

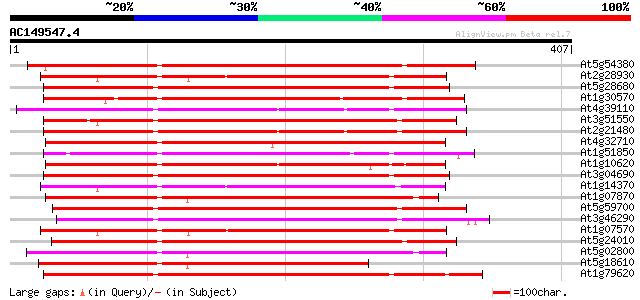
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.4 + phase: 0
(407 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g54380 receptor-protein kinase-like protein 248 3e-66
At2g28930 protein kinase like protein 235 3e-62
At5g28680 receptor-like protein kinase precursor - like 235 4e-62
At1g30570 putative serine/threonine protein kinase 234 6e-62
At4g39110 putative receptor-like protein kinase 233 1e-61
At3g51550 receptor protein kinase like protein 233 1e-61
At2g21480 putative protein kinase 233 1e-61
At4g32710 putative protein kinase 233 2e-61
At1g51850 Putative protein kinase 232 3e-61
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 231 4e-61
At3g04690 putative protein kinase 231 7e-61
At1g14370 putative protein 228 4e-60
At1g07870 putative protein kinase (At1g07870) 228 4e-60
At5g59700 receptor-like protein kinase precursor - like 228 5e-60
At3g46290 receptor protein kinase -like 228 5e-60
At1g07570 protein kinase APK1A 228 5e-60
At5g24010 receptor-protein kinase-like protein 228 6e-60
At5g02800 protein kinase - like 227 8e-60
At5g18610 protein kinase -like protein 227 1e-59
At1g79620 unknown protein 227 1e-59
>At5g54380 receptor-protein kinase-like protein
Length = 855
Score = 248 bits (634), Expect = 3e-66
Identities = 136/331 (41%), Positives = 202/331 (60%), Gaps = 12/331 (3%)
Query: 14 YKSSKYDKISL------RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIK 67
+KS+ ISL RCF +E+ AT F + LLG G F VYKG E +A+K
Sbjct: 479 HKSATASCISLASTHLGRCFMFQEIMDATNKFDESSLLGVGGFGRVYKGTLEDGTKVAVK 538
Query: 68 RPHSESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM 127
R + S + EFR E+ +LS ++H++L+ L+GYC+E ILVYEY+ NG L ++
Sbjct: 539 RGNPRSEQGMAEFRTEIEMLSKLRHRHLVSLIGYCDERSE---MILVYEYMANGPLRSHL 595
Query: 128 MGNRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGL 187
G L+WKQR+ I IGAA+G+ YLH SIIHRD+K +NILL E+ AKV+DFGL
Sbjct: 596 YGADLPPLSWKQRLEICIGAARGLHYLHTGASQSIIHRDVKTTNILLDENLVAKVADFGL 655
Query: 188 VKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAE 247
K+GP+ DQ+HVS+ +KG+ GYLDP Y LT+ SDVYSFGV+L++++ RPA++
Sbjct: 656 SKTGPSLDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLMEVLCCRPALNPVL 715
Query: 248 NPSNQHIIDWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPT 307
+I +WA +KG++ +IMD+NL + P +++ K G+ +C+A+ RP+
Sbjct: 716 PREQVNIAEWAMAWQKKGLLDQIMDSNLTGKVNPASLK---KFGETAEKCLAEYGVDRPS 772
Query: 308 MTQVCREIEHALYSDDSFTSKDSETLGSVQH 338
M V +E+AL +++ ++ S H
Sbjct: 773 MGDVLWNLEYALQLEETSSALMEPDDNSTNH 803
>At2g28930 protein kinase like protein
Length = 412
Score = 235 bits (600), Expect = 3e-62
Identities = 128/308 (41%), Positives = 190/308 (61%), Gaps = 20/308 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ FT EL+ AT+NF D +LG G F +V+KG + + ++A+K+ + +
Sbjct: 53 NLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQD 112
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMM--GN 130
+ +E+ EV L H NL+ L+GYC E E ++LVYE++P GSL ++ G+
Sbjct: 113 GWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEH---RLLVYEFMPRGSLENHLFRRGS 169
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ L+W R+ +A+GAAKG+A+LH + S+I+RD K SNILL + AK+SDFGL K
Sbjct: 170 YFQPLSWTLRLKVALGAAKGLAFLHN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKD 228
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
GPTGD+SHVS++I GT GY P Y ++ HLT SDVYS+GV+LL+++S R AVD P
Sbjct: 229 GPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPG 288
Query: 251 NQHIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMT 309
Q +++WARP + K + ++D L + +ME KV L +RC+ E K RP M
Sbjct: 289 EQKLVEWARPLLANKRKLFRVIDNRL---QDQYSMEEACKVATLALRCLTFEIKLRPNMN 345
Query: 310 QVCREIEH 317
+V +EH
Sbjct: 346 EVVSHLEH 353
>At5g28680 receptor-like protein kinase precursor - like
Length = 858
Score = 235 bits (599), Expect = 4e-62
Identities = 123/295 (41%), Positives = 183/295 (61%), Gaps = 6/295 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F++ E++ T NF + ++G G F VYKG+ + +AIK+ + S + EF E+
Sbjct: 507 RRFSLSEIKHGTHNFDESNVIGVGGFGKVYKGVIDGGTKVAIKKSNPNSEQGLNEFETEI 566
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
LLS ++HK+L+ L+GYC+E G L+Y+Y+ G+L E++ +R LTWK+R+ IA
Sbjct: 567 ELLSRLRHKHLVSLIGYCDE---GGEMCLIYDYMSLGTLREHLYNTKRPQLTWKRRLEIA 623
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH K +IIHRD+K +NILL E++ AKVSDFGL K+GP + HV++ +K
Sbjct: 624 IGAARGLHYLHTGAKYTIIHRDVKTTNILLDENWVAKVSDFGLSKTGPNMNGGHVTTVVK 683
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
G+ GYLDP Y LT+ SDVYSFGV+L +++ ARPA++ + + + DWA K
Sbjct: 684 GSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEVLCARPALNPSLSKEQVSLGDWAMNCKRK 743
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHAL 319
G + +I+D NL N E + K +C++ RPTM V +E AL
Sbjct: 744 GTLEDIIDPNL---KGKINPECLKKFADTAEKCLSDSGLDRPTMGDVLWNLEFAL 795
>At1g30570 putative serine/threonine protein kinase
Length = 849
Score = 234 bits (597), Expect = 6e-62
Identities = 130/309 (42%), Positives = 196/309 (63%), Gaps = 12/309 (3%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKR--PHSESFLSVEEFRN 82
R FT+ E+ ATKNF +G G F VY+G E ++AIKR PHS+ L+ EF
Sbjct: 506 RKFTLAEIRAATKNFDDGLAIGVGGFGKVYRGELEDGTLIAIKRATPHSQQGLA--EFET 563
Query: 83 EVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRIN 142
E+ +LS ++H++L+ L+G+C+E ILVYEY+ NG+L ++ G+ L+WKQR+
Sbjct: 564 EIVMLSRLRHRHLVSLIGFCDEHNE---MILVYEYMANGTLRSHLFGSNLPPLSWKQRLE 620
Query: 143 IAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQ 202
IG+A+G+ YLH + IIHRD+K +NILL E+F AK+SDFGL K+GP+ D +HVS+
Sbjct: 621 ACIGSARGLHYLHTGSERGIIHRDVKTTNILLDENFVAKMSDFGLSKAGPSMDHTHVSTA 680
Query: 203 IKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQ-HIIDWARPS 261
+KG+ GYLDP Y LT+ SDVYSFGV+L + + AR AV N P +Q ++ +WA
Sbjct: 681 VKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEAVCAR-AVINPTLPKDQINLAEWALSW 739
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYS 321
++ + I+D+NL P ++E K G++ +C+A E K+RP M +V +E+ L
Sbjct: 740 QKQRNLESIIDSNLRGNYSPESLE---KYGEIAEKCLADEGKNRPMMGEVLWSLEYVLQI 796
Query: 322 DDSFTSKDS 330
+++ K +
Sbjct: 797 HEAWLRKQN 805
>At4g39110 putative receptor-like protein kinase
Length = 878
Score = 233 bits (595), Expect = 1e-61
Identities = 134/329 (40%), Positives = 199/329 (59%), Gaps = 11/329 (3%)
Query: 6 FSFDAGKEYKSSKYDK-ISL-RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGI 63
F G KS+ Y+ + L R F++ EL+ ATKNF ++G G F NVY G +
Sbjct: 491 FMTSKGGSQKSNFYNSTLGLGRYFSLSELQEATKNFEASQIIGVGGFGNVYIGTLDDGTK 550
Query: 64 LAIKRPHSESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSL 123
+A+KR + +S + EF+ E+++LS ++H++L+ L+GYC+E + ILVYE++ NG
Sbjct: 551 VAVKRGNPQSEQGITEFQTEIQMLSKLRHRHLVSLIGYCDE---NSEMILVYEFMSNGPF 607
Query: 124 LEYMMGNRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVS 183
+++ G LTWKQR+ I IG+A+G+ YLH IIHRD+K +NILL E+ AKV+
Sbjct: 608 RDHLYGKNLAPLTWKQRLEICIGSARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVA 667
Query: 184 DFGLVKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAV 243
DFGL K G Q+HVS+ +KG+ GYLDP Y LT SDVYSFGV+LL+ + ARPA+
Sbjct: 668 DFGLSKDVAFG-QNHVSTAVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAI 726
Query: 244 DNAENPSNQ-HIIDWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEP 302
N + P Q ++ +WA KG++ +I+D +L + N E M K + +C+
Sbjct: 727 -NPQLPREQVNLAEWAMQWKRKGLLEKIIDPHL---AGTINPESMKKFAEAAEKCLEDYG 782
Query: 303 KHRPTMTQVCREIEHALYSDDSFTSKDSE 331
RPTM V +E+AL ++FT +E
Sbjct: 783 VDRPTMGDVLWNLEYALQLQEAFTQGKAE 811
>At3g51550 receptor protein kinase like protein
Length = 895
Score = 233 bits (595), Expect = 1e-61
Identities = 129/303 (42%), Positives = 190/303 (62%), Gaps = 11/303 (3%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG---ILAIKRPHSESFLSVEEFR 81
R F+ E++ ATKNF + +LG G F VY+G E++G +AIKR + S V EF+
Sbjct: 522 RHFSFAEIKAATKNFDESRVLGVGGFGKVYRG--EIDGGTTKVAIKRGNPMSEQGVHEFQ 579
Query: 82 NEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRI 141
E+ +LS ++H++L+ L+GYCEE + ILVY+Y+ +G++ E++ + SL WKQR+
Sbjct: 580 TEIEMLSKLRHRHLVSLIGYCEE---NCEMILVYDYMAHGTMREHLYKTQNPSLPWKQRL 636
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
I IGAA+G+ YLH K +IIHRD+K +NILL E + AKVSDFGL K+GPT D +HVS+
Sbjct: 637 EICIGAARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDHTHVST 696
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPS 261
+KG+ GYLDP Y LT+ SDVYSFGV+L + + ARPA++ + +WA
Sbjct: 697 VVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEALCARPALNPTLAKEQVSLAEWAPYC 756
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYS 321
+KG++ +I+D L + P E K + ++CV + RP+M V +E AL
Sbjct: 757 YKKGMLDQIVDPYLKGKITP---ECFKKFAETAMKCVLDQGIERPSMGDVLWNLEFALQL 813
Query: 322 DDS 324
+S
Sbjct: 814 QES 816
>At2g21480 putative protein kinase
Length = 871
Score = 233 bits (595), Expect = 1e-61
Identities = 130/308 (42%), Positives = 189/308 (61%), Gaps = 9/308 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F++ EL+ TKNF ++G G F NVY G + +AIKR + +S + EF E+
Sbjct: 511 RYFSLSELQEVTKNFDASEIIGVGGFGNVYIGTIDDGTQVAIKRGNPQSEQGITEFHTEI 570
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
++LS ++H++L+ L+GYC+E + ILVYEY+ NG +++ G LTWKQR+ I
Sbjct: 571 QMLSKLRHRHLVSLIGYCDE---NAEMILVYEYMSNGPFRDHLYGKNLSPLTWKQRLEIC 627
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH IIHRD+K +NILL E+ AKV+DFGL K G Q+HVS+ +K
Sbjct: 628 IGAARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVADFGLSKDVAFG-QNHVSTAVK 686
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQ-HIIDWARPSIE 263
G+ GYLDP Y LT SDVYSFGV+LL+ + ARPA+ N + P Q ++ +WA +
Sbjct: 687 GSFGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAI-NPQLPREQVNLAEWAMLWKQ 745
Query: 264 KGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDD 323
KG++ +I+D +L P E M K + +C+A RPTM V +E+AL +
Sbjct: 746 KGLLEKIIDPHLVGAVNP---ESMKKFAEAAEKCLADYGVDRPTMGDVLWNLEYALQLQE 802
Query: 324 SFTSKDSE 331
+F+ +E
Sbjct: 803 AFSQGKAE 810
>At4g32710 putative protein kinase
Length = 731
Score = 233 bits (593), Expect = 2e-61
Identities = 128/293 (43%), Positives = 182/293 (61%), Gaps = 6/293 (2%)
Query: 27 FTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRL 86
F+ EEL +AT FS++ LLG G F V+KG+ + +A+K+ S+ EF+ EV
Sbjct: 377 FSYEELSKATGGFSEENLLGEGGFGYVHKGVLKNGTEVAVKQLKIGSYQGEREFQAEVDT 436
Query: 87 LSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIG 146
+S V HK+L+ LVGYC ++ ++LVYE+VP +L ++ NR L W+ R+ IA+G
Sbjct: 437 ISRVHHKHLVSLVGYCVNGDK---RLLVYEFVPKDTLEFHLHENRGSVLEWEMRLRIAVG 493
Query: 147 AAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVK--SGPTGDQSHVSSQIK 204
AAKG+AYLHE P+IIHRDIK +NILL FEAKVSDFGL K S +H+S+++
Sbjct: 494 AAKGLAYLHEDCSPTIIHRDIKAANILLDSKFEAKVSDFGLAKFFSDTNSSFTHISTRVV 553
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
GT GY+ P Y SS +T SDVYSFGV+LL+LI+ RP++ ++ +NQ ++DWARP + K
Sbjct: 554 GTFGYMAPEYASSGKVTDKSDVYSFGVVLLELITGRPSIFAKDSSTNQSLVDWARPLLTK 613
Query: 265 GIIAEIMDANLFCQSEP-CNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
I E D + + E + M + C+ Q RP M+QV R +E
Sbjct: 614 AISGESFDFLVDSRLEKNYDTTQMANMAACAAACIRQSAWLRPRMSQVVRALE 666
>At1g51850 Putative protein kinase
Length = 875
Score = 232 bits (591), Expect = 3e-61
Identities = 131/316 (41%), Positives = 188/316 (59%), Gaps = 13/316 (4%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R FT ++ T NF + +LG G F VY G +A+K S +EF+ EV
Sbjct: 556 RRFTYSQVAIMTNNFQR--ILGKGGFGMVYHGFVNGTEQVAVKILSHSSSQGYKEFKAEV 613
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRR-SLTWKQRINI 143
LL V HKNL+GLVGYC+E E L+YEY+ NG L E+M G R R +L W R+ I
Sbjct: 614 ELLLRVHHKNLVGLVGYCDEGEN---MALIYEYMANGDLKEHMSGTRNRFTLNWGTRLKI 670
Query: 144 AIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQI 203
+ +A+G+ YLH KP ++HRD+K +NILL E F+AK++DFGL +S P ++HVS+ +
Sbjct: 671 VVESAQGLEYLHNGCKPPMVHRDVKTTNILLNEHFQAKLADFGLSRSFPIEGETHVSTVV 730
Query: 204 KGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIE 263
GTPGYLDP Y + LT+ SDVYSFG++LL+LI+ RP +D + HI +W +
Sbjct: 731 AGTPGYLDPEYYKTNWLTEKSDVYSFGIVLLELITNRPVIDKSR--EKPHIAEWVGVMLT 788
Query: 264 KGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDD 323
KG I IMD NL +E + + K +L + C+ RPTM+QV E+ + S++
Sbjct: 789 KGDINSIMDPNL---NEDYDSGSVWKAVELAMSCLNPSSARRPTMSQVVIELNECIASEN 845
Query: 324 S--FTSKDSETLGSVQ 337
S S+D ++ S++
Sbjct: 846 SRGGASRDMDSKSSIE 861
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 231 bits (590), Expect = 4e-61
Identities = 131/294 (44%), Positives = 183/294 (61%), Gaps = 11/294 (3%)
Query: 27 FTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRL 86
FT EEL + T+ F + ++G G F VYKGI +AIK+ S S EF+ EV +
Sbjct: 358 FTYEELSQITEGFCKSFVVGEGGFGCVYKGILFEGKPVAIKQLKSVSAEGYREFKAEVEI 417
Query: 87 LSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIG 146
+S V H++L+ LVGYC + + L+YE+VPN +L ++ G L W +R+ IAIG
Sbjct: 418 ISRVHHRHLVSLVGYCISEQH---RFLIYEFVPNNTLDYHLHGKNLPVLEWSRRVRIAIG 474
Query: 147 AAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGT 206
AAKG+AYLHE P IIHRDIK SNILL + FEA+V+DFGL + T QSH+S+++ GT
Sbjct: 475 AAKGLAYLHEDCHPKIIHRDIKSSNILLDDEFEAQVADFGLARLNDTA-QSHISTRVMGT 533
Query: 207 PGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARP----SI 262
GYL P Y SS LT SDV+SFGV+LL+LI+ R VD ++ + +++WARP +I
Sbjct: 534 FGYLAPEYASSGKLTDRSDVFSFGVVLLELITGRKPVDTSQPLGEESLVEWARPRLIEAI 593
Query: 263 EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
EKG I+E++D L +++ EV K+ + CV RP M QV R ++
Sbjct: 594 EKGDISEVVDPRL--ENDYVESEV-YKMIETAASCVRHSALKRPRMVQVVRALD 644
>At3g04690 putative protein kinase
Length = 850
Score = 231 bits (588), Expect = 7e-61
Identities = 121/295 (41%), Positives = 181/295 (61%), Gaps = 6/295 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F++ E++ T+NF ++G G F VYKG+ + +A+K+ + S + EF E+
Sbjct: 503 RRFSLPEIKHGTQNFDDSNVIGVGGFGKVYKGVIDGTTKVAVKKSNPNSEQGLNEFETEI 562
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
LLS ++HK+L+ L+GYC+E G LVY+Y+ G+L E++ ++ LTWK+R+ IA
Sbjct: 563 ELLSRLRHKHLVSLIGYCDE---GGEMCLVYDYMAFGTLREHLYNTKKPQLTWKRRLEIA 619
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
IGAA+G+ YLH K +IIHRD+K +NIL+ E++ AKVSDFGL K+GP + HV++ +K
Sbjct: 620 IGAARGLHYLHTGAKYTIIHRDVKTTNILVDENWVAKVSDFGLSKTGPNMNGGHVTTVVK 679
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
G+ GYLDP Y LT+ SDVYSFGV+L +++ ARPA++ + + DWA K
Sbjct: 680 GSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEILCARPALNPSLPKEQVSLGDWAMNCKRK 739
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHAL 319
G + +I+D NL N E + K +C+ RPTM V +E AL
Sbjct: 740 GNLEDIIDPNL---KGKINAECLKKFADTAEKCLNDSGLERPTMGDVLWNLEFAL 791
>At1g14370 putative protein
Length = 426
Score = 228 bits (582), Expect = 4e-60
Identities = 128/305 (41%), Positives = 182/305 (58%), Gaps = 18/305 (5%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ FT EL+ ATKNF QD LLG G F V+KG + ++A+K+ E
Sbjct: 70 NLKAFTFNELKNATKNFRQDNLLGEGGFGCVFKGWIDQTSLTASRPGSGIVVAVKQLKPE 129
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRR 132
F +E+ EV L + H NL+ LVGYC E E ++LVYE++P GSL ++
Sbjct: 130 GFQGHKEWLTEVNYLGQLSHPNLVLLVGYCAEGEN---RLLVYEFMPKGSLENHLFRRGA 186
Query: 133 RSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGP 192
+ LTW R+ +A+GAAKG+ +LHE K +I+RD K +NILL F AK+SDFGL K+GP
Sbjct: 187 QPLTWAIRMKVAVGAAKGLTFLHE-AKSQVIYRDFKAANILLDADFNAKLSDFGLAKAGP 245
Query: 193 TGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQ 252
TGD +HVS+++ GT GY P Y ++ LT SDVYSFGV+LL+LIS R A+DN+ +
Sbjct: 246 TGDNTHVSTKVIGTHGYAAPEYVATGRLTAKSDVYSFGVVLLELISGRRAMDNSNGGNEY 305
Query: 253 HIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQV 311
++DWA P + +K + IMD L Q + L ++C+ + K RP M++V
Sbjct: 306 SLVDWATPYLGDKRKLFRIMDTKLGGQYP---QKGAFTAANLALQCLNPDAKLRPKMSEV 362
Query: 312 CREIE 316
+E
Sbjct: 363 LVTLE 367
>At1g07870 putative protein kinase (At1g07870)
Length = 423
Score = 228 bits (582), Expect = 4e-60
Identities = 120/289 (41%), Positives = 183/289 (62%), Gaps = 10/289 (3%)
Query: 27 FTIEELERATKNFSQDCLLGSGAFCNVYKGIFE-LEGILAIKRPHSESFLSVEEFRNEVR 85
FT +EL AT NF DC LG G F V+KG E L+ ++AIK+ + EF EV
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVL 150
Query: 86 LLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM--MGNRRRSLTWKQRINI 143
LS H NL+ L+G+C E ++ ++LVYEY+P GSL +++ + + ++ L W R+ I
Sbjct: 151 TLSLADHPNLVKLIGFCAEGDQ---RLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKI 207
Query: 144 AIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQI 203
A GAA+G+ YLH+++ P +I+RD+K SNILLGE ++ K+SDFGL K GP+GD++HVS+++
Sbjct: 208 AAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRV 267
Query: 204 KGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSI- 262
GT GY P Y + LT SD+YSFGV+LL+LI+ R A+DN + +Q+++ WARP
Sbjct: 268 MGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFK 327
Query: 263 EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQV 311
++ +++D L Q + L + + CV ++P RP ++ V
Sbjct: 328 DRRNFPKMVDPLLQGQYPVRGLYQALAISAM---CVQEQPTMRPVVSDV 373
>At5g59700 receptor-like protein kinase precursor - like
Length = 829
Score = 228 bits (581), Expect = 5e-60
Identities = 120/300 (40%), Positives = 185/300 (61%), Gaps = 6/300 (2%)
Query: 32 LERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRLLSAVK 91
++ AT +F ++ +G G F VYKG +A+KR + +S + EFR E+ +LS +
Sbjct: 475 VKEATNSFDENRAIGVGGFGKVYKGELHDGTKVAVKRANPKSQQGLAEFRTEIEMLSQFR 534
Query: 92 HKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIGAAKGI 151
H++L+ L+GYC+E + ILVYEY+ NG+L ++ G+ SL+WKQR+ I IG+A+G+
Sbjct: 535 HRHLVSLIGYCDE---NNEMILVYEYMENGTLKSHLYGSGLLSLSWKQRLEICIGSARGL 591
Query: 152 AYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGTPGYLD 211
YLH +IHRD+K +NILL E+ AKV+DFGL K+GP DQ+HVS+ +KG+ GYLD
Sbjct: 592 HYLHTGDAKPVIHRDVKSANILLDENLMAKVADFGLSKTGPEIDQTHVSTAVKGSFGYLD 651
Query: 212 PAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEKGIIAEIM 271
P Y LT+ SDVYSFGV++ +++ ARP +D ++ +WA +KG + I+
Sbjct: 652 PEYFRRQQLTEKSDVYSFGVVMFEVLCARPVIDPTLTREMVNLAEWAMKWQKKGQLEHII 711
Query: 272 DANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDSFTSKDSE 331
D +L + P + + K G+ G +C+A RP+M V +E+AL ++ D E
Sbjct: 712 DPSLRGKIRP---DSLRKFGETGEKCLADYGVDRPSMGDVLWNLEYALQLQEAVVDGDPE 768
>At3g46290 receptor protein kinase -like
Length = 830
Score = 228 bits (581), Expect = 5e-60
Identities = 128/321 (39%), Positives = 192/321 (58%), Gaps = 13/321 (4%)
Query: 35 ATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRLLSAVKHKN 94
AT NF + +G G F VYKG +A+KR + +S + EFR E+ +LS +H++
Sbjct: 481 ATNNFDESRNIGVGGFGKVYKGELNDGTKVAVKRGNPKSQQGLAEFRTEIEMLSQFRHRH 540
Query: 95 LIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIGAAKGIAYL 154
L+ L+GYC+E + IL+YEY+ NG++ ++ G+ SLTWKQR+ I IGAA+G+ YL
Sbjct: 541 LVSLIGYCDE---NNEMILIYEYMENGTVKSHLYGSGLPSLTWKQRLEICIGAARGLHYL 597
Query: 155 HEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGTPGYLDPAY 214
H +IHRD+K +NILL E+F AKV+DFGL K+GP DQ+HVS+ +KG+ GYLDP Y
Sbjct: 598 HTGDSKPVIHRDVKSANILLDENFMAKVADFGLSKTGPELDQTHVSTAVKGSFGYLDPEY 657
Query: 215 CSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEKGIIAEIMDAN 274
LT SDVYSFGV+L +++ ARP +D ++ +WA +KG + +I+D +
Sbjct: 658 FRRQQLTDKSDVYSFGVVLFEVLCARPVIDPTLPREMVNLAEWAMKWQKKGQLDQIIDQS 717
Query: 275 LFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDSFTSKDSE--- 331
L P + + K + G +C+A RP+M V +E+AL ++ + E
Sbjct: 718 LRGNIRP---DSLRKFAETGEKCLADYGVDRPSMGDVLWNLEYALQLQEAVIDGEPEDNS 774
Query: 332 --TLGSV--QHCDFSQGFVSI 348
+G + Q +FSQG S+
Sbjct: 775 TNMIGELPPQINNFSQGDTSV 795
>At1g07570 protein kinase APK1A
Length = 410
Score = 228 bits (581), Expect = 5e-60
Identities = 124/308 (40%), Positives = 189/308 (61%), Gaps = 20/308 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ F+ EL+ AT+NF D +LG G F V+KG + + ++A+K+ + +
Sbjct: 52 NLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQD 111
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMM--GN 130
+ +E+ EV L H++L+ L+GYC E E ++LVYE++P GSL ++ G
Sbjct: 112 GWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEH---RLLVYEFMPRGSLENHLFRRGL 168
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ L+WK R+ +A+GAAKG+A+LH + +I+RD K SNILL + AK+SDFGL K
Sbjct: 169 YFQPLSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKD 227
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
GP GD+SHVS+++ GT GY P Y ++ HLT SDVYSFGV+LL+L+S R AVD
Sbjct: 228 GPIGDKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSG 287
Query: 251 NQHIIDWARP-SIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMT 309
+++++WA+P + K I ++D L + +ME KV L +RC+ E K RP M+
Sbjct: 288 ERNLVEWAKPYLVNKRKIFRVIDNRL---QDQYSMEEACKVATLSLRCLTTEIKLRPNMS 344
Query: 310 QVCREIEH 317
+V +EH
Sbjct: 345 EVVSHLEH 352
>At5g24010 receptor-protein kinase-like protein
Length = 824
Score = 228 bits (580), Expect = 6e-60
Identities = 120/294 (40%), Positives = 179/294 (60%), Gaps = 6/294 (2%)
Query: 31 ELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRLLSAV 90
EL+ T NF + ++G G F V++G + +A+KR S + EF +E+ +LS +
Sbjct: 481 ELQSGTNNFDRSLVIGVGGFGMVFRGSLKDNTKVAVKRGSPGSRQGLPEFLSEITILSKI 540
Query: 91 KHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIGAAKG 150
+H++L+ LVGYCEE ILVYEY+ G L ++ G+ L+WKQR+ + IGAA+G
Sbjct: 541 RHRHLVSLVGYCEEQSE---MILVYEYMDKGPLKSHLYGSTNPPLSWKQRLEVCIGAARG 597
Query: 151 IAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGTPGYL 210
+ YLH IIHRDIK +NILL ++ AKV+DFGL +SGP D++HVS+ +KG+ GYL
Sbjct: 598 LHYLHTGSSQGIIHRDIKSTNILLDNNYVAKVADFGLSRSGPCIDETHVSTGVKGSFGYL 657
Query: 211 DPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEKGIIAEI 270
DP Y LT SDVYSFGV+L +++ ARPAVD ++ +WA KG++ +I
Sbjct: 658 DPEYFRRQQLTDKSDVYSFGVVLFEVLCARPAVDPLLVREQVNLAEWAIEWQRKGMLDQI 717
Query: 271 MDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDS 324
+D N+ + +PC+++ K + +C A RPT+ V +EH L +S
Sbjct: 718 VDPNIADEIKPCSLK---KFAETAEKCCADYGVDRPTIGDVLWNLEHVLQLQES 768
>At5g02800 protein kinase - like
Length = 395
Score = 227 bits (579), Expect = 8e-60
Identities = 125/309 (40%), Positives = 185/309 (59%), Gaps = 10/309 (3%)
Query: 13 EYKSSKYDKISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKG-IFELEGILAIKRPHS 71
E KS D I + FT EL AT+NF ++CL+G G F VYKG + AIK+
Sbjct: 47 ESKSKGSDHIVAQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTSQTAAIKQLDH 106
Query: 72 ESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM--MG 129
EF EV +LS + H NL+ L+GYC + ++ ++LVYEY+P GSL +++ +
Sbjct: 107 NGLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQ---RLLVYEYMPLGSLEDHLHDIS 163
Query: 130 NRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVK 189
++ L W R+ IA GAAKG+ YLH+K P +I+RD+K SNILL + + K+SDFGL K
Sbjct: 164 PGKQPLDWNTRMKIAAGAAKGLEYLHDKTMPPVIYRDLKCSNILLDDDYFPKLSDFGLAK 223
Query: 190 SGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENP 249
GP GD+SHVS+++ GT GY P Y + LT SDVYSFGV+LL++I+ R A+D++ +
Sbjct: 224 LGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDSSRST 283
Query: 250 SNQHIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTM 308
Q+++ WARP ++ +++ D L Q P + L V + CV ++P RP +
Sbjct: 284 GEQNLVAWARPLFKDRRKFSQMADPMLQGQYPPRGLYQALAVAAM---CVQEQPNLRPLI 340
Query: 309 TQVCREIEH 317
V + +
Sbjct: 341 ADVVTALSY 349
>At5g18610 protein kinase -like protein
Length = 513
Score = 227 bits (578), Expect = 1e-59
Identities = 115/242 (47%), Positives = 158/242 (64%), Gaps = 6/242 (2%)
Query: 22 ISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG-ILAIKRPHSESFLSVEEF 80
I+ + FT EL ATKNF +CLLG G F VYKG E G I+A+K+ EF
Sbjct: 66 IAAQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLETTGQIVAVKQLDRNGLQGNREF 125
Query: 81 RNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM--MGNRRRSLTWK 138
EV +LS + H NL+ L+GYC + ++ ++LVYEY+P GSL +++ + + L W
Sbjct: 126 LVEVLMLSLLHHPNLVNLIGYCADGDQ---RLLVYEYMPLGSLEDHLHDLPPDKEPLDWS 182
Query: 139 QRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSH 198
R+ IA GAAKG+ YLH+K P +I+RD+K SNILLG+ + K+SDFGL K GP GD++H
Sbjct: 183 TRMTIAAGAAKGLEYLHDKANPPVIYRDLKSSNILLGDGYHPKLSDFGLAKLGPVGDKTH 242
Query: 199 VSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWA 258
VS+++ GT GY P Y + LT SDVYSFGV+ L+LI+ R A+DNA P +++ WA
Sbjct: 243 VSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVFLELITGRKAIDNARAPGEHNLVAWA 302
Query: 259 RP 260
RP
Sbjct: 303 RP 304
>At1g79620 unknown protein
Length = 971
Score = 227 bits (578), Expect = 1e-59
Identities = 129/319 (40%), Positives = 193/319 (60%), Gaps = 8/319 (2%)
Query: 25 RCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEV 84
R F+ EEL++ T NFS LG G + VYKG+ + ++AIKR S EF+ E+
Sbjct: 624 RWFSYEELKKITNNFSVSSELGYGGYGKVYKGMLQDGHMVAIKRAQQGSTQGGLEFKTEI 683
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIA 144
LLS V HKNL+GLVG+C E G +ILVYEY+ NGSL + + G +L WK+R+ +A
Sbjct: 684 ELLSRVHHKNLVGLVGFCFE---QGEQILVYEYMSNGSLKDSLTGRSGITLDWKRRLRVA 740
Query: 145 IGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIK 204
+G+A+G+AYLHE P IIHRD+K +NILL E+ AKV+DFGL K + HVS+Q+K
Sbjct: 741 LGSARGLAYLHELADPPIIHRDVKSTNILLDENLTAKVADFGLSKLVSDCTKGHVSTQVK 800
Query: 205 GTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK 264
GT GYLDP Y ++ LT+ SDVYSFGV++++LI+A+ ++ + + + + +
Sbjct: 801 GTLGYLDPEYYTTQKLTEKSDVYSFGVVMMELITAKQPIEKGKYIVREIKLVMNKSDDDF 860
Query: 265 GIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALYSDDS 324
+ + MD +L + + + + +L ++CV + RPTM++V +EIE + +S
Sbjct: 861 YGLRDKMDRSL---RDVGTLPELGRYMELALKCVDETADERPTMSEVVKEIE--IIIQNS 915
Query: 325 FTSKDSETLGSVQHCDFSQ 343
S S S DF +
Sbjct: 916 GASSSSSASASSSATDFGE 934
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,527,383
Number of Sequences: 26719
Number of extensions: 359414
Number of successful extensions: 4317
Number of sequences better than 10.0: 1014
Number of HSP's better than 10.0 without gapping: 850
Number of HSP's successfully gapped in prelim test: 164
Number of HSP's that attempted gapping in prelim test: 920
Number of HSP's gapped (non-prelim): 1083
length of query: 407
length of database: 11,318,596
effective HSP length: 102
effective length of query: 305
effective length of database: 8,593,258
effective search space: 2620943690
effective search space used: 2620943690
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149547.4


