

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.6 + phase: 0
(160 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
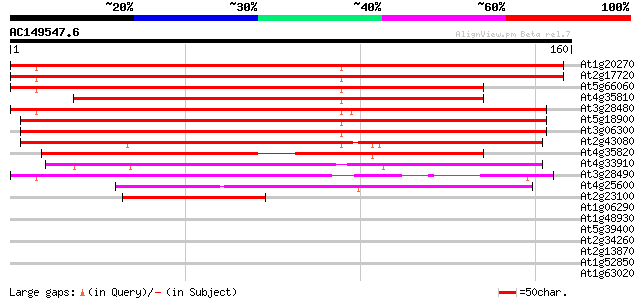
Score E
Sequences producing significant alignments: (bits) Value
At1g20270 unknown protein 243 4e-65
At2g17720 similar to prolyl 4-hydroxylase alpha subunit 228 1e-60
At5g66060 prolyl 4-hydroxylase, alpha subunit-like protein 207 2e-54
At4g35810 putative protein 179 4e-46
At3g28480 prolyl 4-hydroxylase like protein 170 3e-43
At5g18900 unknown protein 167 2e-42
At3g06300 unknown protein 162 6e-41
At2g43080 unknown protein 151 1e-37
At4g35820 putative protein 138 1e-33
At4g33910 unknown protein 120 3e-28
At3g28490 prolyl 4-hydroxylase, putative 103 3e-23
At4g25600 unknown protein 60 4e-10
At2g23100 hypothetical protein 41 3e-04
At1g06290 acyl-CoA oxidase, putative, 3' partial 28 2.1
At1g48930 putative glucanase gb|AAB91971.1; similar to ESTs gb|A... 28 2.8
At5g39400 PTEN -like protein 27 4.7
At2g34260 unknown protein 27 4.7
At2g13870 En/Spm-like transposon protein 27 4.7
At1g52850 hypothetical protein 27 4.7
At1g63020 RNA polymerase IIA largest subunit, putative 27 6.2
>At1g20270 unknown protein
Length = 287
Score = 243 bits (619), Expect = 4e-65
Identities = 114/180 (63%), Positives = 137/180 (75%), Gaps = 22/180 (12%)
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
M KSTV D ETGKS D+ RTSSGTF+ RG DKI++ IE+RIAD+TFIP ++GE + +LH
Sbjct: 108 MVKSTVVDSETGKSKDSRVRTSSGTFLRRGRDKIIKTIEKRIADYTFIPADHGEGLQVLH 167
Query: 60 YEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL---------------------PWWNE 98
YE GQKYEPH D+F DE NTKNGG+R+ATMLMYL PW+NE
Sbjct: 168 YEAGQKYEPHYDYFVDEFNTKNGGQRMATMLMYLSDVEEGGETVFPAANMNFSSVPWYNE 227
Query: 99 LSDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRVGKWSI 158
LS+CGKKGLS+KP+MGDALLFWSM+PD TLDP S+HG CPVI+G+KWS TKWM VG++ I
Sbjct: 228 LSECGKKGLSVKPRMGDALLFWSMRPDATLDPTSLHGGCPVIRGNKWSSTKWMHVGEYKI 287
>At2g17720 similar to prolyl 4-hydroxylase alpha subunit
Length = 291
Score = 228 bits (580), Expect = 1e-60
Identities = 106/180 (58%), Positives = 133/180 (73%), Gaps = 22/180 (12%)
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
M KSTV D++TG S D+ RTSSGTF+ RGHD+++ IE+RI+DFTFIPVENGE + +LH
Sbjct: 112 MVKSTVVDEKTGGSKDSRVRTSSGTFLRRGHDEVVEVIEKRISDFTFIPVENGEGLQVLH 171
Query: 60 YEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL---------------------PWWNE 98
Y+VGQKYEPH D+F DE NTKNGG+R+AT+LMYL PWWNE
Sbjct: 172 YQVGQKYEPHYDYFLDEFNTKNGGQRIATVLMYLSDVDDGGETVFPAARGNISAVPWWNE 231
Query: 99 LSDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRVGKWSI 158
LS CGK+GLS+ PK DALLFW+M+PD +LDP S+HG CPV+KG+KWS TKW V ++ +
Sbjct: 232 LSKCGKEGLSVLPKKRDALLFWNMRPDASLDPSSLHGGCPVVKGNKWSSTKWFHVHEFKV 291
>At5g66060 prolyl 4-hydroxylase, alpha subunit-like protein
Length = 267
Score = 207 bits (527), Expect = 2e-54
Identities = 99/157 (63%), Positives = 118/157 (75%), Gaps = 22/157 (14%)
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
M KSTV D++TGKS D+ RTSSGTF+ RG DK +R IE+RI+DFTFIPVE+GE + +LH
Sbjct: 110 MEKSTVVDEKTGKSTDSRVRTSSGTFLARGRDKTIREIEKRISDFTFIPVEHGEGLQVLH 169
Query: 60 YEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL---------------------PWWNE 98
YE+GQKYEPH D+F DE NT+NGG+R+AT+LMYL PWWNE
Sbjct: 170 YEIGQKYEPHYDYFMDEYNTRNGGQRIATVLMYLSDVEEGGETVFPAAKGNYSAVPWWNE 229
Query: 99 LSDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHG 135
LS+CGK GLS+KPKMGDALLFWSM PD TLDP S+HG
Sbjct: 230 LSECGKGGLSVKPKMGDALLFWSMTPDATLDPSSLHG 266
>At4g35810 putative protein
Length = 307
Score = 179 bits (455), Expect = 4e-46
Identities = 83/138 (60%), Positives = 100/138 (72%), Gaps = 21/138 (15%)
Query: 19 RTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPHPDFFTDEIN 78
RTSSGTF+NRGHD+I+ IE RI+DFTFIP ENGE + +LHYEVGQ+YEPH D+F DE N
Sbjct: 162 RTSSGTFLNRGHDEIVEEIENRISDFTFIPPENGEGLQVLHYEVGQRYEPHHDYFFDEFN 221
Query: 79 TKNGGERVATMLMYL---------------------PWWNELSDCGKKGLSIKPKMGDAL 117
+ GG+R+AT+LMYL PWW+ELS CGK+GLS+ PK DAL
Sbjct: 222 VRKGGQRIATVLMYLSDVDEGGETVFPAAKGNVSDVPWWDELSQCGKEGLSVLPKKRDAL 281
Query: 118 LFWSMKPDGTLDPLSMHG 135
LFWSMKPD +LDP S+HG
Sbjct: 282 LFWSMKPDASLDPSSLHG 299
>At3g28480 prolyl 4-hydroxylase like protein
Length = 316
Score = 170 bits (430), Expect = 3e-43
Identities = 80/174 (45%), Positives = 120/174 (67%), Gaps = 21/174 (12%)
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
+ KS V D+++G+SV++ RTSSG F+++ D I+ N+E ++A +TF+P ENGES+ ILH
Sbjct: 88 LEKSMVADNDSGESVESEVRTSSGMFLSKRQDDIVSNVEAKLAAWTFLPEENGESMQILH 147
Query: 60 YEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL-----------PWW---------NEL 99
YE GQKYEPH D+F D+ N + GG R+AT+LMYL P W +
Sbjct: 148 YENGQKYEPHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLKDDSW 207
Query: 100 SDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRV 153
++C K+G ++KP+ GDALLF+++ P+ T D S+HG+CPV++G+KWS T+W+ V
Sbjct: 208 TECAKQGYAVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWIHV 261
>At5g18900 unknown protein
Length = 298
Score = 167 bits (424), Expect = 2e-42
Identities = 79/173 (45%), Positives = 113/173 (64%), Gaps = 23/173 (13%)
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVG 63
+ D+++G+S + RTSSGTFI++G D I+ IE +I+ +TF+P ENGE + +L YE G
Sbjct: 73 AVADNDSGESKFSEVRTSSGTFISKGKDPIVSGIEDKISTWTFLPKENGEDIQVLRYEHG 132
Query: 64 QKYEPHPDFFTDEINTKNGGERVATMLMYL-----------------------PWWNELS 100
QKY+ H D+F D++N GG R+AT+LMYL +LS
Sbjct: 133 QKYDAHFDYFHDKVNIVRGGHRMATILMYLSNVTKGGETVFPDAEIPSRRVLSENKEDLS 192
Query: 101 DCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRV 153
DC K+G+++KP+ GDALLF+++ PD DPLS+HG CPVI+G+KWS TKW+ V
Sbjct: 193 DCAKRGIAVKPRKGDALLFFNLHPDAIPDPLSLHGGCPVIEGEKWSATKWIHV 245
>At3g06300 unknown protein
Length = 299
Score = 162 bits (411), Expect = 6e-41
Identities = 76/173 (43%), Positives = 112/173 (63%), Gaps = 23/173 (13%)
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVG 63
+ D++ G+S + RTSSGTFI++G D I+ IE +++ +TF+P ENGE + +L YE G
Sbjct: 74 AVADNDNGESQVSDVRTSSGTFISKGKDPIVSGIEDKLSTWTFLPKENGEDLQVLRYEHG 133
Query: 64 QKYEPHPDFFTDEINTKNGGERVATMLMYL-----------------------PWWNELS 100
QKY+ H D+F D++N GG R+AT+L+YL ++LS
Sbjct: 134 QKYDAHFDYFHDKVNIARGGHRIATVLLYLSNVTKGGETVFPDAQEFSRRSLSENKDDLS 193
Query: 101 DCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRV 153
DC KKG+++KPK G+ALLF++++ D DP S+HG CPVI+G+KWS TKW+ V
Sbjct: 194 DCAKKGIAVKPKKGNALLFFNLQQDAIPDPFSLHGGCPVIEGEKWSATKWIHV 246
>At2g43080 unknown protein
Length = 283
Score = 151 bits (382), Expect = 1e-37
Identities = 83/167 (49%), Positives = 104/167 (61%), Gaps = 19/167 (11%)
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDK--ILRNIEQRIADFTFIPVENGESVNILHYE 61
+ VD +TGK V + RTSSG F+ I++ IE+RIA F+ +P ENGE + +L YE
Sbjct: 113 TVVDVKTGKGVKSDVRTSSGMFLTHVERSYPIIQAIEKRIAVFSQVPAENGELIQVLRYE 172
Query: 62 VGQKYEPHPDFFTDEINTKNGGERVATMLMYL-----------PWWNELSDC---GK--K 105
Q Y+PH D+F D N K GG+RVATMLMYL P + DC GK K
Sbjct: 173 PQQFYKPHHDYFADTFNLKRGGQRVATMLMYLTDDVEGGETYFPLAGD-GDCTCGGKIMK 231
Query: 106 GLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMR 152
G+S+KP GDA+LFWSM DG DP S+HG C V+ G+KWS TKWMR
Sbjct: 232 GISVKPTKGDAVLFWSMGLDGQSDPRSIHGGCEVLSGEKWSATKWMR 278
>At4g35820 putative protein
Length = 272
Score = 138 bits (348), Expect = 1e-33
Identities = 70/135 (51%), Positives = 95/135 (69%), Gaps = 19/135 (14%)
Query: 10 TGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPH 69
TG ++S+RTSSGTFI GHDKI++ IE+RI++FTFIP ENGE++ +++YEVGQK+EPH
Sbjct: 138 TGLGEESSSRTSSGTFIRSGHDKIVKEIEKRISEFTFIPQENGETLQVINYEVGQKFEPH 197
Query: 70 PDFFTDEINTKNGGERVATMLMYLPWWNELSDC---------GKKGLSIKPKMGDALLFW 120
D G +R+AT+LMYL ++ + KKG+S++PK GDALLFW
Sbjct: 198 FD----------GFQRIATVLMYLSDVDKGGETVFPEAKGIKSKKGVSVRPKKGDALLFW 247
Query: 121 SMKPDGTLDPLSMHG 135
SM+PDG+ DP S HG
Sbjct: 248 SMRPDGSRDPSSKHG 262
>At4g33910 unknown protein
Length = 288
Score = 120 bits (301), Expect = 3e-28
Identities = 66/166 (39%), Positives = 97/166 (57%), Gaps = 27/166 (16%)
Query: 11 GKSVDNS--ARTSSGTFINRGHDKI--LRNIEQRIADFTFIPVENGESVNILHYEVGQKY 66
G++ +N+ RTSSGTFI+ + L +E++IA T IP +GES NIL YE+GQKY
Sbjct: 120 GETAENTKGTRTSSGTFISASEESTGALDFVERKIARATMIPRSHGESFNILRYELGQKY 179
Query: 67 EPHPDFFTDEINTKNGGERVATMLMYLPWWNELSDCGKK--------------------G 106
+ H D F +R+A+ L+YL +++ + G+ G
Sbjct: 180 DSHYDVFNPTEYGPQSSQRIASFLLYL---SDVEEGGETMFPFENGSNMGIGYDYKQCIG 236
Query: 107 LSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMR 152
L +KP+ GD LLF+S+ P+GT+D S+HG+CPV KG+KW TKW+R
Sbjct: 237 LKVKPRKGDGLLFYSVFPNGTIDQTSLHGSCPVTKGEKWVATKWIR 282
>At3g28490 prolyl 4-hydroxylase, putative
Length = 213
Score = 103 bits (258), Expect = 3e-23
Identities = 63/158 (39%), Positives = 88/158 (54%), Gaps = 29/158 (18%)
Query: 1 MHKSTV--DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNIL 58
+ KS V D ++G+S D+ RTSSG F+ + D I+ N+E ++A +TF+P ENGE++ IL
Sbjct: 64 LEKSMVVADVDSGESEDSEVRTSSGMFLTKRQDDIVANVEAKLAAWTFLPEENGEALQIL 123
Query: 59 HYEVGQKYEPHPDFFTDEINTKNGGERVATMLMYLPWWNELSDCGKKGLSIKPKMGDALL 118
HYE GQKY+PH D+F D+ + GG R+AT+LMY LS+ K G ++ P
Sbjct: 124 HYENGQKYDPHFDYFYDKKALELGGHRIATVLMY------LSNVTKGGETVFPN------ 171
Query: 119 FWSMKPDGTLDPLSMHGACPVIKGDKWS-CTKWMRVGK 155
W G P +K D WS C K GK
Sbjct: 172 -WK-------------GKTPQLKDDSWSKCAKQGYAGK 195
>At4g25600 unknown protein
Length = 291
Score = 60.5 bits (145), Expect = 4e-10
Identities = 37/132 (28%), Positives = 69/132 (52%), Gaps = 14/132 (10%)
Query: 31 DKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPHPDFFTDEINTKNGGERVATML 90
D ++ IE++++ +TF+P ENG S+ + Y +K D+F +E ++ +AT++
Sbjct: 105 DPVVAGIEEKVSAWTFLPGENGGSIKVRSY-TSEKSGKKLDYFGEEPSSVLHESLLATVV 163
Query: 91 MYLPWWNE-------------LSDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGAC 137
+YL + + C + G ++P G+A+LF++ + +LD S H C
Sbjct: 164 LYLSNTTQGGELLFPNSEMKPKNSCLEGGNILRPVKGNAILFFTRLLNASLDGKSTHLRC 223
Query: 138 PVIKGDKWSCTK 149
PV+KG+ TK
Sbjct: 224 PVVKGELLVATK 235
>At2g23100 hypothetical protein
Length = 1036
Score = 40.8 bits (94), Expect = 3e-04
Identities = 20/41 (48%), Positives = 27/41 (65%)
Query: 33 ILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPHPDFF 73
+L IE++IA T P + ES NIL Y++GQKY+ H D F
Sbjct: 861 VLAAIEEKIALATRFPKDYYESFNILRYQLGQKYDSHYDAF 901
>At1g06290 acyl-CoA oxidase, putative, 3' partial
Length = 289
Score = 28.1 bits (61), Expect = 2.1
Identities = 17/52 (32%), Positives = 23/52 (43%), Gaps = 2/52 (3%)
Query: 28 RGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPHPDFFTDEINT 79
R H+K L+N E + F E G N+ E Y+P + F INT
Sbjct: 168 RHHEKWLKNTEDYVVKGCFAMTELGHGSNVRGIETVTTYDPKTEEFV--INT 217
>At1g48930 putative glucanase gb|AAB91971.1; similar to ESTs
gb|AI995074.1, gb|AA651396.1
Length = 627
Score = 27.7 bits (60), Expect = 2.8
Identities = 14/39 (35%), Positives = 20/39 (50%), Gaps = 3/39 (7%)
Query: 64 QKYEPHPDFFTDEINTKNGGERVATM---LMYLPWWNEL 99
++Y+ D+F KNGG + T LMY+ WN L
Sbjct: 308 KQYQTKADYFACACLKKNGGYNIQTTPGGLMYVREWNNL 346
>At5g39400 PTEN -like protein
Length = 412
Score = 26.9 bits (58), Expect = 4.7
Identities = 11/44 (25%), Positives = 19/44 (43%)
Query: 63 GQKYEPHPDFFTDEINTKNGGERVATMLMYLPWWNELSDCGKKG 106
G E + + T N G + + Y+ +W++L KKG
Sbjct: 171 GMSAEEALEMYASRRTTNNNGVSIPSQRRYVKYWSDLLSFSKKG 214
>At2g34260 unknown protein
Length = 353
Score = 26.9 bits (58), Expect = 4.7
Identities = 19/63 (30%), Positives = 29/63 (45%), Gaps = 11/63 (17%)
Query: 65 KYEPHPDFFTDEINT----KNG-----GERVATMLMYLPWWNELSDCGKKGLSIKPKMGD 115
K + +F DE+ + KNG G + T+L+Y W DC + + + P D
Sbjct: 168 KVQSQSEFSEDELLSVVIMKNGRKVICGTQNGTLLLYS--WGFFKDCSDRFVDLAPNSVD 225
Query: 116 ALL 118
ALL
Sbjct: 226 ALL 228
>At2g13870 En/Spm-like transposon protein
Length = 441
Score = 26.9 bits (58), Expect = 4.7
Identities = 10/30 (33%), Positives = 19/30 (63%)
Query: 14 VDNSARTSSGTFINRGHDKILRNIEQRIAD 43
+D + R S GTFI+R +++ + + RI +
Sbjct: 266 LDETHRKSDGTFIDRKSEEVYKEVSSRIRE 295
>At1g52850 hypothetical protein
Length = 447
Score = 26.9 bits (58), Expect = 4.7
Identities = 10/30 (33%), Positives = 19/30 (63%)
Query: 14 VDNSARTSSGTFINRGHDKILRNIEQRIAD 43
+D + R S GTFI+R +++ + + RI +
Sbjct: 266 LDETHRKSDGTFIDRKSEEVYKEVSSRIQE 295
>At1g63020 RNA polymerase IIA largest subunit, putative
Length = 1453
Score = 26.6 bits (57), Expect = 6.2
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 5/52 (9%)
Query: 103 GKKGLSI-KPKMGDALLFWSMKPDGTLDPLSMH----GACPVIKGDKWSCTK 149
G KG+ + K K GD+ F ++ DGT + S H GA +I K + K
Sbjct: 1384 GVKGIRVAKSKHGDSCCFEVVRIDGTFEDFSYHKCVLGATKIIAPKKMNFYK 1435
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,156,296
Number of Sequences: 26719
Number of extensions: 176592
Number of successful extensions: 370
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 337
Number of HSP's gapped (non-prelim): 24
length of query: 160
length of database: 11,318,596
effective HSP length: 91
effective length of query: 69
effective length of database: 8,887,167
effective search space: 613214523
effective search space used: 613214523
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149547.6


