

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.3 + phase: 0
(489 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
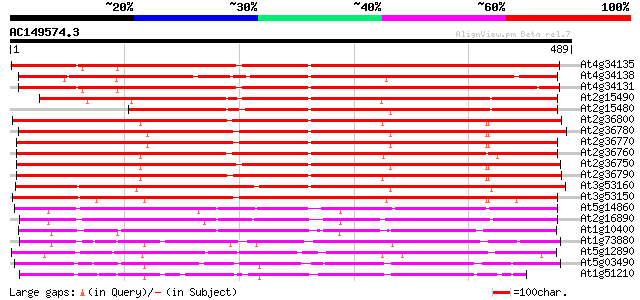
Score E
Sequences producing significant alignments: (bits) Value
At4g34135 glucosyltransferase like protein 530 e-151
At4g34138 glucosyltransferase -like protein 522 e-148
At4g34131 glucosyltransferase -like protein 518 e-147
At2g15490 putative glucosyltransferase 468 e-132
At2g15480 glucosyltransferase like protein 419 e-117
At2g36800 putative glucosyl transferase 402 e-112
At2g36780 putative glucosyl transferase 394 e-110
At2g36770 putative glucosyl transferase 393 e-109
At2g36760 putative glucosyl transferase 390 e-109
At2g36750 putative glucosyl transferase 389 e-108
At2g36790 putative glucosyl transferase 386 e-107
At3g53160 glucosyltransferase - like protein 379 e-105
At3g53150 glucosyltransferase - like protein 364 e-101
At5g14860 glucosyltransferase -like protein 281 8e-76
At2g16890 glucosyltransferase like protein 264 9e-71
At1g10400 glucosyl transferase - like protein 250 1e-66
At1g73880 putative glucosyltransferase (At1g73880) 236 2e-62
At5g12890 glucosyltransferase -like protein 228 6e-60
At5g03490 UDPG glucosyltransferase - like protein 221 9e-58
At1g51210 putative glucosyl transferase 219 3e-57
>At4g34135 glucosyltransferase like protein
Length = 483
Score = 530 bits (1366), Expect = e-151
Identities = 264/488 (54%), Positives = 349/488 (71%), Gaps = 18/488 (3%)
Query: 2 DSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHF 61
D LH++ FPFM +GH IPT+DMAKLF+S+G + TI+TT LN + K ++ K +
Sbjct: 4 DHHHRKLHVMFFPFMAYGHMIPTLDMAKLFSSRGAKSTILTTSLNSKILQKPIDTFK-NL 62
Query: 62 N---NIDIQTIKFPCVEAGLPEGCENVDSIPSVS------FVPAFFAAIRLLQQPFEELL 112
N IDIQ FPCVE GLPEGCENVD S + + FF + R + E+LL
Sbjct: 63 NPGLEIDIQIFNFPCVELGLPEGCENVDFFTSNNNDDKNEMIVKFFFSTRFFKDQLEKLL 122
Query: 113 LQQKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDL 172
+P C++ADMFFPWAT++A KF +PR+VFHGT +FSLCA C+ ++P K V+S ++
Sbjct: 123 GTTRPDCLIADMFFPWATEAAGKFNVPRLVFHGTGYFSLCAGYCIGVHKPQKRVASSSEP 182
Query: 173 FEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVY 232
F I +LPGNI +T Q+ + E+D K E+++SEV+S GV++NSFYELE+ Y
Sbjct: 183 FVIPELPGNIVITEEQIIDGDGESD-----MGKFMTEVRESEVKSSGVVLNSFYELEHDY 237
Query: 233 ADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMC 292
AD+Y+ + + WHIGP S++NR EE+ + RGK+A+ID+ ECLKWLD+K NSV+Y+
Sbjct: 238 ADFYKSCVQKRAWHIGPLSVYNRGFEEK--AERGKKANIDEAECLKWLDSKKPNSVIYVS 295
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWS 352
FGS+ F N QL EIA GLEASG +FIWVVR +D +EWLPEGFEER +GKG+IIRGW+
Sbjct: 296 FGSVAFFKNEQLFEIAAGLEASGTSFIWVVRKTKDDREEWLPEGFEERVKGKGMIIRGWA 355
Query: 353 PQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVG 412
PQV+IL+H+A G FVTHCGWNS+LEGV AG+PM+TWPV AEQFYNEKLVT+VL+TGV VG
Sbjct: 356 PQVLILDHQATGGFVTHCGWNSLLEGVAAGLPMVTWPVGAEQFYNEKLVTQVLRTGVSVG 415
Query: 413 VKKWV-MKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLN 471
K + + +GD + + V+KAV+ V+ GE A E R +AK LA MAK AVEE GSS++ LN
Sbjct: 416 ASKHMKVMMGDFISREKVDKAVREVLAGEAAEERRRRAKKLAAMAKAAVEEGGSSFNDLN 475
Query: 472 ALIEELRS 479
+ +EE S
Sbjct: 476 SFMEEFSS 483
>At4g34138 glucosyltransferase -like protein
Length = 488
Score = 522 bits (1345), Expect = e-148
Identities = 268/483 (55%), Positives = 346/483 (71%), Gaps = 27/483 (5%)
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLN-----KPPISKALEQSKIHFN 62
LH L+FPFM HGH IPT+DMAKLFA+KG + TI+TTPLN + PI K+ Q
Sbjct: 10 LHFLLFPFMAHGHMIPTLDMAKLFATKGAKSTILTTPLNAKLFFEKPI-KSFNQDNPGLE 68
Query: 63 NIDIQTIKFPCVEAGLPEGCENVDSIPSV------SFVPAFFAAIRLLQQPFEELLLQQK 116
+I IQ + FPC E GLP+GCEN D I S F A++ ++P EELL+ +
Sbjct: 69 DITIQILNFPCTELGLPDGCENTDFIFSTPDLNVGDLSQKFLLAMKYFEEPLEELLVTMR 128
Query: 117 PHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEIT 176
P C+V +MFFPW+T A KFG+PR+VFHGT +FSLCAS C++ KNV++ ++ F I
Sbjct: 129 PDCLVGNMFFPWSTKVAEKFGVPRLVFHGTGYFSLCASHCIRLP---KNVATSSEPFVIP 185
Query: 177 DLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYY 236
DLPG+I +T Q+ T E + + F K I+DSE S+GV+VNSFYELE Y+DY+
Sbjct: 186 DLPGDILITEEQVMET--EEESVMGRFMKA---IRDSERDSFGVLVNSFYELEQAYSDYF 240
Query: 237 REVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSM 296
+ + + WHIGP S+ NR EE+ + RGK+ASID+HECLKWLD+K +SV+YM FG+M
Sbjct: 241 KSFVAKRAWHIGPLSLGNRKFEEK--AERGKKASIDEHECLKWLDSKKCDSVIYMAFGTM 298
Query: 297 THFLNSQLKEIAMGLEASGHNFIWVVRTQTE--DGDEWLPEGFEERTEGKGLIIRGWSPQ 354
+ F N QL EIA GL+ SGH+F+WVV + + ++WLPEGFEE+T+GKGLIIRGW+PQ
Sbjct: 299 SSFKNEQLIEIAAGLDMSGHDFVWVVNRKGSQVEKEDWLPEGFEEKTKGKGLIIRGWAPQ 358
Query: 355 VMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK 414
V+ILEH+AIG F+THCGWNS+LEGV AG+PM+TWPV AEQFYNEKLVT+VLKTGV VGVK
Sbjct: 359 VLILEHKAIGGFLTHCGWNSLLEGVAAGLPMVTWPVGAEQFYNEKLVTQVLKTGVSVGVK 418
Query: 415 KWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALI 474
K + VGD + + VE AV+ VM GE E R +AK LAEMAK AV+E GSS +++ L+
Sbjct: 419 KMMQVVGDFISREKVEGAVREVMVGE---ERRKRAKELAEMAKNAVKEGGSSDLEVDRLM 475
Query: 475 EEL 477
EEL
Sbjct: 476 EEL 478
>At4g34131 glucosyltransferase -like protein
Length = 481
Score = 518 bits (1334), Expect = e-147
Identities = 267/482 (55%), Positives = 344/482 (70%), Gaps = 19/482 (3%)
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFN---NI 64
LH++ FPFM +GH IPT+DMAKLF+S+G + TI+TTPLN K +E+ K + N I
Sbjct: 9 LHVVFFPFMAYGHMIPTLDMAKLFSSRGAKSTILTTPLNSKIFQKPIERFK-NLNPSFEI 67
Query: 65 DIQTIKFPCVEAGLPEGCENVDSIPSVS------FVPAFFAAIRLLQQPFEELLLQQKPH 118
DIQ FPCV+ GLPEGCENVD S + FF + R + E+LL +P
Sbjct: 68 DIQIFDFPCVDLGLPEGCENVDFFTSNNNDDRQYLTLKFFKSTRFFKDQLEKLLETTRPD 127
Query: 119 CVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDL 178
C++ADMFFPWAT++A KF +PR+VFHGT +FSLC+ C++ + P V+S + F I DL
Sbjct: 128 CLIADMFFPWATEAAEKFNVPRLVFHGTGYFSLCSEYCIRVHNPQNIVASRYEPFVIPDL 187
Query: 179 PGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYRE 238
PGNI +T+ Q+ + E++ K E+K+S+V+S GVIVNSFYELE YAD+Y+
Sbjct: 188 PGNIVITQEQIADRDEESE-----MGKFMIEVKESDVKSSGVIVNSFYELEPDYADFYKS 242
Query: 239 VLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTH 298
V+ + WHIGP S++NR EE+ + RGK+ASI++ ECLKWLD+K +SV+Y+ FGS+
Sbjct: 243 VVLKRAWHIGPLSVYNRGFEEK--AERGKKASINEVECLKWLDSKKPDSVIYISFGSVAC 300
Query: 299 FLNSQLKEIAMGLEASGHNFIWVVRTQTE-DGDEWLPEGFEERTEGKGLIIRGWSPQVMI 357
F N QL EIA GLE SG NFIWVVR + +EWLPEGFEER +GKG+IIRGW+PQV+I
Sbjct: 301 FKNEQLFEIAAGLETSGANFIWVVRKNIGIEKEEWLPEGFEERVKGKGMIIRGWAPQVLI 360
Query: 358 LEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWV 417
L+H+A FVTHCGWNS+LEGV AG+PM+TWPVAAEQFYNEKLVT+VL+TGV VG KK V
Sbjct: 361 LDHQATCGFVTHCGWNSLLEGVAAGLPMVTWPVAAEQFYNEKLVTQVLRTGVSVGAKKNV 420
Query: 418 MKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEEL 477
GD + + V KAV+ V+ GEEA E R +AK LAEMAK AV E GSS++ LN+ IEE
Sbjct: 421 RTTGDFISREKVVKAVREVLVGEEADERRERAKKLAEMAKAAV-EGGSSFNDLNSFIEEF 479
Query: 478 RS 479
S
Sbjct: 480 TS 481
>At2g15490 putative glucosyltransferase
Length = 460
Score = 468 bits (1205), Expect = e-132
Identities = 245/464 (52%), Positives = 320/464 (68%), Gaps = 21/464 (4%)
Query: 27 MAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI--QTIKFPCVEAGLPEGCEN 84
MAKLFA +G + T++TTP+N + K +E K+ +++I + + FPCVE GLPEGCEN
Sbjct: 1 MAKLFARRGAKSTLLTTPINAKILEKPIEAFKVQNPDLEIGIKILNFPCVELGLPEGCEN 60
Query: 85 VDSIPSVSFVPAFFAAIRLL------QQPFEELLLQQKPHCVVADMFFPWATDSAAKFGI 138
D I S +F ++ L +Q E + KP +VADMFFPWAT+SA K G+
Sbjct: 61 RDFINSYQKSDSFDLFLKFLFSTKYMKQQLESFIETTKPSALVADMFFPWATESAEKIGV 120
Query: 139 PRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTRLQLPNTLTENDP 198
PR+VFHGTS F+LC S M+ ++P+K V+S + F I LPG+I +T Q N E P
Sbjct: 121 PRLVFHGTSSFALCCSYNMRIHKPHKKVASSSTPFVIPGLPGDIVITEDQA-NVTNEETP 179
Query: 199 ISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIKEWHIGPFSIHNRNKE 258
F K ++E+++SE S+GV+VNSFYELE+ YAD+YR + K WHIGP S+ NR
Sbjct: 180 ----FGKFWKEVRESETSSFGVLVNSFYELESSYADFYRSFVAKKAWHIGPLSLSNRGIA 235
Query: 259 EEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNF 318
E+ + RGK+A+ID+ ECLKWLD+K SVVY+ FGS T N QL EIA GLE SG NF
Sbjct: 236 EK--AGRGKKANIDEQECLKWLDSKTPGSVVYLSFGSGTGLPNEQLLEIAFGLEGSGQNF 293
Query: 319 IWVV-----RTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWN 373
IWVV + T + ++WLP+GFEER +GKGLIIRGW+PQV+IL+H+AIG FVTHCGWN
Sbjct: 294 IWVVSKNENQVGTGENEDWLPKGFEERNKGKGLIIRGWAPQVLILDHKAIGGFVTHCGWN 353
Query: 374 SVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAV 433
S LEG+ AG+PM+TWP+ AEQFYNEKL+T+VL+ GV VG + V K G + VEKAV
Sbjct: 354 STLEGIAAGLPMVTWPMGAEQFYNEKLLTKVLRIGVNVGATELVKK-GKLISRAQVEKAV 412
Query: 434 KRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEEL 477
+ V+ GE+A E R +AK L EMAK AVEE GSSY+ +N +EEL
Sbjct: 413 REVIGGEKAEERRLRAKELGEMAKAAVEEGGSSYNDVNKFMEEL 456
>At2g15480 glucosyltransferase like protein
Length = 372
Score = 419 bits (1076), Expect = e-117
Identities = 212/376 (56%), Positives = 270/376 (71%), Gaps = 10/376 (2%)
Query: 104 LQQPFEELLLQQKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPY 163
++Q E + KP +VADMFFPWAT+SA K G+PR+VFHGTSFFSLC S M+ ++P+
Sbjct: 1 MKQQLESFIETTKPSALVADMFFPWATESAEKLGVPRLVFHGTSFFSLCCSYNMRIHKPH 60
Query: 164 KNVSSDTDLFEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVN 223
K V++ + F I LPG+I +T Q N E P+ K +E+++SE S+GV+VN
Sbjct: 61 KKVATSSTPFVIPGLPGDIVITEDQA-NVAKEETPMG----KFMKEVRESETNSFGVLVN 115
Query: 224 SFYELENVYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTK 283
SFYELE+ YAD+YR + + WHIGP S+ NR E+ + RGK+A+ID+ ECLKWLD+K
Sbjct: 116 SFYELESAYADFYRSFVAKRAWHIGPLSLSNRELGEK--ARRGKKANIDEQECLKWLDSK 173
Query: 284 NINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGD--EWLPEGFEERT 341
SVVY+ FGS T+F N QL EIA GLE SG +FIWVVR GD EWLPEGF+ERT
Sbjct: 174 TPGSVVYLSFGSGTNFTNDQLLEIAFGLEGSGQSFIWVVRKNENQGDNEEWLPEGFKERT 233
Query: 342 EGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLV 401
GKGLII GW+PQV+IL+H+AIG FVTHCGWNS +EG+ AG+PM+TWP+ AEQFYNEKL+
Sbjct: 234 TGKGLIIPGWAPQVLILDHKAIGGFVTHCGWNSAIEGIAAGLPMVTWPMGAEQFYNEKLL 293
Query: 402 TEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVE 461
T+VL+ GV VG + V K G + VEKAV+ V+ GE+A E R AK L EMAK AVE
Sbjct: 294 TKVLRIGVNVGATELVKK-GKLISRAQVEKAVREVIGGEKAEERRLWAKKLGEMAKAAVE 352
Query: 462 EDGSSYSQLNALIEEL 477
E GSSY+ +N +EEL
Sbjct: 353 EGGSSYNDVNKFMEEL 368
>At2g36800 putative glucosyl transferase
Length = 495
Score = 402 bits (1034), Expect = e-112
Identities = 213/491 (43%), Positives = 318/491 (64%), Gaps = 17/491 (3%)
Query: 3 SQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFN 62
++S+PLH ++FPFM GH IP +D+A+L A +GV +TIVTTP N L ++
Sbjct: 6 TKSSPLHFVLFPFMAQGHMIPMVDIARLLAQRGVIITIVTTPHNAARFKNVLNRAIESGL 65
Query: 63 NIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKPHCV 120
I++ +KFP +EAGL EG EN+DS+ ++ + FF A+ L++P ++L+ + +P C+
Sbjct: 66 PINLVQVKFPYLEAGLQEGQENIDSLDTMERMIPFFKAVNFLEEPVQKLIEEMNPRPSCL 125
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSL-CASQCMKKYQPYKNVSSDTDLFEITDLP 179
++D P+ + A KF IP+I+FHG F L C K + N+ SD +LF + D P
Sbjct: 126 ISDFCLPYTSKIAKKFNIPKILFHGMGCFCLLCMHVLRKNREILDNLKSDKELFTVPDFP 185
Query: 180 GNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREV 239
++ TR Q+P E + + +F+ + ++ SYGVIVNSF ELE YA Y+EV
Sbjct: 186 DRVEFTRTQVP---VETYVPAGDWKDIFDGMVEANETSYGVIVNSFQELEPAYAKDYKEV 242
Query: 240 LGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHF 299
K W IGP S+ N+ ++ + RG ++ ID+ ECLKWLD+K SV+Y+C GS+ +
Sbjct: 243 RSGKAWTIGPVSLCNKVGADK--AERGNKSDIDQDECLKWLDSKKHGSVLYVCLGSICNL 300
Query: 300 LNSQLKEIAMGLEASGHNFIWVVR--TQTEDGDEWLPE-GFEERTEGKGLIIRGWSPQVM 356
SQLKE+ +GLE S FIWV+R + ++ EW E GFE+R + +GL+I+GWSPQ++
Sbjct: 301 PLSQLKELGLGLEESQRPFIWVIRGWEKYKELVEWFSESGFEDRIQDRGLLIKGWSPQML 360
Query: 357 ILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK-- 414
IL H ++G F+THCGWNS LEG+ AG+P++TWP+ A+QF NEKLV EVLK GV GV+
Sbjct: 361 ILSHPSVGGFLTHCGWNSTLEGITAGLPLLTWPLFADQFCNEKLVVEVLKAGVRSGVEQP 420
Query: 415 -KW--VMKVGDNVEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQL 470
KW K+G V+ + V+KAV+ +M E ++A E R +AK L + A KAVEE GSS+S +
Sbjct: 421 MKWGEEEKIGVLVDKEGVKKAVEELMGESDDAKERRRRAKELGDSAHKAVEEGGSSHSNI 480
Query: 471 NALIEELRSLS 481
+ L++++ L+
Sbjct: 481 SFLLQDIMELA 491
>At2g36780 putative glucosyl transferase
Length = 496
Score = 394 bits (1011), Expect = e-110
Identities = 209/490 (42%), Positives = 311/490 (62%), Gaps = 18/490 (3%)
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQ 67
LH ++FPFM GH IP ID+A+L A +GV +TIVTTP N L ++ I+I
Sbjct: 13 LHFVLFPFMAQGHMIPMIDIARLLAQRGVTITIVTTPHNAARFKNVLNRAIESGLAINIL 72
Query: 68 TIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPH--CVVADMF 125
+KFP E GLPEG EN+DS+ S + FF A+ LL+ P +L+ + KP C+++D
Sbjct: 73 HVKFPYQEFGLPEGKENIDSLDSTELMVPFFKAVNLLEDPVMKLMEEMKPRPSCLISDWC 132
Query: 126 FPWATDSAAKFGIPRIVFHGTSFFSL-CASQCMKKYQPYKNVSSDTDLFEITDLPGNIKM 184
P+ + A F IP+IVFHG F+L C + + +NV SD + F + P ++
Sbjct: 133 LPYTSIIAKNFNIPKIVFHGMGCFNLLCMHVLRRNLEILENVKSDEEYFLVPSFPDRVEF 192
Query: 185 TRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIKE 244
T+LQLP S + ++ +E+ +E SYGVIVN+F ELE Y Y+E + K
Sbjct: 193 TKLQLPVKANA----SGDWKEIMDEMVKAEYTSYGVIVNTFQELEPPYVKDYKEAMDGKV 248
Query: 245 WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQL 304
W IGP S+ N+ ++ + RG +A+ID+ ECL+WLD+K SV+Y+C GS+ + SQL
Sbjct: 249 WSIGPVSLCNKAGADK--AERGSKAAIDQDECLQWLDSKEEGSVLYVCLGSICNLPLSQL 306
Query: 305 KEIAMGLEASGHNFIWVVRTQTEDGD--EWLPE-GFEERTEGKGLIIRGWSPQVMILEHE 361
KE+ +GLE S +FIWV+R + + EW+ E GFEER + +GL+I+GW+PQV+IL H
Sbjct: 307 KELGLGLEESRRSFIWVIRGSEKYKELFEWMLESGFEERIKERGLLIKGWAPQVLILSHP 366
Query: 362 AIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK---KW-- 416
++G F+THCGWNS LEG+ +G+P+ITWP+ +QF N+KLV +VLK GV GV+ KW
Sbjct: 367 SVGGFLTHCGWNSTLEGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEEVMKWGE 426
Query: 417 VMKVGDNVEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIE 475
K+G V+ + V+KAV+ +M + ++A E R + K L E+A KAVE+ GSS+S + L++
Sbjct: 427 EDKIGVLVDKEGVKKAVEELMGDSDDAKERRRRVKELGELAHKAVEKGGSSHSNITLLLQ 486
Query: 476 ELRSLSHHQH 485
++ L+ ++
Sbjct: 487 DIMQLAQFKN 496
>At2g36770 putative glucosyl transferase
Length = 496
Score = 393 bits (1010), Expect = e-109
Identities = 209/483 (43%), Positives = 303/483 (62%), Gaps = 18/483 (3%)
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PLH ++FPFM GH IP ID+A+L A +G VTIVTT N L ++ I+I
Sbjct: 12 PLHFILFPFMAQGHMIPMIDIARLLAQRGATVTIVTTRYNAGRFENVLSRAMESGLPINI 71
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPH--CVVADM 124
+ FP E GLPEG EN+DS S+ + FF A+ +L+ P +L+ + KP C+++D+
Sbjct: 72 VHVNFPYQEFGLPEGKENIDSYDSMELMVPFFQAVNMLEDPVMKLMEEMKPRPSCIISDL 131
Query: 125 FFPWATDSAAKFGIPRIVFHGTSFFSL-CASQCMKKYQPYKNVSSDTDLFEITDLPGNIK 183
P+ + A KF IP+IVFHGT F+L C + + KN+ SD D F + P ++
Sbjct: 132 LLPYTSKIARKFSIPKIVFHGTGCFNLLCMHVLRRNLEILKNLKSDKDYFLVPSFPDRVE 191
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIK 243
T+ Q+P T S + +E+ ++E SYGVIVN+F ELE Y Y + K
Sbjct: 192 FTKPQVPVETTA----SGDWKAFLDEMVEAEYTSYGVIVNTFQELEPAYVKDYTKARAGK 247
Query: 244 EWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQ 303
W IGP S+ N+ ++ + RG +A+ID+ ECL+WLD+K SV+Y+C GS+ + SQ
Sbjct: 248 VWSIGPVSLCNKAGADK--AERGNQAAIDQDECLQWLDSKEDGSVLYVCLGSICNLPLSQ 305
Query: 304 LKEIAMGLEASGHNFIWVVRTQTEDGD--EWLPE-GFEERTEGKGLIIRGWSPQVMILEH 360
LKE+ +GLE S +FIWV+R + + EW+ E GFEER + +GL+I+GWSPQV+IL H
Sbjct: 306 LKELGLGLEKSQRSFIWVIRGWEKYNELYEWMMESGFEERIKERGLLIKGWSPQVLILSH 365
Query: 361 EAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK---KW- 416
++G F+THCGWNS LEG+ +G+P+ITWP+ +QF N+KLV +VLK GV GV+ KW
Sbjct: 366 PSVGGFLTHCGWNSTLEGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEEVMKWG 425
Query: 417 -VMKVGDNVEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALI 474
K+G V+ + V+KAV+ +M ++A E R + K L E A KAVEE GSS+S + L+
Sbjct: 426 EEEKIGVLVDKEGVKKAVEELMGASDDAKERRRRVKELGESAHKAVEEGGSSHSNITYLL 485
Query: 475 EEL 477
+++
Sbjct: 486 QDI 488
>At2g36760 putative glucosyl transferase
Length = 496
Score = 390 bits (1003), Expect = e-109
Identities = 204/484 (42%), Positives = 313/484 (64%), Gaps = 20/484 (4%)
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PLH ++FPFM GH IP +D+A++ A +GV +TIVTTP N L ++ +I +
Sbjct: 12 PLHFVLFPFMAQGHMIPMVDIARILAQRGVTITIVTTPHNAARFKDVLNRAIQSGLHIRV 71
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKPHCVVADM 124
+ +KFP EAGL EG ENVD + S+ + FF A+ +L+ P +L+ ++ KP C+++D
Sbjct: 72 EHVKFPFQEAGLQEGQENVDFLDSMELMVHFFKAVNMLENPVMKLMEEMKPKPSCLISDF 131
Query: 125 FFPWATDSAAKFGIPRIVFHGTSFFSLCASQCM-KKYQPYKNVSSDTDLFEITDLPGNIK 183
P+ + A +F IP+IVFHG S F L + + + + + SD + F + P ++
Sbjct: 132 CLPYTSKIAKRFNIPKIVFHGVSCFCLLSMHILHRNHNILHALKSDKEYFLVPSFPDRVE 191
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIK 243
T+LQ+ T S + ++ +E D++ SYGVIVN+F +LE+ Y Y E K
Sbjct: 192 FTKLQV----TVKTNFSGDWKEIMDEQVDADDTSYGVIVNTFQDLESAYVKNYTEARAGK 247
Query: 244 EWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQ 303
W IGP S+ N+ E++ + RG +A+ID+ EC+KWLD+K++ SV+Y+C GS+ + +Q
Sbjct: 248 VWSIGPVSLCNKVGEDK--AERGNKAAIDQDECIKWLDSKDVESVLYVCLGSICNLPLAQ 305
Query: 304 LKEIAMGLEASGHNFIWVVRT--QTEDGDEWLPE-GFEERTEGKGLIIRGWSPQVMILEH 360
L+E+ +GLEA+ FIWV+R + + EW+ E GFEERT+ + L+I+GWSPQ++IL H
Sbjct: 306 LRELGLGLEATKRPFIWVIRGGGKYHELAEWILESGFEERTKERSLLIKGWSPQMLILSH 365
Query: 361 EAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKV 420
A+G F+THCGWNS LEG+ +GVP+ITWP+ +QF N+KL+ +VLK GV VGV++ VMK
Sbjct: 366 PAVGGFLTHCGWNSTLEGITSGVPLITWPLFGDQFCNQKLIVQVLKAGVSVGVEE-VMKW 424
Query: 421 GDN------VEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNAL 473
G+ V+ + V+KAV +M E +EA E R + + L E+A KAVEE GSS+S + L
Sbjct: 425 GEEESIGVLVDKEGVKKAVDEIMGESDEAKERRKRVRELGELAHKAVEEGGSSHSNIIFL 484
Query: 474 IEEL 477
++++
Sbjct: 485 LQDI 488
>At2g36750 putative glucosyl transferase
Length = 491
Score = 389 bits (1000), Expect = e-108
Identities = 205/486 (42%), Positives = 305/486 (62%), Gaps = 19/486 (3%)
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PLH ++FPFM GH IP +D+A+L A +GV +TIVTTP N L ++ I++
Sbjct: 8 PLHFVLFPFMAQGHMIPMVDIARLLAQRGVTITIVTTPQNAGRFKNVLSRAIQSGLPINL 67
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKPHCVVADM 124
+KFP E+G PEG EN+D + S+ FF A LL++P E+LL +Q +P+C++ADM
Sbjct: 68 VQVKFPSQESGSPEGQENLDLLDSLGASLTFFKAFSLLEEPVEKLLKEIQPRPNCIIADM 127
Query: 125 FFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPY-KNVSSDTDLFEITDLPGNIK 183
P+ A GIP+I+FHG F+L + M + + + + SD + F I + P ++
Sbjct: 128 CLPYTNRIAKNLGIPKIIFHGMCCFNLLCTHIMHQNHEFLETIESDKEYFPIPNFPDRVE 187
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIK 243
T+ QLP L D + + + + + SYGVIVN+F ELE Y Y++V K
Sbjct: 188 FTKSQLPMVLVAGD-----WKDFLDGMTEGDNTSYGVIVNTFEELEPAYVRDYKKVKAGK 242
Query: 244 EWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQ 303
W IGP S+ N+ E++ + RG +A ID+ EC+KWLD+K SV+Y+C GS+ + SQ
Sbjct: 243 IWSIGPVSLCNKLGEDQ--AERGNKADIDQDECIKWLDSKEEGSVLYVCLGSICNLPLSQ 300
Query: 304 LKEIAMGLEASGHNFIWVVRTQTEDGD--EWLPE-GFEERTEGKGLIIRGWSPQVMILEH 360
LKE+ +GLE S FIWV+R + + EW+ E G++ER + +GL+I GWSPQ++IL H
Sbjct: 301 LKELGLGLEESQRPFIWVIRGWEKYNELLEWISESGYKERIKERGLLITGWSPQMLILTH 360
Query: 361 EAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK---KW- 416
A+G F+THCGWNS LEG+ +GVP++TWP+ +QF NEKL ++LK GV GV+ +W
Sbjct: 361 PAVGGFLTHCGWNSTLEGITSGVPLLTWPLFGDQFCNEKLAVQILKAGVRAGVEESMRWG 420
Query: 417 -VMKVGDNVEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALI 474
K+G V+ + V+KAV+ +M + +A E R + K L E+A KAVEE GSS+S + L+
Sbjct: 421 EEEKIGVLVDKEGVKKAVEELMGDSNDAKERRKRVKELGELAHKAVEEGGSSHSNITFLL 480
Query: 475 EELRSL 480
+++ L
Sbjct: 481 QDIMQL 486
>At2g36790 putative glucosyl transferase
Length = 495
Score = 386 bits (991), Expect = e-107
Identities = 205/487 (42%), Positives = 310/487 (63%), Gaps = 18/487 (3%)
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PLH ++FPFM GH IP +D+A+L A +GV +TIVTTP N L ++ I++
Sbjct: 11 PLHFVLFPFMAQGHMIPMVDIARLLAQRGVLITIVTTPHNAARFKNVLNRAIESGLPINL 70
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKPHCVVADM 124
+KFP EAGL EG EN+D + ++ + +FF A+ LL++P + L+ + +P C+++DM
Sbjct: 71 VQVKFPYQEAGLQEGQENMDLLTTMEQITSFFKAVNLLKEPVQNLIEEMSPRPSCLISDM 130
Query: 125 FFPWATDSAAKFGIPRIVFHGTSFFSL-CASQCMKKYQPYKNVSSDTDLFEITDLPGNIK 183
+ ++ A KF IP+I+FHG F L C + K + N+ SD + F + P ++
Sbjct: 131 CLSYTSEIAKKFKIPKILFHGMGCFCLLCVNVLRKNREILDNLKSDKEYFIVPYFPDRVE 190
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIK 243
TR Q+P + + ++ E++ +++ SYGVIVNSF ELE YA ++E K
Sbjct: 191 FTRPQVP----VETYVPAGWKEILEDMVEADKTSYGVIVNSFQELEPAYAKDFKEARSGK 246
Query: 244 EWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQ 303
W IGP S+ N+ ++ + RG ++ ID+ ECL+WLD+K SV+Y+C GS+ + SQ
Sbjct: 247 AWTIGPVSLCNKVGVDK--AERGNKSDIDQDECLEWLDSKEPGSVLYVCLGSICNLPLSQ 304
Query: 304 LKEIAMGLEASGHNFIWVVR--TQTEDGDEWLPE-GFEERTEGKGLIIRGWSPQVMILEH 360
L E+ +GLE S FIWV+R + ++ EW E GFE+R + +GL+I+GWSPQ++IL H
Sbjct: 305 LLELGLGLEESQRPFIWVIRGWEKYKELVEWFSESGFEDRIQDRGLLIKGWSPQMLILSH 364
Query: 361 EAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK---KW- 416
++G F+THCGWNS LEG+ AG+PM+TWP+ A+QF NEKLV ++LK GV VK KW
Sbjct: 365 PSVGGFLTHCGWNSTLEGITAGLPMLTWPLFADQFCNEKLVVQILKVGVSAEVKEVMKWG 424
Query: 417 -VMKVGDNVEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALI 474
K+G V+ + V+KAV+ +M E ++A E R +AK L E A KAVEE GSS+S + L+
Sbjct: 425 EEEKIGVLVDKEGVKKAVEELMGESDDAKERRRRAKELGESAHKAVEEGGSSHSNITFLL 484
Query: 475 EELRSLS 481
+++ L+
Sbjct: 485 QDIMQLA 491
>At3g53160 glucosyltransferase - like protein
Length = 490
Score = 379 bits (972), Expect = e-105
Identities = 206/492 (41%), Positives = 309/492 (61%), Gaps = 20/492 (4%)
Query: 6 NPLHILVFPFMGHGHTIPTIDMAKLFASK-GVRVTIVTTPLNKPPISKALEQSKIHFNNI 64
+PLH +V PFM GH IP +D+++L + + GV V I+TT N I +L S + F I
Sbjct: 5 DPLHFVVIPFMAQGHMIPLVDISRLLSQRQGVTVCIITTTQNVAKIKTSLSFSSL-FATI 63
Query: 65 DIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFE---ELLLQQKPHCVV 121
+I +KF + GLPEGCE++D + S+ + FF A L++ E E ++Q +P C++
Sbjct: 64 NIVEVKFLSQQTGLPEGCESLDMLASMGDMVKFFDAANSLEEQVEKAMEEMVQPRPSCII 123
Query: 122 ADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGN 181
DM P+ + A KF IP+++FHG S FSL + Q +++ K + S+ + F++ LP
Sbjct: 124 GDMSLPFTSRLAKKFKIPKLIFHGFSCFSLMSIQVVRESGILKMIESNDEYFDLPGLPDK 183
Query: 182 IKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLG 241
++ T+ Q+ + +S AK+ E DS YGVIVN+F ELE YA YR+
Sbjct: 184 VEFTKPQVSVLQPVEGNMKESTAKIIEADNDS----YGVIVNTFEELEVDYAREYRKARA 239
Query: 242 IKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLN 301
K W +GP S+ NR ++ + RG +ASI + +CL+WLD++ SV+Y+C GS+ +
Sbjct: 240 GKVWCVGPVSLCNRLGLDK--AKRGDKASIGQDQCLQWLDSQETGSVLYVCLGSLCNLPL 297
Query: 302 SQLKEIAMGLEASGHNFIWVVRTQTEDGD--EWLPE-GFEERTEGKGLIIRGWSPQVMIL 358
+QLKE+ +GLEAS FIWV+R + GD W+ + GFEER + +GL+I+GW+PQV IL
Sbjct: 298 AQLKELGLGLEASNKPFIWVIREWGKYGDLANWMQQSGFEERIKDRGLVIKGWAPQVFIL 357
Query: 359 EHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVM 418
H +IG F+THCGWNS LEG+ AGVP++TWP+ AEQF NEKLV ++LK G+ +GV+K +
Sbjct: 358 SHASIGGFLTHCGWNSTLEGITAGVPLLTWPLFAEQFLNEKLVVQILKAGLKIGVEKLMK 417
Query: 419 -----KVGDNVEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNA 472
++G V + V KAV +M + EEA E R K L+++A KA+E+ GSS S +
Sbjct: 418 YGKEEEIGAMVSRECVRKAVDELMGDSEEAEERRRKVTELSDLANKALEKGGSSDSNITL 477
Query: 473 LIEELRSLSHHQ 484
LI+++ S +Q
Sbjct: 478 LIQDIMEQSQNQ 489
>At3g53150 glucosyltransferase - like protein
Length = 507
Score = 364 bits (934), Expect = e-101
Identities = 201/499 (40%), Positives = 302/499 (60%), Gaps = 29/499 (5%)
Query: 3 SQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFN 62
S++ LH ++ P M GH IP +D++K+ A +G VTIVTTP N +K ++++++ +
Sbjct: 7 SKAKRLHFVLIPLMAQGHLIPMVDISKILARQGNIVTIVTTPQNASRFAKTVDRARLE-S 65
Query: 63 NIDIQTIKFPCV--EAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQK--PH 118
++I +KFP E GLP+ CE +D++PS + F+ A+ LQ+P E L QQ P
Sbjct: 66 GLEINVVKFPIPYKEFGLPKDCETLDTLPSKDLLRRFYDAVDKLQEPMERFLEQQDIPPS 125
Query: 119 CVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDL 178
C+++D W + +A +F IPRIVFHG FSL +S + + P+ +VSS + F I +
Sbjct: 126 CIISDKCLFWTSRTAKRFKIPRIVFHGMCCFSLLSSHNIHLHSPHLSVSSAVEPFPIPGM 185
Query: 179 PGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYRE 238
P I++ R QLP + + + E++++SE ++GVIVNSF ELE YA+ Y E
Sbjct: 186 PHRIEIARAQLPGAFEK----LANMDDVREKMRESESEAFGVIVNSFQELEPGYAEAYAE 241
Query: 239 VLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTH 298
+ K W +GP S+ N + +I + ECL++LD+ SV+Y+ GS+
Sbjct: 242 AINKKVWFVGPVSLCNDRMADLFDRGSNGNIAISETECLQFLDSMRPRSVLYVSLGSLCR 301
Query: 299 FLNSQLKEIAMGLEASGHNFIWVVRTQTE---DGDEWLP-EGFEERTEGKGLIIRGWSPQ 354
+ +QL E+ +GLE SG FIWV++T+ + + DEWL E FEER G+G++I+GWSPQ
Sbjct: 302 LIPNQLIELGLGLEESGKPFIWVIKTEEKHMIELDEWLKRENFEERVRGRGIVIKGWSPQ 361
Query: 355 VMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK 414
MIL H + G F+THCGWNS +E + GVPMITWP+ AEQF NEKL+ EVL GV VGV+
Sbjct: 362 AMILSHGSTGGFLTHCGWNSTIEAICFGVPMITWPLFAEQFLNEKLIVEVLNIGVRVGVE 421
Query: 415 ---KW--VMKVGDNVEWDAVEKAVKRVMEGE-----------EAYEMRNKAKMLAEMAKK 458
+W ++G V+ +V KA+K +M+ + E R + + LA MAKK
Sbjct: 422 IPVRWGDEERLGVLVKKPSVVKAIKLLMDQDCQRVDENDDDNEFVRRRRRIQELAVMAKK 481
Query: 459 AVEEDGSSYSQLNALIEEL 477
AVEE GSS ++ LI+++
Sbjct: 482 AVEEKGSSSINVSILIQDV 500
>At5g14860 glucosyltransferase -like protein
Length = 492
Score = 281 bits (718), Expect = 8e-76
Identities = 180/492 (36%), Positives = 268/492 (53%), Gaps = 39/492 (7%)
Query: 5 SNPLHILVFPFMGHGHTIPTIDMAKLFA-----------SKGVRVTIVTTPLNKPPISKA 53
S+ H ++FP+M GHTIP + A+L + VT+ TTP N+P +S
Sbjct: 4 SSSHHAVLFPYMSKGHTIPLLQFARLLLRHRRIVSVDDEEPTISVTVFTTPKNQPFVSNF 63
Query: 54 LEQSKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFE-ELL 112
L ++I + ++ FP AG+P G E+ D +PS+S F A + LQ FE EL
Sbjct: 64 LSDVA---SSIKVISLPFPENIAGIPPGVESTDMLPSISLYVPFTRATKSLQPFFEAELK 120
Query: 113 LQQKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPY---KNVSSD 169
+K +V+D F W ++SAAKF IPR+ F+G + ++ + ++ + ++V SD
Sbjct: 121 NLEKVSFMVSDGFLWWTSESAAKFEIPRLAFYGMNSYASAMCSAISVHELFTKPESVKSD 180
Query: 170 TDLFEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE 229
T+ + D P I + + + LTE D +F L + + ++ +S GVIVNSFYELE
Sbjct: 181 TEPVTVPDFPW-ICVKKCEFDPVLTEPDQSDPAFELLIDHLMSTK-KSRGVIVNSFYELE 238
Query: 230 NVYADY-YREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINS- 287
+ + DY R+ K W +GP + N K E DK + + WLD K
Sbjct: 239 STFVDYRLRDNDEPKPWCVGPLCLVNPPKPES-----------DKPDWIHWLDRKLEERC 287
Query: 288 -VVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGL 346
V+Y+ FG+ N QLKEIA+GLE S NF+WV R E+ L GFE+R + G+
Sbjct: 288 PVMYVAFGTQAEISNEQLKEIALGLEDSKVNFLWVTRKDLEEVTGGL--GFEKRVKEHGM 345
Query: 347 IIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLK 406
I+R W Q IL H+++ F++HCGWNS E + AGVP++ WP+ AEQ N KLV E LK
Sbjct: 346 IVRDWVDQWEILSHKSVKGFLSHCGWNSAQESICAGVPLLAWPMMAEQPLNAKLVVEELK 405
Query: 407 TGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEE-DGS 465
GV + + + V V + + + VK++MEGE K A+MAKKA+ + GS
Sbjct: 406 IGVRIETED--VSVKGFVTREELSRKVKQLMEGEMGKTTMKNVKEYAKMAKKAMAQGTGS 463
Query: 466 SYSQLNALIEEL 477
S+ L++L+EEL
Sbjct: 464 SWKSLDSLLEEL 475
>At2g16890 glucosyltransferase like protein
Length = 478
Score = 264 bits (674), Expect = 9e-71
Identities = 170/481 (35%), Positives = 250/481 (51%), Gaps = 34/481 (7%)
Query: 9 HILVFPFMGHGHTIPTIDMAKLFA-----SKGVRVTIVTTPLNKPPISKALEQSKIHFNN 63
H+++FPFM GH IP + +L + VT+ TTP N+P IS L +
Sbjct: 9 HVVLFPFMSKGHIIPLLQFGRLLLRHHRKEPTITVTVFTTPKNQPFISDFLSDTP----E 64
Query: 64 IDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQ-QKPHCVVA 122
I + ++ FP G+P G EN + +PS+S F A +LLQ FEE L K +V+
Sbjct: 65 IKVISLPFPENITGIPPGVENTEKLPSMSLFVPFTRATKLLQPFFEETLKTLPKVSFMVS 124
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSS--DTDLFEITDLPG 180
D F W ++SAAKF IPR V +G + +S S + K++ + S DT+ + D P
Sbjct: 125 DGFLWWTSESAAKFNIPRFVSYGMNSYSAAVSISVFKHELFTEPESKSDTEPVTVPDFPW 184
Query: 181 NIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVL 240
IK+ + + TE + + ++IK S S+G +VNSFYELE+ + DY
Sbjct: 185 -IKVKKCDFDHGTTEPEESGAALELSMDQIK-STTTSHGFLVNSFYELESAFVDYNNNSG 242
Query: 241 GI-KEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINS--VVYMCFGSMT 297
K W +GP + + K+ K + WLD K V+Y+ FG+
Sbjct: 243 DKPKSWCVGPLCLTDPPKQGSA-----------KPAWIHWLDQKREEGRPVLYVAFGTQA 291
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMI 357
N QL E+A GLE S NF+WV R +D +E + EGF +R G+I+R W Q I
Sbjct: 292 EISNKQLMELAFGLEDSKVNFLWVTR---KDVEEIIGEGFNDRIRESGMIVRDWVDQWEI 348
Query: 358 LEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWV 417
L HE++ F++HCGWNS E + GVP++ WP+ AEQ N K+V E +K GV V +
Sbjct: 349 LSHESVKGFLSHCGWNSAQESICVGVPLLAWPMMAEQPLNAKMVVEEIKVGVRVETEDGS 408
Query: 418 MKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKA-VEEDGSSYSQLNALIEE 476
+K V + + +K +MEGE R K ++MAK A VE GSS+ L+ +++E
Sbjct: 409 VK--GFVTREELSGKIKELMEGETGKTARKNVKEYSKMAKAALVEGTGSSWKNLDMILKE 466
Query: 477 L 477
L
Sbjct: 467 L 467
>At1g10400 glucosyl transferase - like protein
Length = 467
Score = 250 bits (639), Expect = 1e-66
Identities = 167/481 (34%), Positives = 259/481 (53%), Gaps = 37/481 (7%)
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASKG----VRVTIVTTPLNKPPISKALEQSKIHFNN 63
+H+++FP++ GH IP + +A+L S + VT+ TTPLN+P I +L +K
Sbjct: 6 VHVVLFPYLSKGHMIPMLQLARLLLSHSFAGDISVTVFTTPLNRPFIVDSLSGTKA---- 61
Query: 64 IDIQTIKFPCVEAGLPEGCENVDSIPSVS---FVPAFFAAIRLLQQPFE-ELLLQQKPHC 119
I + FP +P G E D +P++S FVP F A + +Q FE EL+ +
Sbjct: 62 -TIVDVPFPDNVPEIPPGVECTDKLPALSSSLFVP-FTRATKSMQADFERELMSLPRVSF 119
Query: 120 VVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLP 179
+V+D F W +SA K G PR+VF G + S + + Q NV S+T+ + + P
Sbjct: 120 MVSDGFLWWTQESARKLGFPRLVFFGMNCASTVICDSVFQNQLLSNVKSETEPVSVPEFP 179
Query: 180 GNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREV 239
IK+ + + + + KL + S +S G+I N+F +LE V+ D+Y+
Sbjct: 180 W-IKVRKCDFVKDMFDPKTTTDPGFKLILDQVTSMNQSQGIIFNTFDDLEPVFIDFYKRK 238
Query: 240 LGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNIN--SVVYMCFGSMT 297
+K W +GP N ++E+ E + K +KWLD K +V+Y+ FGS
Sbjct: 239 RKLKLWAVGPLCYVNNFLDDEV------EEKV-KPSWMKWLDEKRDKGCNVLYVAFGSQA 291
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRG-WSPQVM 356
QL+EIA+GLE S NF+WVV+ G+E + +GFEER +G+++R W Q
Sbjct: 292 EISREQLEEIALGLEESKVNFLWVVK-----GNE-IGKGFEERVGERGMMVRDEWVDQRK 345
Query: 357 ILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKW 416
ILEHE++ F++HCGWNS+ E + + VP++ +P+AAEQ N LV E L+ ++
Sbjct: 346 ILEHESVRGFLSHCGWNSLTESICSEVPILAFPLAAEQPLNAILVVEELRV-----AERV 400
Query: 417 VMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEED-GSSYSQLNALIE 475
V V + + + VK +MEGE+ E+R + +MAKKA+EE GSS L+ LI
Sbjct: 401 VAASEGVVRREEIAEKVKELMEGEKGKELRRNVEAYGKMAKKALEEGIGSSRKNLDNLIN 460
Query: 476 E 476
E
Sbjct: 461 E 461
>At1g73880 putative glucosyltransferase (At1g73880)
Length = 473
Score = 236 bits (603), Expect = 2e-62
Identities = 161/488 (32%), Positives = 241/488 (48%), Gaps = 48/488 (9%)
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASKG---VRVTIVTTPLNKPPISKALEQSKIHFNNID 65
H+L+FPF GH IP +D A +G +++T++ TP N P +S L NI+
Sbjct: 14 HVLIFPFPAQGHMIPLLDFTHRLALRGGAALKITVLVTPKNLPFLSPLLSAVV----NIE 69
Query: 66 IQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQK--PHCVVAD 123
+ FP +P G ENV +P F P A+ L P + P +V+D
Sbjct: 70 PLILPFPS-HPSIPSGVENVQDLPPSGF-PLMIHALGNLHAPLISWITSHPSPPVAIVSD 127
Query: 124 MFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYK-NVSSDTDLFEITDLPGNI 182
F W + GIPR F ++ + C + P K N D ++ +P
Sbjct: 128 FFLGWTKN----LGIPRFDFSPSAAITCCILNTLWIEMPTKINEDDDNEILHFPKIPNCP 183
Query: 183 KMTRLQLPN---TLTENDPISQSFAKLFEEIKDS---EVRSYGVIVNSFYELENVYADYY 236
K Q+ + + DP +E I+DS V S+G++VNSF +E VY ++
Sbjct: 184 KYRFDQISSLYRSYVHGDPA-------WEFIRDSFRDNVASWGLVVNSFTAMEGVYLEHL 236
Query: 237 REVLGI-KEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGS 295
+ +G + W +GP + + RG S+ + WLD + N VVY+CFGS
Sbjct: 237 KREMGHDRVWAVGPIIPLSGDN-------RGGPTSVSVDHVMSWLDAREDNHVVYVCFGS 289
Query: 296 MTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEW--LPEGFEERTEGKGLIIRGWSP 353
Q +A GLE SG +FIW V+ E + +GF++R G+GL+IRGW+P
Sbjct: 290 QVVLTKEQTLALASGLEKSGVHFIWAVKEPVEKDSTRGNILDGFDDRVAGRGLVIRGWAP 349
Query: 354 QVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGV 413
QV +L H A+GAF+THCGWNSV+E VVAGV M+TWP+ A+Q+ + LV + LK GV
Sbjct: 350 QVAVLRHRAVGAFLTHCGWNSVVEAVVAGVLMLTWPMRADQYTDASLVVDELKVGVR--- 406
Query: 414 KKWVMKVGDNV-EWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNA 472
+ D V + D + + + G + R KA L + A A++E GSS + L+
Sbjct: 407 ---ACEGPDTVPDPDELARVFADSVTGNQT--ERIKAVELRKAALDAIQERGSSVNDLDG 461
Query: 473 LIEELRSL 480
I+ + SL
Sbjct: 462 FIQHVVSL 469
>At5g12890 glucosyltransferase -like protein
Length = 488
Score = 228 bits (581), Expect = 6e-60
Identities = 154/500 (30%), Positives = 257/500 (50%), Gaps = 47/500 (9%)
Query: 2 DSQSNPLHILVFPFMGHGHTIPTIDMAK-------LFASKGVRVTIVTTPLNKPPISKAL 54
+++ L I++FPFMG GH IP + +A + + ++++ TP N P I L
Sbjct: 3 EAKPRNLRIVMFPFMGQGHIIPFVALALRLEKIMIMNRANKTTISMINTPSNIPKIRSNL 62
Query: 55 EQSKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQ 114
++I + + F + GLP EN DS+P S V + A R L++PF + + +
Sbjct: 63 PPE----SSISLIELPFNSSDHGLPHDGENFDSLP-YSLVISLLEASRSLREPFRDFMTK 117
Query: 115 ------QKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSS 168
Q V+ D F W + G+ ++F + F L + + P+K
Sbjct: 118 ILKEEGQSSVIVIGDFFLGWIGKVCKEVGVYSVIFSASGAFGLGCYRSIWLNLPHKETKQ 177
Query: 169 DTDLFEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYEL 228
D F + D P ++ + QL + + E D + ++ ++I G + N+ E+
Sbjct: 178 DQ--FLLDDFPEAGEIEKTQLNSFMLEADG-TDDWSVFMKKIIPGWSDFDGFLFNTVAEI 234
Query: 229 ENVYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSV 288
+ + Y+R + G+ W +GP ++ ++++ S +EA WLD+K +SV
Sbjct: 235 DQMGLSYFRRITGVPVWPVGPVL---KSPDKKVGSRSTEEA------VKSWLDSKPDHSV 285
Query: 289 VYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVR------TQTE-DGDEWLPEGFEERT 341
VY+CFGSM L + + E+AM LE+S NFIWVVR ++E D +LPEGFEER
Sbjct: 286 VYVCFGSMNSILQTHMLELAMALESSEKNFIWVVRPPIGVEVKSEFDVKGYLPEGFEERI 345
Query: 342 --EGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEK 399
+GL+++ W+PQV IL H+A F++HCGWNS+LE + GVP++ WP+AAEQF+N
Sbjct: 346 TRSERGLLVKKWAPQVDILSHKATCVFLSHCGWNSILESLSHGVPLLGWPMAAEQFFNSI 405
Query: 400 LVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEE-AYEMRNKAKMLAEMAKK 458
L+ + + V V K ++ D + +K VME E E+R KA+ + E+ ++
Sbjct: 406 LMEKHIGVSVEVARGKRC-----EIKCDDIVSKIKLVMEETEVGKEIRKKAREVKELVRR 460
Query: 459 AVEE--DGSSYSQLNALIEE 476
A+ + GSS L +++
Sbjct: 461 AMVDGVKGSSVIGLEEFLDQ 480
>At5g03490 UDPG glucosyltransferase - like protein
Length = 465
Score = 221 bits (562), Expect = 9e-58
Identities = 154/485 (31%), Positives = 238/485 (48%), Gaps = 43/485 (8%)
Query: 5 SNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNI 64
S P HI+VFPF GH +P +D+ +G V+++ TP N +S L H +++
Sbjct: 15 SKPPHIVVFPFPAQGHLLPLLDLTHQLCLRGFNVSVIVTPGNLTYLSPLLSA---HPSSV 71
Query: 65 DIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQK--PHCVVA 122
FP L G ENV + + +P A++R L++P P +++
Sbjct: 72 TSVVFPFP-PHPSLSPGVENVKDVGNSGNLP-IMASLRQLREPIINWFQSHPNPPIALIS 129
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQ-CMKKYQPYKNVSSDTDLFEITDLPGN 181
D F W D + GIPR F SFF + Q C + K+ TD + DLP
Sbjct: 130 DFFLGWTHDLCNQIGIPRFAFFSISFFLVSVLQFCFENIDLIKS----TDPIHLLDLPRA 185
Query: 182 IKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVR--SYGVIVNSFYELENVYADYYREV 239
LP+ + + Q+ + E IKD + SYG + NS LE+ Y Y ++
Sbjct: 186 PIFKEEHLPSIVRRS---LQTPSPDLESIKDFSMNLLSYGSVFNSSEILEDDYLQYVKQR 242
Query: 240 LGI-KEWHIGPF-SIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMT 297
+G + + IGP SI + + S+D L WLD SV+Y+CFGS
Sbjct: 243 MGHDRVYVIGPLCSIGS--------GLKSNSGSVDP-SLLSWLDGSPNGSVLYVCFGSQK 293
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMI 357
Q +A+GLE S F+WVV+ +P+GFE+R G+GL++RGW Q+ +
Sbjct: 294 ALTKDQCDALALGLEKSMTRFVWVVKKDP------IPDGFEDRVSGRGLVVRGWVSQLAV 347
Query: 358 LEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWV 417
L H A+G F++HCGWNSVLEG+ +G ++ WP+ A+QF N +L+ E L GV V
Sbjct: 348 LRHVAVGGFLSHCGWNSVLEGITSGAVILGWPMEADQFVNARLLVEHL------GVAVRV 401
Query: 418 MKVGDNV-EWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAV-EEDGSSYSQLNALIE 475
+ G+ V + D + + + M GE E+ +A+ + + AV E +GSS + L++
Sbjct: 402 CEGGETVPDSDELGRVIAETM-GEGGREVAARAEEIRRKTEAAVTEANGSSVENVQRLVK 460
Query: 476 ELRSL 480
E +
Sbjct: 461 EFEKV 465
>At1g51210 putative glucosyl transferase
Length = 433
Score = 219 bits (558), Expect = 3e-57
Identities = 145/447 (32%), Positives = 229/447 (50%), Gaps = 41/447 (9%)
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQT 68
HI+VFP+ GH +P +D+ +G+ V+I+ TP N P +S L H + + + T
Sbjct: 20 HIMVFPYPAQGHLLPLLDLTHQLCLRGLTVSIIVTPKNLPYLSPLLSA---HPSAVSVVT 76
Query: 69 IKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQK--PHCVVADMFF 126
+ FP +P G ENV + P A++R L++P L P +++D F
Sbjct: 77 LPFPHHPL-IPSGVENVKDLGGYGN-PLIMASLRQLREPIVNWLSSHPNPPVALISDFFL 134
Query: 127 PWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTR 186
W D GIPR F + F + + ++ T+ ++DLP +
Sbjct: 135 GWTKD----LGIPRFAFFSSGAF---LASILHFVSDKPHLFESTEPVCLSDLPRSPVFKT 187
Query: 187 LQLPNTLTENDPISQSFAKLFEEIKDSEVR--SYGVIVNSFYELENVYADYYRE-VLGIK 243
LP+ + ++ P+SQ E +KDS + SYG I N+ LE Y +Y ++ V +
Sbjct: 188 EHLPSLIPQS-PLSQDL----ESVKDSTMNFSSYGCIFNTCECLEEDYMEYVKQKVSENR 242
Query: 244 EWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQ 303
+ +GP S +KE+ + +++D L WLD +SV+Y+CFGS Q
Sbjct: 243 VFGVGPLSSVGLSKEDSV-------SNVDAKALLSWLDGCPDDSVLYICFGSQKVLTKEQ 295
Query: 304 LKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMILEHEAI 363
++A+GLE S F+WVV+ +P+GFE+R G+G+I+RGW+PQV +L H A+
Sbjct: 296 CDDLALGLEKSMTRFVWVVKKDP------IPDGFEDRVAGRGMIVRGWAPQVAMLSHVAV 349
Query: 364 GAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDN 423
G F+ HCGWNSVLE + +G ++ WP+ A+QF + +LV E + GV V V + V D
Sbjct: 350 GGFLIHCGWNSVLEAMASGTMILAWPMEADQFVDARLVVEHM--GVAVSVCEGGKTVPDP 407
Query: 424 VEWDAVEKAVKRVMEGEEAYEMRNKAK 450
E + + + M GE E R +AK
Sbjct: 408 YE---MGRIIADTM-GESGGEARARAK 430
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,146,687
Number of Sequences: 26719
Number of extensions: 482009
Number of successful extensions: 1900
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1338
Number of HSP's gapped (non-prelim): 161
length of query: 489
length of database: 11,318,596
effective HSP length: 103
effective length of query: 386
effective length of database: 8,566,539
effective search space: 3306684054
effective search space used: 3306684054
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149574.3


