEDEHGDTLCGSCGGHYTN 202
>At5g05610 nucleic acid binding protein-like
Length = 241
Score = 57.4 bits (137), Expect = 1e-09
Identities = 38/85 (44%), Positives = 51/85 (59%), Gaps = 6/85 (7%)
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKE-KPSVSSHNSNKSKSGSKARGSELAKYSKPPAK 59
KRLF+ INDLPT+FEVVTG +K K+ KPS+ +KS++G K R E S P
Sbjct: 118 KRLFSLINDLPTLFEVVTG--RKPIKDGKPSMDL--GSKSRNGVK-RSIEGQTKSTPKLM 172
Query: 60 EDDEEVDDEEEYQGECTACGENYVS 84
E+ E +D+E C +CG NY +
Sbjct: 173 EESYEDEDDEHGDTLCGSCGGNYTN 197
>At5g20510 zinc finger protein - like
Length = 232
Score = 33.5 bits (75), Expect = 0.019
Identities = 17/32 (53%), Positives = 23/32 (71%), Gaps = 4/32 (12%)
Query: 59 KEDDEEVDDEEEYQGE--CTACGENYVSASDE 88
+E +EE D++E+ GE C ACG+NY ASDE
Sbjct: 161 EEGEEEEDEDEDEHGETLCGACGDNY--ASDE 190
>At3g54540 ABC transporter - like protein
Length = 723
Score = 33.1 bits (74), Expect = 0.025
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Query: 12 TIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEY 71
++ ++ G +K K K SS KS S G +L PP+ E+D+ DEEE
Sbjct: 32 SVSAMLAGMDQKDDKPKKGSSSRTKAAPKSTSYTDGIDL-----PPSDEEDDGESDEEER 86
Query: 72 QGE 74
Q E
Sbjct: 87 QKE 89
>At4g26600 unknown protein
Length = 671
Score = 31.6 bits (70), Expect = 0.072
Identities = 21/72 (29%), Positives = 30/72 (41%), Gaps = 1/72 (1%)
Query: 18 TGSAKKQTKEKPSVSSHNSNKSKSGSKARG-SELAKYSKPPAKEDDEEVDDEEEYQGECT 76
T KQTK P + + K + + SE KP D+EE ++E E E +
Sbjct: 16 TPPLNKQTKASPLKKAAKTQKPPLKKQRKCISEKKPLKKPEVSTDEEEEEEENEQSDEGS 75
Query: 77 ACGENYVSASDE 88
G + S DE
Sbjct: 76 ESGSDLFSDGDE 87
Score = 26.2 bits (56), Expect = 3.0
Identities = 15/68 (22%), Positives = 32/68 (47%), Gaps = 3/68 (4%)
Query: 20 SAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACG 79
S ++ +E+ + S ++S S + G E +DD++ DD++E +
Sbjct: 58 STDEEEEEEENEQSDEGSESGSDLFSDGDEEGNNDSDDDDDDDDDDDDDDE---DAEPLA 114
Query: 80 ENYVSASD 87
E+++ SD
Sbjct: 115 EDFLDGSD 122
>At4g20160 Glu-rich protein
Length = 1188
Score = 31.6 bits (70), Expect = 0.072
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Query: 24 QTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACGENYV 83
Q +E S S S K K+ S+ + E + + P K+ +E +DEE + E GEN
Sbjct: 846 QGEESASHGSRESAKEKNSSQ-QDDETSTHRNPNDKKGIKEPEDEESKKVEREETGENVE 904
Query: 84 SASDE 88
AS E
Sbjct: 905 EASVE 909
>At2g23300 putative receptor-like protein kinase
Length = 773
Score = 31.6 bits (70), Expect = 0.072
Identities = 16/58 (27%), Positives = 29/58 (49%)
Query: 24 QTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACGEN 81
+ K+ S+S +S + S S + S AK+S ++ +E ++E+E GEN
Sbjct: 376 EAKDTTSLSPSSSTTTSSSSPEQSSRFAKWSCLRKNQETDETEEEDEENQRSGEIGEN 433
>At1g32530 unknown protein
Length = 711
Score = 30.8 bits (68), Expect = 0.12
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query: 30 SVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACGENYVS 84
S SSH SN + K++G +AK + K + D+E Y EC C ++ VS
Sbjct: 609 SDSSHISNNAWKPKKSQGENIAKLLEEIDKLEG-SYDNEANYDRECIICMKDEVS 662
>At1g71760 hypothetical protein
Length = 259
Score = 30.4 bits (67), Expect = 0.16
Identities = 17/58 (29%), Positives = 28/58 (47%)
Query: 15 EVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQ 72
E+ S +++ +KP S K + R E + +KP +K D +DDE E+Q
Sbjct: 160 EIFKPSFSRRSIKKPDFRSDEVVTRKKDMEERNLESERVTKPASKWDAYLIDDEGEHQ 217
>At5g54460 unknown protein
Length = 141
Score = 30.0 bits (66), Expect = 0.21
Identities = 18/62 (29%), Positives = 30/62 (48%)
Query: 9 DLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDE 68
DL E G+ + + ++K + N+SKS K G+E K K ++ E ++E
Sbjct: 75 DLQKGREKYKGATRLKKEKKKWERKNKRNQSKSPVKEEGAEPVKEEKEKEEQGTENEEEE 134
Query: 69 EE 70
EE
Sbjct: 135 EE 136
>At5g49930 putative protein
Length = 1080
Score = 30.0 bits (66), Expect = 0.21
Identities = 16/53 (30%), Positives = 28/53 (52%)
Query: 18 TGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEE 70
TG K+Q KEK S + KS +K G ++++ + K+ E+ D++E
Sbjct: 827 TGQEKQQRKEKDVSSLSQATKSIPDNKPAGEKVSRGQRGKLKKMKEKYADQDE 879
>At1g65130 hypothetical protein
Length = 1088
Score = 30.0 bits (66), Expect = 0.21
Identities = 18/49 (36%), Positives = 24/49 (48%)
Query: 20 SAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDE 68
S KKQ K K S S ++KS + A S AKED E++D+
Sbjct: 693 SEKKQEKAKKSGSKKRNHKSTKRTSASMSSHLDQDVEQAKEDSMELEDD 741
>At1g04300 unknown protein
Length = 1082
Score = 30.0 bits (66), Expect = 0.21
Identities = 14/49 (28%), Positives = 25/49 (50%)
Query: 22 KKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEE 70
+K++K+K + N NK K K A ++K + ++ +DEEE
Sbjct: 452 EKKSKKKQAKQKRNKNKGKDKRKEEKVSFATHAKDLEENQNQNQNDEEE 500
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 29.6 bits (65), Expect = 0.27
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 4/73 (5%)
Query: 19 GSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE---DDEEVDD-EEEYQGE 74
G+ K P S NS+ + S SK GSE + K KE D + +E++Q E
Sbjct: 145 GNTAKSPPVAPKKSGLNSSSTSSKSKKEGSENVRIKKASDKEIALDSASMSSAQEDHQEE 204
Query: 75 CTACGENYVSASD 87
+++ SD
Sbjct: 205 ILKVESDHLQVSD 217
>At3g15340 hypothetical protein
Length = 478
Score = 29.6 bits (65), Expect = 0.27
Identities = 18/53 (33%), Positives = 29/53 (53%), Gaps = 5/53 (9%)
Query: 16 VVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDE 68
V+T KK+T++K + + S S E + Y KP K+++EEVD+E
Sbjct: 384 VITDKRKKETRKK---AMDFNRSSAEESDVTDLEFSVYEKP--KKEEEEVDEE 431
>At2g07260 putative protein on FARE2.1 (CDS2)
Length = 300
Score = 29.6 bits (65), Expect = 0.27
Identities = 18/61 (29%), Positives = 27/61 (43%), Gaps = 5/61 (8%)
Query: 28 KPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACGENYVSASD 87
KP + K G + E K K P +E+ EE D+ +E + + E YV SD
Sbjct: 52 KPKEETEKQENPKQGDEEMEREEGKEEKVPKEENVEEHDEHDETEDQ-----EAYVILSD 106
Query: 88 E 88
+
Sbjct: 107 D 107
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.299 0.121 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,246,044
Number of Sequences: 26719
Number of extensions: 96182
Number of successful extensions: 1046
Number of sequences better than 10.0: 190
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 79
Number of HSP's that attempted gapping in prelim test: 788
Number of HSP's gapped (non-prelim): 299
length of query: 88
length of database: 11,318,596
effective HSP length: 64
effective length of query: 24
effective length of database: 9,608,580
effective search space: 230605920
effective search space used: 230605920
T: 11
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC149574.9


