

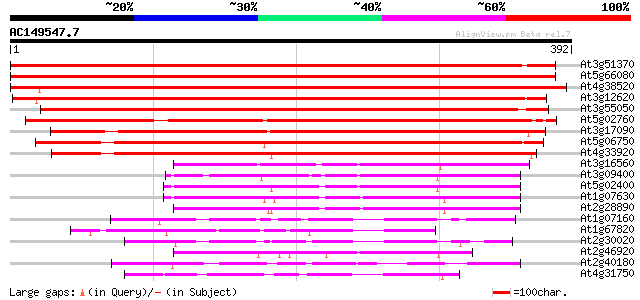
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.7 - phase: 0
(392 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g51370 protein phosphatase 2C like protein 607 e-174
At5g66080 protein phosphatase 2C-like protein 580 e-166
At4g38520 putative protein phosphatase-2c 571 e-163
At3g12620 unknown protein 501 e-142
At3g55050 protein phosphatase 2C - like protein 470 e-133
At5g02760 protein phosphatase - like protein 439 e-123
At3g17090 protein phosphatase-2c, putative 411 e-115
At5g06750 protein phosphatase 2C-like 384 e-107
At4g33920 unknown protein 375 e-104
At3g16560 unknown protein 151 7e-37
At3g09400 unknown protein 147 1e-35
At5g02400 putative protein 145 3e-35
At1g07630 unknown protein 140 1e-33
At2g28890 unknown protein 132 4e-31
At1g07160 unknown protein 110 1e-24
At1g67820 protein phosphatase-like protein 107 1e-23
At2g30020 putative protein phosphatase 2C 102 4e-22
At2g46920 unknown protein 99 3e-21
At2g40180 protein phosphatase 2C (AthPP2C5) 99 3e-21
At4g31750 unknown protein 93 2e-19
>At3g51370 protein phosphatase 2C like protein
Length = 379
Score = 607 bits (1565), Expect = e-174
Identities = 295/381 (77%), Positives = 337/381 (88%), Gaps = 3/381 (0%)
Query: 1 MLSTLMDFLTACWRRRSSDRKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQ 60
MLSTLM L+AC SS KSSD GK++GLLWYKD GQHL G++SMAVVQANNLLEDQ
Sbjct: 1 MLSTLMKLLSACLWPSSSSGKSSDSTGKQDGLLWYKDFGQHLVGEFSMAVVQANNLLEDQ 60
Query: 61 SQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRK 120
SQ+ESGPLS LD+GPYGTF+G+YDGHGGPETSRF+ DHLFQHLKRFA E SMSV+VI+K
Sbjct: 61 SQVESGPLSTLDSGPYGTFIGIYDGHGGPETSRFVNDHLFQHLKRFAAEQASMSVDVIKK 120
Query: 121 AYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATG 180
AY+ATEEGFLGVVTK WP PQIAAVGSCCLVGVICGG LYIAN+GDSRAVLGRA++ATG
Sbjct: 121 AYEATEEGFLGVVTKQWPTKPQIAAVGSCCLVGVICGGMLYIANVGDSRAVLGRAMKATG 180
Query: 181 EVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKA 240
EV+A+QLS EHNV+IESVRQEMHSLHPDD IV+LKHNVWRVKGLIQISRSIGDVYLKKA
Sbjct: 181 EVIALQLSAEHNVSIESVRQEMHSLHPDDSHIVMLKHNVWRVKGLIQISRSIGDVYLKKA 240
Query: 241 EFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIV 300
EFN+EPLY K+R+RE FK PILS +P+I+ HE+Q D+FLIFASDGLWE +SNQ+AVDIV
Sbjct: 241 EFNKEPLYTKYRIREPFKRPILSGEPTITEHEIQPQDKFLIFASDGLWEQMSNQEAVDIV 300
Query: 301 QNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSR 360
QNHP +G AR+L+K+AL EAAKKREMRYSDLKKI+RGVRRHFHDDITVV+IFLD+N V
Sbjct: 301 QNHPRNGIARRLVKMALQEAAKKREMRYSDLKKIERGVRRHFHDDITVVIIFLDTNQV-- 358
Query: 361 ASTVTGPPVSLRGAGVPLPSR 381
S+V GPP+S+RG G+ P +
Sbjct: 359 -SSVKGPPLSIRGGGMTFPKK 378
>At5g66080 protein phosphatase 2C-like protein
Length = 385
Score = 580 bits (1495), Expect = e-166
Identities = 284/384 (73%), Positives = 330/384 (84%), Gaps = 3/384 (0%)
Query: 1 MLSTLMDFLTAC-WRRRSSDRKS-SDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLE 58
MLS +FLT+C W S+ + SD GK++GLLWYKD+ HLFGD+SMAVVQANNLLE
Sbjct: 1 MLSLFFNFLTSCLWPSSSTTSHTYSDSKGKQDGLLWYKDSAHHLFGDFSMAVVQANNLLE 60
Query: 59 DQSQIESGPLSFLDT-GPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEV 117
DQSQ+ESGPL+ L + GPYGTFVGVYDGHGGPETSRF+ DHLF HLKRFA E SMSV+V
Sbjct: 61 DQSQVESGPLTTLSSSGPYGTFVGVYDGHGGPETSRFVNDHLFHHLKRFAAEQDSMSVDV 120
Query: 118 IRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVR 177
IRKAY+ATEEGFLGVV K W + P IAAVGSCCL+GV+C G LY+AN+GDSRAVLG+ ++
Sbjct: 121 IRKAYEATEEGFLGVVAKQWAVKPHIAAVGSCCLIGVVCDGKLYVANVGDSRAVLGKVIK 180
Query: 178 ATGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYL 237
ATGEV A+QLS EHNV+IESVRQEMHSLHPDD IVVLKHNVWRVKG+IQ+SRSIGDVYL
Sbjct: 181 ATGEVNALQLSAEHNVSIESVRQEMHSLHPDDSHIVVLKHNVWRVKGIIQVSRSIGDVYL 240
Query: 238 KKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAV 297
KK+EFN+EPLY K+RLRE K PILS +PSI+VH+LQ DQFLIFASDGLWE LSNQ+AV
Sbjct: 241 KKSEFNKEPLYTKYRLREPMKRPILSWEPSITVHDLQPDDQFLIFASDGLWEQLSNQEAV 300
Query: 298 DIVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNL 357
+IVQNHP +G AR+L+K AL EAAKKREMRYSDL KI+RGVRRHFHDDITVVV+FLD+NL
Sbjct: 301 EIVQNHPRNGIARRLVKAALQEAAKKREMRYSDLNKIERGVRRHFHDDITVVVLFLDTNL 360
Query: 358 VSRASTVTGPPVSLRGAGVPLPSR 381
+SRAS++ P VS+RG G+ LP +
Sbjct: 361 LSRASSLKTPSVSIRGGGITLPKK 384
>At4g38520 putative protein phosphatase-2c
Length = 400
Score = 571 bits (1472), Expect = e-163
Identities = 278/391 (71%), Positives = 327/391 (83%), Gaps = 2/391 (0%)
Query: 1 MLSTLMDFLTACWRRRSSD--RKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLE 58
MLS LM+FL AC RS R +SD G++EGLLW++D+GQH+FGD+SMAVVQAN+LLE
Sbjct: 1 MLSGLMNFLNACLWPRSDQQARSASDSGGRQEGLLWFRDSGQHVFGDFSMAVVQANSLLE 60
Query: 59 DQSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVI 118
DQSQ+ESG LS D+GP+GTFVGVYDGHGGPETSRFI DH+F HLKRF E + MS EVI
Sbjct: 61 DQSQLESGSLSSHDSGPFGTFVGVYDGHGGPETSRFINDHMFHHLKRFTAEQQCMSSEVI 120
Query: 119 RKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRA 178
+KA+QATEEGFL +VT + PQIA VGSCCLV VIC G LY+AN GDSRAVLG+ +R
Sbjct: 121 KKAFQATEEGFLSIVTNQFQTRPQIATVGSCCLVSVICDGKLYVANAGDSRAVLGQVMRV 180
Query: 179 TGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLK 238
TGE A QLS EHN +IESVR+E+ +LHPD P IVVLKHNVWRVKG+IQ+SRSIGDVYLK
Sbjct: 181 TGEAHATQLSAEHNASIESVRRELQALHPDHPDIVVLKHNVWRVKGIIQVSRSIGDVYLK 240
Query: 239 KAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVD 298
++EFNREPLYAKFRLR F P+LS++P+I+VH L+ HDQF+I ASDGLWEH+SNQ+AVD
Sbjct: 241 RSEFNREPLYAKFRLRSPFSKPLLSAEPAITVHTLEPHDQFIICASDGLWEHMSNQEAVD 300
Query: 299 IVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLV 358
IVQNHP +G A++L+KVAL EAAKKREMRYSDLKKIDRGVRRHFHDDITV+V+F D+NLV
Sbjct: 301 IVQNHPRNGIAKRLVKVALQEAAKKREMRYSDLKKIDRGVRRHFHDDITVIVVFFDTNLV 360
Query: 359 SRASTVTGPPVSLRGAGVPLPSRSLAPMELP 389
SR S + GP VS+RGAGV LP +LAP P
Sbjct: 361 SRGSMLRGPAVSVRGAGVNLPHNTLAPCTTP 391
>At3g12620 unknown protein
Length = 385
Score = 501 bits (1291), Expect = e-142
Identities = 243/375 (64%), Positives = 299/375 (78%), Gaps = 3/375 (0%)
Query: 3 STLMDFLTACWRRRS--SDRKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQ 60
+T++ + CWRR S D + D G+ +GLLWYKD+G H+ G++SM+V+QANNLLED
Sbjct: 5 ATILRMVAPCWRRPSVKGDHSTRDANGRCDGLLWYKDSGNHVAGEFSMSVIQANNLLEDH 64
Query: 61 SQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRK 120
S++ESGP+S D+GP TFVGVYDGHGGPE +RF+ HLF ++++F +E+ MS VI K
Sbjct: 65 SKLESGPVSMFDSGPQATFVGVYDGHGGPEAARFVNKHLFDNIRKFTSENHGMSANVITK 124
Query: 121 AYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATG 180
A+ ATEE FL +V + W + PQIA+VG+CCLVG+IC G LYIAN GDSR VLGR +A
Sbjct: 125 AFLATEEDFLSLVRRQWQIKPQIASVGACCLVGIICSGLLYIANAGDSRVVLGRLEKAFK 184
Query: 181 EVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKA 240
V A+QLS EHN ++ESVR+E+ SLHP+DP+IVVLKH VWRVKG+IQ+SRSIGD YLKKA
Sbjct: 185 IVKAVQLSSEHNASLESVREELRSLHPNDPQIVVLKHKVWRVKGIIQVSRSIGDAYLKKA 244
Query: 241 EFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIV 300
EFNREPL AKFR+ E F PIL ++P+I+VH++ DQFLIFASDGLWEHLSNQ+AVDIV
Sbjct: 245 EFNREPLLAKFRVPEVFHKPILRAEPAITVHKIHPEDQFLIFASDGLWEHLSNQEAVDIV 304
Query: 301 QNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSR 360
P +G ARKLIK AL EAAKKREMRYSDLKKIDRGVRRHFHDDITV+V+FLDS+LVSR
Sbjct: 305 NTCPRNGIARKLIKTALREAAKKREMRYSDLKKIDRGVRRHFHDDITVIVVFLDSHLVSR 364
Query: 361 ASTVTGPPVSLRGAG 375
ST P +S+ G G
Sbjct: 365 -STSRRPLLSISGGG 378
>At3g55050 protein phosphatase 2C - like protein
Length = 409
Score = 470 bits (1210), Expect = e-133
Identities = 221/355 (62%), Positives = 282/355 (79%), Gaps = 5/355 (1%)
Query: 22 SSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGPYGTFVG 81
S D G+ +GLLWYKD+G H+ G++SMAVVQANNLLED SQ+ESGP+S ++GP TFVG
Sbjct: 52 SDDTNGRLDGLLWYKDSGNHITGEFSMAVVQANNLLEDHSQLESGPISLHESGPEATFVG 111
Query: 82 VYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNP 141
VYDGHGGPE +RF+ D LF ++KR+ +E + MS +VI + + ATEE FLG+V + W P
Sbjct: 112 VYDGHGGPEAARFVNDRLFYNIKRYTSEQRGMSPDVITRGFVATEEEFLGLVQEQWKTKP 171
Query: 142 QIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQE 201
QIA+VG+CCLVG++C G LY+AN GDSR VLG+ E+ A+QLS EHN +IESVR+E
Sbjct: 172 QIASVGACCLVGIVCNGLLYVANAGDSRVVLGKVANPFKELKAVQLSTEHNASIESVREE 231
Query: 202 MHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPI 261
+ LHPDDP IVVLKH VWRVKG+IQ+SRSIGD YLK+AEFN+EPL KFR+ E F+ PI
Sbjct: 232 LRLLHPDDPNIVVLKHKVWRVKGIIQVSRSIGDAYLKRAEFNQEPLLPKFRVPERFEKPI 291
Query: 262 LSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAA 321
+ ++P+I+VH++ DQFLIFASDGLWEHLSNQ+AVDIV + P +G ARKL+K AL EAA
Sbjct: 292 MRAEPTITVHKIHPEDQFLIFASDGLWEHLSNQEAVDIVNSCPRNGVARKLVKAALQEAA 351
Query: 322 KKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSLRGAGV 376
KKREMRYSDL+KI+RG+RRHFHDDITV+V+FL + + T P+S++G G+
Sbjct: 352 KKREMRYSDLEKIERGIRRHFHDDITVIVVFLHA-----TNFATRTPISVKGGGL 401
>At5g02760 protein phosphatase - like protein
Length = 361
Score = 439 bits (1130), Expect = e-123
Identities = 223/371 (60%), Positives = 276/371 (74%), Gaps = 14/371 (3%)
Query: 12 CWRRRSSDRKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFL 71
CWR + +S K +GL WYKD G H FG++SMA++QAN+++EDQ QIESGPL+F
Sbjct: 5 CWRIGAGMERSKINPTKVDGLTWYKDLGLHTFGEFSMAMIQANSVMEDQCQIESGPLTFN 64
Query: 72 DTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLG 131
+ GTFVGVYDGHGGPE SRFI D++F K +S +VI KA+ T++ FL
Sbjct: 65 NPTVQGTFVGVYDGHGGPEASRFIADNIFP---------KEISEQVISKAFAETDKDFLK 115
Query: 132 VVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEH 191
VTK WP NPQ+A+VGSCCL GVIC G +YIAN GDSRAVLGR+ R G V A+QLS EH
Sbjct: 116 TVTKQWPTNPQMASVGSCCLAGVICNGLVYIANTGDSRAVLGRSER--GGVRAVQLSVEH 173
Query: 192 NVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKF 251
N +ES RQE+ SLHP+DP I+V+KH +WRVKG+IQ++RSIGD YLK+AEFNREPL KF
Sbjct: 174 NANLESARQELWSLHPNDPTILVMKHRLWRVKGVIQVTRSIGDAYLKRAEFNREPLLPKF 233
Query: 252 RLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARK 311
RL E F PILS+DPS+++ L D+F+I ASDGLWEHLSNQ+AVDIV N P G AR+
Sbjct: 234 RLPEHFTKPILSADPSVTITRLSPQDEFIILASDGLWEHLSNQEAVDIVHNSPRQGIARR 293
Query: 312 LIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSL 371
L+K AL EAAKKREMRYSDL +I GVRRHFHDDITV+V++L+ + V S + P+S+
Sbjct: 294 LLKAALKEAAKKREMRYSDLTEIHPGVRRHFHDDITVIVVYLNPHPVKTNSWAS--PLSI 351
Query: 372 RGAGVPLPSRS 382
RG G P+ S S
Sbjct: 352 RG-GYPMHSTS 361
>At3g17090 protein phosphatase-2c, putative
Length = 384
Score = 411 bits (1056), Expect = e-115
Identities = 199/349 (57%), Positives = 263/349 (75%), Gaps = 12/349 (3%)
Query: 29 KEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGG 88
K+GLLW++D G++ GD+SMAV+QAN +LEDQSQ+ESG +GTFVGVYDGHGG
Sbjct: 42 KDGLLWFRDLGKYCGGDFSMAVIQANQVLEDQSQVESGN--------FGTFVGVYDGHGG 93
Query: 89 PETSRFICDHLFQHLKRFATEHKSMSV-EVIRKAYQATEEGFLGVVTKHWPMNPQIAAVG 147
PE +R++CDHLF H + + E + + E I +A+ ATEEGF +V++ W P +A VG
Sbjct: 94 PEAARYVCDHLFNHFREISAETQGVVTRETIERAFHATEEGFASIVSELWQEIPNLATVG 153
Query: 148 SCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQEMHSLHP 207
+CCLVGVI +L++A+LGDSR VLG+ G + AIQLS EHN E +R E+ LHP
Sbjct: 154 TCCLVGVIYQNTLFVASLGDSRVVLGKKGNCGG-LSAIQLSTEHNANNEDIRWELKDLHP 212
Query: 208 DDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDPS 267
DDP+IVV +H VWRVKG+IQ+SRSIGD+Y+K+ EFN+EP+ KFR+ E K P++S+ P+
Sbjct: 213 DDPQIVVFRHGVWRVKGIIQVSRSIGDMYMKRPEFNKEPISQKFRIAEPMKRPLMSATPT 272
Query: 268 ISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAAKKREMR 327
I H L +D FLIFASDGLWEHL+N+ AV+IV NHP +GSA++LIK AL EAA+KREMR
Sbjct: 273 ILSHPLHPNDSFLIFASDGLWEHLTNEKAVEIVHNHPRAGSAKRLIKAALHEAARKREMR 332
Query: 328 YSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRA--STVTGPPVSLRGA 374
YSDL+KID+ VRRHFHDDITV+V+FL+ +L+SR ++ VS+R A
Sbjct: 333 YSDLRKIDKKVRRHFHDDITVIVVFLNHDLISRGHINSTQDTTVSIRSA 381
>At5g06750 protein phosphatase 2C-like
Length = 393
Score = 384 bits (987), Expect = e-107
Identities = 194/358 (54%), Positives = 251/358 (69%), Gaps = 12/358 (3%)
Query: 19 DRKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGPYGT 78
D D + LLW ++ +H FGD+S+AVVQAN ++ED SQ+E TG
Sbjct: 30 DDHDGDSSSSGDSLLWSRELERHSFGDFSIAVVQANEVIEDHSQVE--------TGNGAV 81
Query: 79 FVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWP 138
FVGVYDGHGGPE SR+I DHLF HL R + E +S E +R A+ ATEEGFL +V +
Sbjct: 82 FVGVYDGHGGPEASRYISDHLFSHLMRVSRERSCISEEALRAAFSATEEGFLTLVRRTCG 141
Query: 139 MNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAV---RATGEVLAIQLSPEHNVAI 195
+ P IAAVGSCCLVGVI G+L IAN+GDSRAVLG + +++A QL+ +HN A+
Sbjct: 142 LKPLIAAVGSCCLVGVIWKGTLLIANVGDSRAVLGSMGSNNNRSNKIVAEQLTSDHNAAL 201
Query: 196 ESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRE 255
E VRQE+ SLHPDD IVVLKH VWR+KG+IQ+SRSIGD YLK+ EF+ +P + +F L E
Sbjct: 202 EEVRQELRSLHPDDSHIVVLKHGVWRIKGIIQVSRSIGDAYLKRPEFSLDPSFPRFHLAE 261
Query: 256 TFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKV 315
+ P+LS++P + LQ D+F+IFASDGLWE ++NQ AV+IV HP G AR+L++
Sbjct: 262 ELQRPVLSAEPCVYTRVLQTSDKFVIFASDGLWEQMTNQQAVEIVNKHPRPGIARRLVRR 321
Query: 316 ALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSLRG 373
A+ AAKKREM Y DLKK++RGVRR FHDDITVVVIF+D+ L+ T P +S++G
Sbjct: 322 AITIAAKKREMNYDDLKKVERGVRRFFHDDITVVVIFIDNELL-MVEKATVPELSIKG 378
>At4g33920 unknown protein
Length = 380
Score = 375 bits (964), Expect = e-104
Identities = 187/349 (53%), Positives = 250/349 (71%), Gaps = 18/349 (5%)
Query: 30 EGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGGP 89
+GLLW + H GDYS+AVVQAN+ LEDQSQ+ T T+VGVYDGHGGP
Sbjct: 20 DGLLWQSELRPHAGGDYSIAVVQANSRLEDQSQVF--------TSSSATYVGVYDGHGGP 71
Query: 90 ETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSC 149
E SRF+ HLF ++ +FA EH +SV+VI+KA++ TEE F G+V + PM PQ+A VGSC
Sbjct: 72 EASRFVNRHLFPYMHKFAREHGGLSVDVIKKAFKETEEEFCGMVKRSLPMKPQMATVGSC 131
Query: 150 CLVGVICGGSLYIANLGDSRAVLGRAVRATGE---VLAIQLSPEHNVAIESVRQEMHSLH 206
CLVG I +LY+ANLGDSRAVLG V +A +LS +HNVA+E VR+E+ +L+
Sbjct: 132 CLVGAISNDTLYVANLGDSRAVLGSVVSGVDSNKGAVAERLSTDHNVAVEEVRKEVKALN 191
Query: 207 PDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDP 266
PDD +IV+ VWR+KG+IQ+SRSIGDVYLKK E+ R+P++ + + P ++++P
Sbjct: 192 PDDSQIVLYTRGVWRIKGIIQVSRSIGDVYLKKPEYYRDPIFQRHGNPIPLRRPAMTAEP 251
Query: 267 SISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAAKKREM 326
SI V +L+ D FLIFASDGLWEHLS++ AV+IV HP +G AR+L++ AL EAAKKREM
Sbjct: 252 SIIVRKLKPQDLFLIFASDGLWEHLSDETAVEIVLKHPRTGIARRLVRAALEEAAKKREM 311
Query: 327 RYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRAST-------VTGPP 368
RY D+KKI +G+RRHFHDDI+V+V++LD N S +++ +T PP
Sbjct: 312 RYGDIKKIAKGIRRHFHDDISVIVVYLDQNKTSSSNSKLVKQGGITAPP 360
>At3g16560 unknown protein
Length = 493
Score = 151 bits (381), Expect = 7e-37
Identities = 89/254 (35%), Positives = 147/254 (57%), Gaps = 11/254 (4%)
Query: 115 VEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGR 174
++ + +A E FL +V + P + +VGSC LV ++ G LY+ NLGDSRAVL
Sbjct: 244 LDCLNRALFQAETDFLRMVEQEMEERPDLVSVGSCVLVTLLVGKDLYVLNLGDSRAVLA- 302
Query: 175 AVRATGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGD 234
++ A+QL+ +H V E + S H DDPKIV+ ++KG ++++R++G
Sbjct: 303 TYNGNKKLQAVQLTEDHTVDNEVEEARLLSEHLDDPKIVI----GGKIKGKLKVTRALGV 358
Query: 235 VYLKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQ 294
YLKK + N + L R+R P +S +PS+ VH++ E D F+I ASDGL++ SN+
Sbjct: 359 GYLKKEKLN-DALMGILRVRNLLSPPYVSVEPSMRVHKITESDHFVIVASDGLFDFFSNE 417
Query: 295 DAVDI----VQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVV 350
+A+ + V ++P A+ L++ + +AA + +L + G RR +HDD+T++V
Sbjct: 418 EAIGLVHSFVSSNPSGDPAKFLLERLVAKAAARAGFTLEELTNVPAGRRRRYHDDVTIMV 477
Query: 351 IFLDSN-LVSRAST 363
I L ++ S+AST
Sbjct: 478 ITLGTDQRTSKAST 491
>At3g09400 unknown protein
Length = 650
Score = 147 bits (370), Expect = 1e-35
Identities = 91/279 (32%), Positives = 155/279 (54%), Gaps = 41/279 (14%)
Query: 110 HKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSR 169
HK + + +++A + TEE F +V + NP++A +GSC LV ++ G +Y+ ++GDSR
Sbjct: 377 HKDV-LRALQQALEKTEESFDLMVNE----NPELALMGSCVLVTLMKGEDVYVMSVGDSR 431
Query: 170 AVLGR---------------------------AVRATGEVLAIQLSPEHNVAIESVRQEM 202
AVL R R ++ +QL+ EH+ ++E + +
Sbjct: 432 AVLARRPNVEKMKMQKELERVKEESPLETLFITERGLSLLVPVQLNKEHSTSVEEEVRRI 491
Query: 203 HSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPIL 262
HPDD I+ +++N RVKG ++++R+ G +LK+ ++N E L FR+ +P +
Sbjct: 492 KKEHPDD--ILAIENN--RVKGYLKVTRAFGAGFLKQPKWN-EALLEMFRIDYVGTSPYI 546
Query: 263 SSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAV----DIVQNHPHSGSARKLIKVALL 318
+ PS+ H L D+FLI +SDGL+E+ SN++A+ + P A+ LI+ LL
Sbjct: 547 TCSPSLHHHRLSSRDKFLILSSDGLYEYFSNEEAIFEVDSFISAFPEGDPAQHLIQEVLL 606
Query: 319 EAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNL 357
AAKK M + +L +I +G RR +HDD++V+VI L+ +
Sbjct: 607 RAAKKYGMDFHELLEIPQGDRRRYHDDVSVIVISLEGRI 645
>At5g02400 putative protein
Length = 674
Score = 145 bits (367), Expect = 3e-35
Identities = 93/284 (32%), Positives = 153/284 (53%), Gaps = 40/284 (14%)
Query: 108 TEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGD 167
T HK + ++ + +A + TE+ +L + + NP++A +GSC LV ++ G +Y+ N+GD
Sbjct: 392 TNHKDV-LKALLQALRKTEDAYLELADQMVKENPELALMGSCVLVTLMKGEDVYVMNVGD 450
Query: 168 SRAVLGRAVR-ATGE-----------------------------VLAIQLSPEHNVAIES 197
SRAVLGR ATG ++ +QL+ EH+ IE
Sbjct: 451 SRAVLGRKPNLATGRKRQKELERIREDSSLEDKEILMNGAMRNTLVPLQLNMEHSTRIEE 510
Query: 198 VRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETF 257
+ + HPDD V RVKG ++++R+ G +LK+ ++N + L FR+
Sbjct: 511 EVRRIKKEHPDDDCAVEND----RVKGYLKVTRAFGAGFLKQPKWN-DALLEMFRIDYIG 565
Query: 258 KTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAV----DIVQNHPHSGSARKLI 313
+P ++ PS+ H+L D+FLI +SDGL+E+ SNQ+A+ + P A+ LI
Sbjct: 566 TSPYITCSPSLCHHKLTSRDKFLILSSDGLYEYFSNQEAIFEVESFISAFPEGDPAQHLI 625
Query: 314 KVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNL 357
+ LL AA K M + +L +I +G RR +HDD++V+VI L+ +
Sbjct: 626 QEVLLRAANKFGMDFHELLEIPQGDRRRYHDDVSVIVISLEGRI 669
Score = 28.9 bits (63), Expect = 5.1
Identities = 9/27 (33%), Positives = 19/27 (70%)
Query: 79 FVGVYDGHGGPETSRFICDHLFQHLKR 105
FVG+YDG GP+ ++ ++L+ +++
Sbjct: 277 FVGIYDGFSGPDAPDYLLNNLYTAVQK 303
>At1g07630 unknown protein
Length = 662
Score = 140 bits (353), Expect = 1e-33
Identities = 91/287 (31%), Positives = 151/287 (51%), Gaps = 43/287 (14%)
Query: 108 TEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGD 167
T H + +E + +A + TEE +L K NP++A +GSC LV ++ G +Y+ N+GD
Sbjct: 377 TNHSEV-LEALSQALRKTEEAYLDTADKMLDENPELALMGSCVLVMLMKGEDIYVMNVGD 435
Query: 168 SRAVLGRAV--------------RATGEVL-------------------AIQLSPEHNVA 194
SRAVLG+ R E + A QL+ +H+
Sbjct: 436 SRAVLGQKSEPDYWLAKIRQDLERINEETMMNDLEGCEGDQSSLVPNLSAFQLTVDHSTN 495
Query: 195 IESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLR 254
IE + + + HPDD V + RVKG ++++R+ G +LK+ ++N L F++
Sbjct: 496 IEEEVERIRNEHPDDVTAVTNE----RVKGSLKVTRAFGAGFLKQPKWNNA-LLEMFQID 550
Query: 255 ETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQN----HPHSGSAR 310
K+P ++ PS+ H L D+FLI +SDGL+++ +N++AV V+ P A+
Sbjct: 551 YVGKSPYINCLPSLYHHRLGSKDRFLILSSDGLYQYFTNEEAVSEVELFITLQPEGDPAQ 610
Query: 311 KLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNL 357
L++ L AAKK M + +L +I +G RR +HDD+++VVI L+ +
Sbjct: 611 HLVQELLFRAAKKAGMDFHELLEIPQGERRRYHDDVSIVVISLEGRM 657
Score = 31.2 bits (69), Expect = 1.0
Identities = 11/27 (40%), Positives = 17/27 (62%)
Query: 79 FVGVYDGHGGPETSRFICDHLFQHLKR 105
FVG+YDG GP+ ++ HL+ + R
Sbjct: 281 FVGIYDGFNGPDAPDYLLSHLYPVVHR 307
>At2g28890 unknown protein
Length = 654
Score = 132 bits (331), Expect = 4e-31
Identities = 86/280 (30%), Positives = 145/280 (51%), Gaps = 42/280 (15%)
Query: 115 VEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGR 174
++ + +A + TEE +L NP++A +GSC LV ++ G +Y+ N+GDSRAVLG+
Sbjct: 375 LKALSQALRKTEEAYLENADMMLDENPELALMGSCVLVMLMKGEDVYLMNVGDSRAVLGQ 434
Query: 175 AVRAT--------------------------GE-------VLAIQLSPEHNVAIESVRQE 201
+ GE + A QL+ +H+ +E
Sbjct: 435 KAESDYWIGKIKQDLERINEETMNDFDGCGDGEGASLVPTLSAFQLTVDHSTNVEEEVNR 494
Query: 202 MHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPI 261
+ HPDD V + RVKG ++++R+ G +LK+ ++N L F++ +P
Sbjct: 495 IRKEHPDDASAVSNE----RVKGSLKVTRAFGAGFLKQPKWNNA-LLEMFQIDYKGTSPY 549
Query: 262 LSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQN----HPHSGSARKLIKVAL 317
++ PS+ H L DQFLI +SDGL+++ +N++AV V+ P A+ L++ L
Sbjct: 550 INCLPSLYHHRLGSKDQFLILSSDGLYQYFTNEEAVSEVELFITLQPEGDPAQHLVQELL 609
Query: 318 LEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNL 357
AAKK M + +L +I +G RR +HDD+++VVI L+ +
Sbjct: 610 FRAAKKAGMDFHELLEIPQGERRRYHDDVSIVVISLEGRM 649
Score = 31.6 bits (70), Expect = 0.79
Identities = 11/27 (40%), Positives = 17/27 (62%)
Query: 79 FVGVYDGHGGPETSRFICDHLFQHLKR 105
FVG+YDG GP+ ++ HL+ + R
Sbjct: 275 FVGIYDGFNGPDAPDYLLSHLYPAVHR 301
>At1g07160 unknown protein
Length = 380
Score = 110 bits (275), Expect = 1e-24
Identities = 87/285 (30%), Positives = 133/285 (46%), Gaps = 55/285 (19%)
Query: 71 LDTGPYGTFVGVYDGHGGPETSRFICDHLFQHL--KRFATEHKSMSVEVIRKAYQATEEG 128
L P GVYDGHGGP + F +L ++ + ++S E +++ Y AT+
Sbjct: 145 LQGDPKQAIFGVYDGHGGPTAAEFAAKNLCSNILGEIVGGRNESKIEEAVKRGYLATDSE 204
Query: 129 FLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLS 188
FL + GSCC+ +I G+L +AN GD RAVL +V E L +
Sbjct: 205 FL---------KEKNVKGGSCCVTALISDGNLVVANAGDCRAVL--SVGGFAEAL----T 249
Query: 189 PEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLY 248
+H + + R + S V ++VWR++G + +SR IGD +LK+
Sbjct: 250 SDHRPSRDDERNRIES----SGGYVDTFNSVWRIQGSLAVSRGIGDAHLKQW-------- 297
Query: 249 AKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGS 308
+ S+P I++ + +FLI ASDGLW+ +SNQ+AVDI
Sbjct: 298 -------------IISEPEINILRINPQHEFLILASDGLWDKVSNQEAVDI--------- 335
Query: 309 ARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFL 353
AR K +KR+ + K +D V R DDI+V++I L
Sbjct: 336 ARPFCK----GTDQKRKPLLACKKLVDLSVSRGSLDDISVMLIQL 376
>At1g67820 protein phosphatase-like protein
Length = 447
Score = 107 bits (267), Expect = 1e-23
Identities = 84/265 (31%), Positives = 124/265 (46%), Gaps = 41/265 (15%)
Query: 43 FGDYSMAVVQANN---LLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHL 99
FG VV N +ED +I + L +F GVYDGHGG + + F+ ++L
Sbjct: 99 FGGNGFGVVSRNGKKKFMEDTHRI----VPCLVGNSKKSFFGVYDGHGGAKAAEFVAENL 154
Query: 100 FQHLKRFAT--EHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICG 157
+++ + K VE + A+ T+ FL V K + ++ G+CC+ VI
Sbjct: 155 HKYVVEMMENCKGKEEKVEAFKAAFLRTDRDFLEKVIKEQSLKGVVS--GACCVTAVIQD 212
Query: 158 GSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQEMHSLHPD-----DPKI 212
+ ++NLGD RAVL RA E L P + E R E SL P
Sbjct: 213 QEMIVSNLGDCRAVLCRA--GVAEALTDDHKPGRDD--EKERIESQSLIPFMTFGLQGGY 268
Query: 213 VVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHE 272
V WRV+G++ +SRSIGD +LKK + ++P V E
Sbjct: 269 VDNHQGAWRVQGILAVSRSIGDAHLKKW---------------------VVAEPETRVLE 307
Query: 273 LQEHDQFLIFASDGLWEHLSNQDAV 297
L++ +FL+ ASDGLW+ +SNQ+AV
Sbjct: 308 LEQDMEFLVLASDGLWDVVSNQEAV 332
>At2g30020 putative protein phosphatase 2C
Length = 396
Score = 102 bits (254), Expect = 4e-22
Identities = 83/277 (29%), Positives = 126/277 (44%), Gaps = 64/277 (23%)
Query: 81 GVYDGHGGPETSRFICDHLFQHLKRFATEHKSMS--VEVIRKAYQATEEGFLGVVTKHWP 138
GVYDGHGG + + F +L +++ + S E ++ Y AT+ FL
Sbjct: 172 GVYDGHGGVKAAEFAAKNLDKNIVEEVVGKRDESEIAEAVKHGYLATDASFL-------- 223
Query: 139 MNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESV 198
+ GSCC+ ++ G+L ++N GD RAV+ + G V A LS +H + +
Sbjct: 224 -KEEDVKGGSCCVTALVNEGNLVVSNAGDCRAVM-----SVGGV-AKALSSDHRPSRDDE 276
Query: 199 RQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFK 258
R+ + + V H VWR++G + +SR IGD LKK
Sbjct: 277 RKRIETTGG----YVDTFHGVWRIQGSLAVSRGIGDAQLKKW------------------ 314
Query: 259 TPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLI----K 314
+ ++P + ++ +FLI ASDGLW+ +SNQ+AVDI AR L K
Sbjct: 315 ---VIAEPETKISRIEHDHEFLILASDGLWDKVSNQEAVDI---------ARPLCLGTEK 362
Query: 315 VALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVI 351
LL A K K +D R DDI+V++I
Sbjct: 363 PLLLAACK---------KLVDLSASRGSSDDISVMLI 390
>At2g46920 unknown protein
Length = 814
Score = 99.4 bits (246), Expect = 3e-21
Identities = 72/266 (27%), Positives = 123/266 (46%), Gaps = 58/266 (21%)
Query: 115 VEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVL-- 172
+ + +A ++TEE ++ +V K +NP++A +GSC LV ++ +Y+ N+GDSRA+L
Sbjct: 541 LRAMARALESTEEAYMDMVEKSLDINPELALMGSCVLVMLMKDQDVYVMNVGDSRAILAQ 600
Query: 173 ------------------GRAVRATGEVLAIQL------SPEHNVA-----------IES 197
G R+ ++ I+L SP HN A + S
Sbjct: 601 ERLHDRHSNPGFGNDEGIGHKSRSRESLVRIELDRISEESPIHNQATPISVSNKNRDVTS 660
Query: 198 VRQEMHSLHPDDPKIVVLKHNVW----------------RVKGLIQISRSIGDVYLKKAE 241
R +M ++ ++ +W RVKG ++++R+ G +LKK
Sbjct: 661 YRLKMRAVQLSSDHSTSVEEEIWRIRSEHPEDDQSILKDRVKGQLKVTRAFGAGFLKKPN 720
Query: 242 FNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVD--- 298
FN E L F++ P ++ +P H L D+F++ +SDGL+E+ SN++ V
Sbjct: 721 FN-EALLEMFQVEYIGTDPYITCEPCTVHHRLTSSDRFMVLSSDGLYEYFSNEEVVAHVT 779
Query: 299 -IVQNHPHSGSARKLIKVALLEAAKK 323
++N P A+ LI L AA K
Sbjct: 780 WFIENVPEGDPAQYLIAELLSRAATK 805
Score = 32.7 bits (73), Expect = 0.35
Identities = 10/27 (37%), Positives = 18/27 (66%)
Query: 79 FVGVYDGHGGPETSRFICDHLFQHLKR 105
F+G+YDG GP+ F+ HL++ + +
Sbjct: 302 FIGIYDGFSGPDAPDFVMSHLYKAIDK 328
>At2g40180 protein phosphatase 2C (AthPP2C5)
Length = 390
Score = 99.4 bits (246), Expect = 3e-21
Identities = 84/292 (28%), Positives = 135/292 (45%), Gaps = 62/292 (21%)
Query: 72 DTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKS----MSVE-VIRKAYQATE 126
D G F GV+DGHGG + + F +L +++ +S S+E IR+ Y T+
Sbjct: 154 DGGYKNAFFGVFDGHGGSKAAEFAAMNLGNNIEAAMASARSGEDGCSMESAIREGYIKTD 213
Query: 127 EGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQ 186
E FL ++ G+CC+ +I G L ++N GD RAV+ R A
Sbjct: 214 EDFLKEGSRG----------GACCVTALISKGELAVSNAGDCRAVMSRGGTAEA------ 257
Query: 187 LSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREP 246
L+ +HN + + + + +L V + VWR++G + +SR IGD YLK+ EP
Sbjct: 258 LTSDHNPSQANELKRIEALGG----YVDCCNGVWRIQGTLAVSRGIGDRYLKEWVI-AEP 312
Query: 247 LYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHS 306
R++ F +FLI ASDGLW+ ++NQ+AVD+V+ +
Sbjct: 313 ETRTLRIKPEF--------------------EFLILASDGLWDKVTNQEAVDVVRPYC-- 350
Query: 307 GSARKLIKVALLEAAKKREMRYSDLKKI-DRGVRRHFHDDITVVVIFLDSNL 357
+ M S KK+ + V+R DDI++++I L + L
Sbjct: 351 -------------VGVENPMTLSACKKLAELSVKRGSLDDISLIIIQLQNFL 389
>At4g31750 unknown protein
Length = 311
Score = 93.2 bits (230), Expect = 2e-19
Identities = 70/236 (29%), Positives = 110/236 (45%), Gaps = 41/236 (17%)
Query: 81 GVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMN 140
GV+DGHGG + ++ +LF +L R + S + I AY T+ FL N
Sbjct: 66 GVFDGHGGARAAEYVKQNLFSNLIRHP-KFISDTTAAIADAYNQTDSEFLK------SEN 118
Query: 141 PQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQ 200
Q GS ++ G L +AN+GDSRAV+ R A I +S +H RQ
Sbjct: 119 SQNRDAGSTASTAILVGDRLLVANVGDSRAVICRGGNA------IAVSRDHKPDQSDERQ 172
Query: 201 EMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTP 260
+ +D V+ WRV G++ +SR+ GD LK+
Sbjct: 173 RI-----EDAGGFVMWAGTWRVGGVLAVSRAFGDRLLKQ--------------------- 206
Query: 261 ILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQ--NHPHSGSARKLIK 314
+ +DP I ++ +FLI ASDGLW+ +SN++AV +++ P G+ R +++
Sbjct: 207 YVVADPEIQEEKVDSSLEFLILASDGLWDVVSNEEAVGMIKAIEDPEEGAKRLMME 262
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,783,373
Number of Sequences: 26719
Number of extensions: 370530
Number of successful extensions: 1169
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 907
Number of HSP's gapped (non-prelim): 126
length of query: 392
length of database: 11,318,596
effective HSP length: 101
effective length of query: 291
effective length of database: 8,619,977
effective search space: 2508413307
effective search space used: 2508413307
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149547.7


