

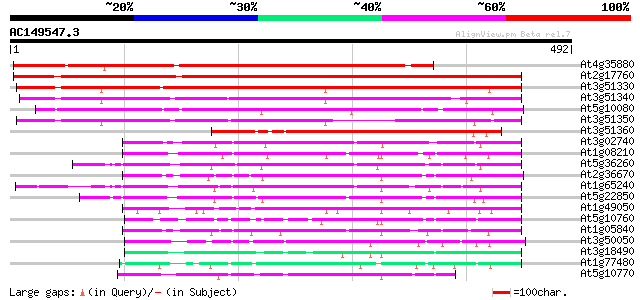
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.3 + phase: 0
(492 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g35880 predicted GPI-anchored protein 516 e-147
At2g17760 putative chloroplast nucleoid DNA-binding protein 483 e-136
At3g51330 predicted GPI-anchored protein 397 e-111
At3g51340 putative protein 338 4e-93
At5g10080 predicted GPI-anchored protein 302 2e-82
At3g51350 predicted GPI-anchored protein 280 2e-75
At3g51360 putative protein 209 3e-54
At3g02740 aspartyl protease - like predicted GPI-anchored protein 132 6e-31
At1g08210 GPI-anchored protein 127 2e-29
At5g36260 aspartyl protease-like predicted GPI-anchored protein 125 5e-29
At2g36670 putative protease 124 1e-28
At1g65240 hypothetical protein 122 4e-28
At5g22850 protease-like protein 118 8e-27
At1g49050 unknown protein 117 2e-26
At5g10760 nucleoid DNA-binding protein cnd41 - like protein 110 1e-24
At1g05840 unknown protein 108 7e-24
At3g50050 unknown protein 105 7e-23
At3g18490 chloroplast nucleoid DNA-binding protein like 103 3e-22
At1g77480 nucellin-like protein 102 4e-22
At5g10770 nucleoid DNA-binding protein cnd41 - like protein 102 5e-22
>At4g35880 predicted GPI-anchored protein
Length = 455
Score = 516 bits (1330), Expect = e-147
Identities = 255/373 (68%), Positives = 301/373 (80%), Gaps = 14/373 (3%)
Query: 4 FTKIIVIILIILHLSM-CCNAHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYA 62
F + +I I++ LS CN IFTF MHHR+S+ VK+WS S + ++P KGS EY+
Sbjct: 6 FKTTLFLIPILMLLSFGSCNGRIFTFEMHHRFSDEVKQWSDSTGRFA-KFPPKGSFEYFN 64
Query: 63 ELADRDRFLRGRRLSQFDA----GLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVAL 118
L RD +RGRRLS+ ++ L FSDGNST RISSLGFLHYTT++LGTPG++FMVAL
Sbjct: 65 ALVLRDWLIRGRRLSESESESESSLTFSDGNSTSRISSLGFLHYTTVKLGTPGMRFMVAL 124
Query: 119 DTGSDLFWVPCDCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQ 178
DTGSDLFWVPCDC +C+ T + +AS +F+LS+YNP S+T+KKVTCNNSLC RNQ
Sbjct: 125 DTGSDLFWVPCDCGKCAPTEGATYAS----EFELSIYNPKVSTTNKKVTCNNSLCAQRNQ 180
Query: 179 CLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLD 238
CLGTFS CPYMVSYVSA+TSTSGIL+EDV+HLT D N + VEA V FGCGQVQSGSFLD
Sbjct: 181 CLGTFSTCPYMVSYVSAQTSTSGILMEDVMHLTTEDKNPERVEAYVTFGCGQVQSGSFLD 240
Query: 239 VAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFGRDGIGRISFGDKGSLDQDETPFNV 298
+AAPNGLFGLGMEKISVPS+L+REG ADSFSMCFG DG+GRISFGDKGS DQ+ETPFN+
Sbjct: 241 IAAPNGLFGLGMEKISVPSVLAREGLVADSFSMCFGHDGVGRISFGDKGSSDQEETPFNL 300
Query: 299 NPSHPTYNITINQVRVGTTLIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRP 358
NPSHP YNIT+ +VRVGTTLID EFTALFD+GTSFTYLVDP Y+ +SES +D+R
Sbjct: 301 NPSHPNYNITVTRVRVGTTLIDDEFTALFDTGTSFTYLVDPMYTTVSES----AQDKRHS 356
Query: 359 PDSRIPFDYCYDM 371
PDSRIPF+YCYDM
Sbjct: 357 PDSRIPFEYCYDM 369
>At2g17760 putative chloroplast nucleoid DNA-binding protein
Length = 513
Score = 483 bits (1243), Expect = e-136
Identities = 239/448 (53%), Positives = 311/448 (69%), Gaps = 11/448 (2%)
Query: 4 FTKIIVIILIILHLSMCCNAHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYAE 63
F +++++ L C F F HHR+S+ V P P + S +YY
Sbjct: 11 FLGLLILLASSWVLDRCEGFGEFGFEFHHRFSDQVV-----GVLPGDGLPNRDSSKYYRV 65
Query: 64 LADRDRFLRGRRLSQFDAGLA-FSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGS 122
+A RDR +RGRRL+ D L FSDGN T R+ +LGFLHY + +GTP FMVALDTGS
Sbjct: 66 MAHRDRLIRGRRLANEDQSLVTFSDGNETVRVDALGFLHYANVTVGTPSDWFMVALDTGS 125
Query: 123 DLFWVPCDCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQCLGT 182
DLFW+PCDCT C + S+L DL++Y+PN SSTS KV CN++LCT ++C
Sbjct: 126 DLFWLPCDCTNCVRELKAPGGSSL----DLNIYSPNASSTSTKVPCNSTLCTRGDRCASP 181
Query: 183 FSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAP 242
S+CPY + Y+S TS++G+LVEDVLHL D + + A V FGCGQVQ+G F D AAP
Sbjct: 182 ESDCPYQIRYLSNGTSSTGVLVEDVLHLVSNDKSSKAIPARVTFGCGQVQTGVFHDGAAP 241
Query: 243 NGLFGLGMEKISVPSMLSREGFTADSFSMCFGRDGIGRISFGDKGSLDQDETPFNVNPSH 302
NGLFGLG+E ISVPS+L++EG A+SFSMCFG DG GRISFGDKGS+DQ ETP N+ H
Sbjct: 242 NGLFGLGLEDISVPSVLAKEGIAANSFSMCFGNDGAGRISFGDKGSVDQRETPLNIRQPH 301
Query: 303 PTYNITINQVRVGTTLIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRR-RPPDS 361
PTYNIT+ ++ VG D+EF A+FDSGTSFTYL D Y+ +SESF+S D+R + DS
Sbjct: 302 PTYNITVTKISVGGNTGDLEFDAVFDSGTSFTYLTDAAYTLISESFNSLALDKRYQTTDS 361
Query: 362 RIPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPIIIISTQSELVYCLAVVKSAELN 421
+PF+YCY +SP+ ++ P+++LTM GGS + VY P+++I + VYCLA++K +++
Sbjct: 362 ELPFEYCYALSPNKDSFQYPAVNLTMKGGSSYPVYHPLVVIPMKDTDVYCLAIMKIEDIS 421
Query: 422 IIGQNFMTGYRVVFDRGKLILGWKKSDC 449
IIGQNFMTGYRVVFDR KLILGWK+SDC
Sbjct: 422 IIGQNFMTGYRVVFDREKLILGWKESDC 449
>At3g51330 predicted GPI-anchored protein
Length = 529
Score = 397 bits (1019), Expect = e-111
Identities = 199/450 (44%), Positives = 290/450 (64%), Gaps = 13/450 (2%)
Query: 7 IIVIILIILHLSMCCNAHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYAELAD 66
++ ++++ L C + F+F +HH +S+ VK+ + PEKGS+EY+ LA
Sbjct: 10 LLSLLVVCWGLERCEASGKFSFEVHHMFSDRVKQ----SLGLDDLVPEKGSLEYFKVLAQ 65
Query: 67 RDRFLRGRRLSQF--DAGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDL 124
RDR +RGR L+ + + F GN T I LGFLHY + +GTP F+VALDTGSDL
Sbjct: 66 RDRLIRGRGLASNNEETPITFMRGNRTISIDLLGFLHYANVSVGTPATWFLVALDTGSDL 125
Query: 125 FWVPCDCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQCLGTFS 184
FW+PC+C S L+ L++Y+PN SSTS + C++ C ++C S
Sbjct: 126 FWLPCNCG--STCIRDLKEVGLSQSRPLNLYSPNTSSTSSSIRCSDDRCFGSSRCSSPAS 183
Query: 185 NCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPNG 244
+CPY + Y+S +T T+G L EDVLHL D+ + V+AN+ GCG+ Q+G AA NG
Sbjct: 184 SCPYQIQYLSKDTFTTGTLFEDVLHLVTEDEGLEPVKANITLGCGKNQTGFLQSSAAVNG 243
Query: 245 LFGLGMEKISVPSMLSREGFTADSFSMCFGR--DGIGRISFGDKGSLDQDETPFNVNPSH 302
L GLG++ SVPS+L++ TA+SFSMCFG D +GRISFGDKG DQ ETP
Sbjct: 244 LLGLGLKDYSVPSILAKAKITANSFSMCFGNIIDVVGRISFGDKGYTDQMETPLLPTEPS 303
Query: 303 PTYNITINQVRVGTTLIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSR 362
PTY +++ +V VG + V+ ALFD+GTSFT+L++P Y ++++F V D+RRP D
Sbjct: 304 PTYAVSVTEVSVGGDAVGVQLLALFDTGTSFTHLLEPEYGLITKAFDDHVTDKRRPIDPE 363
Query: 363 IPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPIIII-STQSELVYCLAVVKSAE-- 419
+PF++CYD+SP+ T L P +++T GGS+ + +P+ I+ + + +YCL ++KS +
Sbjct: 364 LPFEFCYDLSPNKTTILFPRVAMTFEGGSQMFLRNPLFIVWNEDNSAMYCLGILKSVDFK 423
Query: 420 LNIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
+NIIGQNFM+GYR+VFDR ++ILGWK+SDC
Sbjct: 424 INIIGQNFMSGYRIVFDRERMILGWKRSDC 453
>At3g51340 putative protein
Length = 518
Score = 338 bits (867), Expect = 4e-93
Identities = 188/462 (40%), Positives = 268/462 (57%), Gaps = 36/462 (7%)
Query: 9 VIILIILHLSMCCNAHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYAELADRD 68
+++LI L C + F+F +HH +S+ VK+ PE GS+EY+ LA RD
Sbjct: 1 MLVLIFWGLERCEASGKFSFEVHHMFSDVVKQ----TLGFDDLVPENGSLEYFKVLAHRD 56
Query: 69 RFLRGRRLSQF--DAGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDLFW 126
RF+RGR L+ + L N T ++ LGFLHY + LGTP F+VALDTGSDLFW
Sbjct: 57 RFIRGRGLASNNEETPLTSIGSNLTLALNFLGFLHYANVSLGTPATWFLVALDTGSDLFW 116
Query: 127 VPCDC-TRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQCLGTFSN 185
+PC+C T C A + L++Y PN S+TS + C++ C +C S
Sbjct: 117 LPCNCGTTCIHDLKDA---RFSESVPLNLYTPNASTTSSSIRCSDKRCFGSGKCSSPESI 173
Query: 186 CPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPNGL 245
CPY ++ +S+ T T+G L++DVLHL D++ V ANV GCGQ Q+G+F A NG+
Sbjct: 174 CPYQIA-LSSNTVTTGTLLQDVLHLVTEDEDLKPVNANVTLGCGQNQTGAFQTDIAVNGV 232
Query: 246 FGLGMEKISVPSMLSREGFTADSFSMCFGR--DGIGRISFGDKGSLDQDETPFNVNPSHP 303
GL M++ SVPS+L++ TA+SFSMCFGR +GRISFGDKG DQ+ETP +
Sbjct: 233 LGLSMKEYSVPSLLAKANITANSFSMCFGRIISVVGRISFGDKGYTDQEETPLVSLETST 292
Query: 304 TYNITINQVRVGTTLIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRI 363
Y + + V VG +DV ALFD+G+SFT L++ Y +++F +ED+RRP D
Sbjct: 293 AYGVNVTGVSVGGVPVDVPLFALFDTGSSFTLLLESAYGVFTKAFDDLMEDKRRPVDPDF 352
Query: 364 PFDYCYDMSPDS-NTSLIPSMSLTMGGGSRFVVYDPI---------------IIISTQSE 407
PF++CYD+ + N+ P + Y+P + S +
Sbjct: 353 PFEFCYDLREEHLNSDARPRHMQSK-------CYNPCRDDFRWRIQNDSQESVSYSNEGT 405
Query: 408 LVYCLAVVKSAELNIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
+YCL ++KS LNIIGQN M+G+R+VFDR ++ILGWK+S+C
Sbjct: 406 KMYCLGILKSINLNIIGQNLMSGHRIVFDRERMILGWKQSNC 447
>At5g10080 predicted GPI-anchored protein
Length = 528
Score = 302 bits (774), Expect = 2e-82
Identities = 174/440 (39%), Positives = 252/440 (56%), Gaps = 19/440 (4%)
Query: 23 AHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYAELADRDRFLRGRRLSQFDAG 82
A +F+ + HR+S+ + S PS S P K S+EYY LA+ D + L
Sbjct: 22 ASLFSSRLIHRFSDEGRA-SIKTPSSSDSLPNKQSLEYYRLLAESDFRRQRMNLGAKVQS 80
Query: 83 LAFSDGNSTFRISS-LGFLHYTTIELGTPGVKFMVALDTGSDLFWVPCDCTRCSATRSSA 141
L S+G+ T + G+LHYT I++GTP V F+VALDTGS+L W+PC+C +C A +S
Sbjct: 81 LVPSEGSKTISSGNDFGWLHYTWIDIGTPSVSFLVALDTGSNLLWIPCNCVQC-APLTST 139
Query: 142 FASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSG 201
+ S+LA+ DL+ YNP+ SSTSK C++ LC + C CPY V+Y+S TS+SG
Sbjct: 140 YYSSLATK-DLNEYNPSSSSTSKVFLCSHKLCDSASDCESPKEQCPYTVNYLSGNTSSSG 198
Query: 202 ILVEDVLHLTQPDDNHDL-----VEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVP 256
+LVED+LHLT +N + V+A V+ GCG+ QSG +LD AP+GL GLG +ISVP
Sbjct: 199 LLVEDILHLTYNTNNRLMNGSSSVKARVVIGCGKKQSGDYLDGVAPDGLMGLGPAEISVP 258
Query: 257 SMLSREGFTADSFSMCFGRDGIGRISFGDKGSLDQDETPFNV--NPSHPTYNITINQVRV 314
S LS+ G +SFS+CF + GRI FGD G Q TPF N + Y + + +
Sbjct: 259 SFLSKAGLMRNSFSLCFDEEDSGRIYFGDMGPSIQQSTPFLQLDNNKYSGYIVGVEACCI 318
Query: 315 GTT-LIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSP 373
G + L FT DSG SFTYL + Y +++ + + + + ++YCY+ S
Sbjct: 319 GNSCLKQTSFTTFIDSGQSFTYLPEEIYRKVALEIDRHINATSKNFEG-VSWEYCYESSA 377
Query: 374 DSNTSLIPSMSLTMGGGSRFVVYDPIIIISTQSELV-YCLAVVKSAELNI--IGQNFMTG 430
+ +P++ L + FV++ P+ + LV +CL + S + I IGQN+M G
Sbjct: 378 EPK---VPAIKLKFSHNNTFVIHKPLFVFQQSQGLVQFCLPISPSGQEGIGSIGQNYMRG 434
Query: 431 YRVVFDRGKLILGWKKSDCK 450
YR+VFDR + LGW S C+
Sbjct: 435 YRMVFDRENMKLGWSPSKCQ 454
>At3g51350 predicted GPI-anchored protein
Length = 475
Score = 280 bits (715), Expect = 2e-75
Identities = 165/453 (36%), Positives = 236/453 (51%), Gaps = 73/453 (16%)
Query: 7 IIVIILIILHLSMCCNAHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYAELAD 66
++ ++++ C F F +HH +S+ VK+ + PE+GS+EY+ LA
Sbjct: 10 LLSVLVVCWGFERCEATGKFGFEVHHIFSDSVKQ----SLGLGDLVPEQGSLEYFKVLAH 65
Query: 67 RDRFLRGRRLSQF--DAGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDL 124
RDR +RGR L+ + + F GN T + LG L+Y + +GTP F+VALDTGSDL
Sbjct: 66 RDRLIRGRGLASNNDETPITFDGGNLTVSVKLLGSLYYANVSVGTPPSSFLVALDTGSDL 125
Query: 125 FWVPCDC-TRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQCLGTF 183
FW+PC+C T C + L++Y PN S+TS + C++ C +C
Sbjct: 126 FWLPCNCGTTCIRDLEDI---GVPQSVPLNLYTPNASTTSSSIRCSDKRCFGSKKCSSPS 182
Query: 184 SNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPN 243
S CPY +SY S T T G L++DVLHL D+N V+ANV GCGQ Q+G F + N
Sbjct: 183 SICPYQISY-SNSTGTKGTLLQDVLHLATEDENLTPVKANVTLGCGQKQTGLFQRNNSVN 241
Query: 244 GLFGLGMEKISVPSMLSREGFTADSFSMCFGR--DGIGRISFGDKGSLDQDETPFNVNPS 301
G+ GLG++ SVPS+L++ TA+SFSMCFGR +GRISFG
Sbjct: 242 GVLGLGIKGYSVPSLLAKANITANSFSMCFGRVIGNVGRISFG----------------- 284
Query: 302 HPTYNITINQVRVGTTLIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDS 361
D Y+ E+ V RRRP D
Sbjct: 285 ------------------------------------DRGYTDQEETPFISVAPRRRPVDP 308
Query: 362 RIPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPIIIISTQS-----ELVYCLAVVK 416
+PF++CYD+SP++ T P + +T GGS+ ++ +P TQ+ ++YCL V+K
Sbjct: 309 ELPFEFCYDLSPNATTIQFPLVEMTFIGGSKIILNNPFFTARTQARHGEGNVMYCLGVLK 368
Query: 417 SAELNIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
S L I NF+ GYR+VFDR ++ILGWK+S C
Sbjct: 369 SVGLKI--NNFVAGYRIVFDRERMILGWKQSLC 399
>At3g51360 putative protein
Length = 426
Score = 209 bits (532), Expect = 3e-54
Identities = 110/258 (42%), Positives = 167/258 (64%), Gaps = 9/258 (3%)
Query: 178 QCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFL 237
+C+ S+CPY + Y+S + ++G+LVEDV+H++ + + +A + FG + Q G F
Sbjct: 127 RCISPVSDCPYRIRYLSPGSKSTGVLVEDVIHMSTEEG--EARDARITFG--ESQLGLFK 182
Query: 238 DVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFGRDGIGRISFGDKGSLDQDETPFN 297
+VA NG+ GL + I+VP+ML + G +DSFSMCFG +G G ISFGDKGS DQ ETP +
Sbjct: 183 EVAV-NGIMGLAIADIAVPNMLVKAGVASDSFSMCFGPNGKGTISFGDKGSSDQLETPLS 241
Query: 298 VNPSHPTYNITINQVRVGTTLIDVEFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRR 357
S Y+++I + +VG +D EFTA FDSGT+ T+L++P Y+ L+ +FH V DRR
Sbjct: 242 GTISPMFYDVSITKFKVGKVTVDTEFTATFDSGTAVTWLIEPYYTALTTNFHLSVPDRRL 301
Query: 358 PPDSRIPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPIIIISTQ--SELVYCLAVV 415
PF++CY ++ S+ +PS+S M GG+ + V+ PI++ T S VYCLAV+
Sbjct: 302 SKSVDSPFEFCYIITSTSDEDKLPSVSFEMKGGAAYDVFSPILVFDTSDGSFQVYCLAVL 361
Query: 416 K--SAELNIIGQNFMTGY 431
K +A+ +IIG+N G+
Sbjct: 362 KQVNADFSIIGRNDTNGF 379
>At3g02740 aspartyl protease - like predicted GPI-anchored protein
Length = 488
Score = 132 bits (331), Expect = 6e-31
Identities = 109/375 (29%), Positives = 170/375 (45%), Gaps = 44/375 (11%)
Query: 100 LHYTTIELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPN 158
L++ I LGTP F V +DTGSD+ WV C C RC R S +L+ Y+ +
Sbjct: 84 LYFAKIGLGTPSRDFHVQVDTGSDILWVNCAGCIRCP--RKSDLV-------ELTPYDVD 134
Query: 159 GSSTSKKVTCNNSLCTHRNQC--LGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDN 216
SST+K V+C+++ C++ NQ + S C Y++ Y +ST+G LV+DV+HL N
Sbjct: 135 ASSTAKSVSCSDNFCSYVNQRSECHSGSTCQYVIMYGDG-SSTNGYLVKDVVHLDLVTGN 193
Query: 217 HDLVEAN--VIFGCGQVQSGSFLDV-AAPNGLFGLGMEKISVPSMLSREGFTADSFSMCF 273
N +IFGCG QSG + AA +G+ G G S S L+ +G SF+ C
Sbjct: 194 RQTGSTNGTIIFGCGSKQSGQLGESQAAVDGIMGFGQSNSSFISQLASQGKVKRSFAHCL 253
Query: 274 -GRDGIGRISFGDKGSLDQDETPFNVNPSHPTYNITINQVRVGTTLIDVEFTA------- 325
+G G + G+ S TP +H Y++ +N + VG +++++ A
Sbjct: 254 DNNNGGGIFAIGEVVSPKVKTTPMLSKSAH--YSVNLNAIEVGNSVLELSSNAFDSGDDK 311
Query: 326 --LFDSGTSFTYLVDPTYSRL-SESFHSQVE-DRRRPPDSRIPFDYCYDMSPDSNTSLIP 381
+ DSGT+ YL D Y+ L +E S E +S F Y P
Sbjct: 312 GVIIDSGTTLVYLPDAVYNPLLNEILASHPELTLHTVQESFTCFHYT------DKLDRFP 365
Query: 382 SMSLTMGGGSRFVVYDPIIIISTQSELVYC-------LAVVKSAELNIIGQNFMTGYRVV 434
+++ VY + + + +C L A L I+G ++ VV
Sbjct: 366 TVTFQFDKSVSLAVYPREYLFQVRED-TWCFGWQNGGLQTKGGASLTILGDMALSNKLVV 424
Query: 435 FDRGKLILGWKKSDC 449
+D ++GW +C
Sbjct: 425 YDIENQVIGWTNHNC 439
>At1g08210 GPI-anchored protein
Length = 492
Score = 127 bits (318), Expect = 2e-29
Identities = 103/374 (27%), Positives = 171/374 (45%), Gaps = 40/374 (10%)
Query: 100 LHYTTIELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPN 158
L+YT ++LGTP +F V +DTGSD+ WV C C C T LS ++P
Sbjct: 83 LYYTKVKLGTPPREFNVQIDTGSDVLWVSCTSCNGCPKTSE--------LQIQLSFFDPG 134
Query: 159 GSSTSKKVTCNNSLCTHRNQCLGTFSN---CPYMVSYVSAETSTSGILVEDVLHLTQPDD 215
SS++ V+C++ C Q S C Y Y + TSG + D +
Sbjct: 135 VSSSASLVSCSDRRCYSNFQTESGCSPNNLCSYSFKYGDG-SGTSGYYISDFMSFDTVIT 193
Query: 216 NHDLVEANV--IFGCGQVQSGSF-LDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMC 272
+ + ++ +FGC +QSG A +G+FGLG +SV S L+ +G FS C
Sbjct: 194 STLAINSSAPFVFGCSNLQSGDLQRPRRAVDGIFGLGQGSLSVISQLAVQGLAPRVFSHC 253
Query: 273 FGRD--GIGRISFGDKGSLDQDETPFNVNPSHPTYNITINQVRVGTTLIDVE---FT--- 324
D G G + G D TP + PS P YN+ + + V ++ ++ FT
Sbjct: 254 LKGDKSGGGIMVLGQIKRPDTVYTP--LVPSQPHYNVNLQSIAVNGQILPIDPSVFTIAT 311
Query: 325 ---ALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFD--YCYDMSPDSNTSL 379
+ D+GT+ YL D YS ++ + V RP I ++ C++++ + +
Sbjct: 312 GDGTIIDTGTTLAYLPDEAYSPFIQAVANAVSQYGRP----ITYESYQCFEITA-GDVDV 366
Query: 380 IPSMSLTMGGGSRFVVYDP--IIIISTQSELVYCLAVVKSA--ELNIIGQNFMTGYRVVF 435
P +SL+ GG+ V+ + I S+ ++C+ + + + I+G + VV+
Sbjct: 367 FPQVSLSFAGGASMVLGPRAYLQIFSSSGSSIWCIGFQRMSHRRITILGDLVLKDKVVVY 426
Query: 436 DRGKLILGWKKSDC 449
D + +GW + DC
Sbjct: 427 DLVRQRIGWAEYDC 440
>At5g36260 aspartyl protease-like predicted GPI-anchored protein
Length = 478
Score = 125 bits (314), Expect = 5e-29
Identities = 112/417 (26%), Positives = 183/417 (43%), Gaps = 45/417 (10%)
Query: 56 GSVEYYAELADRDRFLRGRRLSQFDAGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFM 115
G + +EL D F R L+ D L G+S R S+G L++T I+LG+P ++
Sbjct: 35 GKEKQLSELKSHDSFRHARMLANIDLPLG---GDS--RADSIG-LYFTKIKLGSPPKEYY 88
Query: 116 VALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCT 174
V +DTGSD+ WV C C +C LS+Y+ SSTSK V C + C+
Sbjct: 89 VQVDTGSDILWVNCAPCPKCPVKTDLG--------IPLSLYDSKTSSTSKNVGCEDDFCS 140
Query: 175 H--RNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDL--VEANVIFGCGQ 230
+++ G C Y V Y TS G ++D + L Q N + V+FGCG+
Sbjct: 141 FIMQSETCGAKKPCSYHVVYGDGSTS-DGDFIKDNITLEQVTGNLRTAPLAQEVVFGCGK 199
Query: 231 VQSGSFLDV-AAPNGLFGLGMEKISVPSMLSREGFTADSFSMCF-GRDGIGRISFGDKGS 288
QSG +A +G+ G G S+ S L+ G T FS C +G G + G+ S
Sbjct: 200 NQSGQLGQTDSAVDGIMGFGQSNTSIISQLAAGGSTKRIFSHCLDNMNGGGIFAVGEVES 259
Query: 289 LDQDETPFNVNPSHPTYNITINQVRVGTTLIDV---------EFTALFDSGTSFTYLVDP 339
TP N H YN+ + + V ID+ + + DSGT+ YL
Sbjct: 260 PVVKTTPIVPNQVH--YNVILKGMDVDGDPIDLPPSLASTNGDGGTIIDSGTTLAYLPQN 317
Query: 340 TYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPI 399
Y+ L E ++ + + C+ + +++ + P ++L + VY
Sbjct: 318 LYNSLIEKITAKQQVKLHMVQETFA---CFSFTSNTDKA-FPVVNLHFEDSLKLSVYPHD 373
Query: 400 IIISTQSELVYC-------LAVVKSAELNIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
+ S + ++ YC + A++ ++G ++ VV+D ++GW +C
Sbjct: 374 YLFSLREDM-YCFGWQSGGMTTQDGADVILLGDLVLSNKLVVYDLENEVIGWADHNC 429
>At2g36670 putative protease
Length = 469
Score = 124 bits (311), Expect = 1e-28
Identities = 107/374 (28%), Positives = 162/374 (42%), Gaps = 37/374 (9%)
Query: 100 LHYTTIELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPN 158
L++T ++LG+P +F V +DTGSD+ WV C C+ C SS DL ++
Sbjct: 99 LYFTKVKLGSPPTEFNVQIDTGSDILWVTCSSCSNC--PHSSGLG------IDLHFFDAP 150
Query: 159 GSSTSKKVTCNNSLC-----THRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQP 213
GS T+ VTC++ +C T QC + C Y Y + TSG + D +
Sbjct: 151 GSLTAGSVTCSDPICSSVFQTTAAQC-SENNQCGYSFRYGDG-SGTSGYYMTDTFYF-DA 207
Query: 214 DDNHDLV---EANVIFGCGQVQSGSFL-DVAAPNGLFGLGMEKISVPSMLSREGFTADSF 269
LV A ++FGC QSG A +G+FG G K+SV S LS G T F
Sbjct: 208 ILGESLVANSSAPIVFGCSTYQSGDLTKSDKAVDGIFGFGKGKLSVVSQLSSRGITPPVF 267
Query: 270 SMCFGRDGIGRISFGDKGSLDQDETPFNVNPSHPTYNITINQVRVGTTLIDVEFT----- 324
S C DG G F L + PS P YN+ + + V ++ ++
Sbjct: 268 SHCLKGDGSGGGVFVLGEILVPGMVYSPLVPSQPHYNLNLLSIGVNGQMLPLDAAVFEAS 327
Query: 325 ----ALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLI 380
+ D+GT+ TYLV Y + + V P S + CY +S S + +
Sbjct: 328 NTRGTIVDTGTTLTYLVKEAYDLFLNAISNSVSQLVTPIISN--GEQCYLVS-TSISDMF 384
Query: 381 PSMSLTMGGGSRFVVYDPIIIIS---TQSELVYCLAVVKS-AELNIIGQNFMTGYRVVFD 436
PS+SL GG+ ++ + ++C+ K+ E I+G + V+D
Sbjct: 385 PSVSLNFAGGASMMLRPQDYLFHYGIYDGASMWCIGFQKAPEEQTILGDLVLKDKVFVYD 444
Query: 437 RGKLILGWKKSDCK 450
+ +GW DCK
Sbjct: 445 LARQRIGWASYDCK 458
>At1g65240 hypothetical protein
Length = 475
Score = 122 bits (306), Expect = 4e-28
Identities = 122/467 (26%), Positives = 203/467 (43%), Gaps = 69/467 (14%)
Query: 6 KIIVIILIILHLSMCCNAHIFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSVEYYAELA 65
K+ +++ + + + +A+ F F H+++ K H + R
Sbjct: 6 KLCIVVAVFVIVIEFASAN-FVFKAQHKFAGKKKNLEHFKSHDTRR-------------- 50
Query: 66 DRDRFLRGRRLSQFDAGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDLF 125
R L+ D L G+S R+ S+G L++T I+LG+P ++ V +DTGSD+
Sbjct: 51 ------HSRMLASIDLPLG---GDS--RVDSVG-LYFTKIKLGSPPKEYHVQVDTGSDIL 98
Query: 126 WVPC-DCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQ---CLG 181
W+ C C +C + +F LS+++ N SSTSKKV C++ C+ +Q C
Sbjct: 99 WINCKPCPKCPTKTN--------LNFRLSLFDMNASSTSKKVGCDDDFCSFISQSDSCQP 150
Query: 182 TFSNCPYMVSYVSAETSTSGILVEDVLHLTQ--PDDNHDLVEANVIFGCGQVQSGSFLD- 238
C Y + Y E+++ G + D+L L Q D + V+FGCG QSG +
Sbjct: 151 AL-GCSYHIVYAD-ESTSDGKFIRDMLTLEQVTGDLKTGPLGQEVVFGCGSDQSGQLGNG 208
Query: 239 VAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFGR-DGIGRISFGDKGSLDQDETPFN 297
+A +G+ G G SV S L+ G FS C G G + G S TP
Sbjct: 209 DSAVDGVMGFGQSNTSVLSQLAATGDAKRVFSHCLDNVKGGGIFAVGVVDSPKVKTTPMV 268
Query: 298 VNPSHPTYNITINQVRVGTTLIDVEFT------ALFDSGTSFTYLVDPTYSRLSESFHSQ 351
N H YN+ + + V T +D+ + + DSGT+ Y Y L E+ +
Sbjct: 269 PNQMH--YNVMLMGMDVDGTSLDLPRSIVRNGGTIVDSGTTLAYFPKVLYDSLIETILA- 325
Query: 352 VEDRRRPPDSRIPFD--YCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPIIIISTQSELV 409
R+P I + C+ S + + + P +S + VY P + T E +
Sbjct: 326 ----RQPVKLHIVEETFQCFSFSTNVDEA-FPPVSFEFEDSVKLTVY-PHDYLFTLEEEL 379
Query: 410 YC-------LAVVKSAELNIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
YC L + +E+ ++G ++ VV+D ++GW +C
Sbjct: 380 YCFGWQAGGLTTDERSEVILLGDLVLSNKLVVYDLDNEVIGWADHNC 426
>At5g22850 protease-like protein
Length = 539
Score = 118 bits (295), Expect = 8e-27
Identities = 115/415 (27%), Positives = 180/415 (42%), Gaps = 45/415 (10%)
Query: 62 AELADRDRFLRGRRLSQFDAGLAFS-DGNSTFRISSLGFLHYTTIELGTPGVKFMVALDT 120
++L RD GR L + F DG TF +G L+YT + LGTP F V +DT
Sbjct: 44 SQLKARDEARHGRLLQSLGGVIDFPVDG--TFDPFVVG-LYYTKLRLGTPPRDFYVQVDT 100
Query: 121 GSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLCTHRNQ- 178
GSD+ WV C C C T L+ ++P S T+ ++C++ C+ Q
Sbjct: 101 GSDVLWVSCASCNGCPQTS--------GLQIQLNFFDPGSSVTASPISCSDQRCSWGIQS 152
Query: 179 ----CLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLV---EANVIFGCGQV 231
C + C Y Y + TSG V DVL + LV A V+FGC
Sbjct: 153 SDSGCSVQNNLCAYTFQYGDG-SGTSGFYVSDVLQFDMIVGS-SLVPNSTAPVVFGCSTS 210
Query: 232 QSGSFL-DVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCF-GRDGIGRI-SFGDKGS 288
Q+G + A +G+FG G + +SV S L+ +G FS C G +G G I G+
Sbjct: 211 QTGDLVKSDRAVDGIFGFGQQGMSVISQLASQGIAPRVFSHCLKGENGGGGILVLGEIVE 270
Query: 289 LDQDETPFNVNPSHPTYNITINQVRVGTTLIDVEFT---------ALFDSGTSFTYLVDP 339
+ TP + PS P YN+ + + V + + + + D+GT+ YL +
Sbjct: 271 PNMVFTP--LVPSQPHYNVNLLSISVNGQALPINPSVFSTSNGQGTIIDTGTTLAYLSEA 328
Query: 340 TYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPI 399
Y E+ + V RP S+ + CY ++ S + P +SL GG+ +
Sbjct: 329 AYVPFVEAITNAVSQSVRPVVSK--GNQCYVIT-TSVGDIFPPVSLNFAGGASMFLNPQD 385
Query: 400 IIISTQS---ELVYCLAV--VKSAELNIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
+I + V+C+ +++ + I+G + V+D +GW DC
Sbjct: 386 YLIQQNNVGGTAVWCIGFQRIQNQGITILGDLVLKDKIFVYDLVGQRIGWANYDC 440
>At1g49050 unknown protein
Length = 583
Score = 117 bits (292), Expect = 2e-26
Identities = 104/387 (26%), Positives = 171/387 (43%), Gaps = 57/387 (14%)
Query: 100 LHYTTIELGTP--GVKFMVALDTGSDLFWVPCD--CTRCSATRSSAFASALASDFDLSVY 155
L+YT I +G P G + + +DTGS+L W+ CD CT C+ + +Y
Sbjct: 202 LYYTRILVGKPEDGQYYHLDIDTGSELTWIQCDAPCTSCAKGANQ-------------LY 248
Query: 156 NPNGSST--SKKVTC----NNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLH 209
P + S + C N L H C C Y + Y S G+L +D H
Sbjct: 249 KPRKDNLVRSSEAFCVEVQRNQLTEHCENC----HQCDYEIEYADHSYSM-GVLTKDKFH 303
Query: 210 LTQPDDNHDLVEANVIFGCGQVQSGSFLD-VAAPNGLFGLGMEKISVPSMLSREGFTADS 268
L N L E++++FGCG Q G L+ + +G+ GL KIS+PS L+ G ++
Sbjct: 304 LKL--HNGSLAESDIVFGCGYDQQGLLLNTLLKTDGILGLSRAKISLPSQLASRGIISNV 361
Query: 269 FSMCFGRD--GIGRISFGDK--GSLDQDETPFNVNPSHPTYNITINQVRVGTTLIDVEFT 324
C D G G I G S P + Y + + ++ G ++ ++
Sbjct: 362 VGHCLASDLNGEGYIFMGSDLVPSHGMTWVPMLHDSRLDAYQMQVTKMSYGQGMLSLDGE 421
Query: 325 ------ALFDSGTSFTYLVDPTYSRLSESFH--SQVEDRRRPPDSRIPFDYCYDMS-PDS 375
LFD+G+S+TY + YS+L S S +E R D +P + + P S
Sbjct: 422 NGRVGKVLFDTGSSYTYFPNQAYSQLVTSLQEVSGLELTRDDSDETLPICWRAKTNFPFS 481
Query: 376 NTSLIPSM--SLTMGGGSRFVVYDPIIIISTQSELVY------CLAVVKSAELN-----I 422
+ S + +T+ GS++++ ++I + L+ CL ++ + ++ I
Sbjct: 482 SLSDVKKFFRPITLQIGSKWLIISRKLLIQPEDYLIISNKGNVCLGILDGSSVHDGSTII 541
Query: 423 IGQNFMTGYRVVFDRGKLILGWKKSDC 449
+G M G+ +V+D K +GW KSDC
Sbjct: 542 LGDISMRGHLIVYDNVKRRIGWMKSDC 568
>At5g10760 nucleoid DNA-binding protein cnd41 - like protein
Length = 464
Score = 110 bits (276), Expect = 1e-24
Identities = 105/360 (29%), Positives = 165/360 (45%), Gaps = 38/360 (10%)
Query: 101 HYTTIELGTPGVKFMVALDTGSDLFWVPCDCTRCSATRSSAFASALASDFDLSVYNPNGS 160
+ TI +GTP + DTGSDL W T+C S ++ +NP+ S
Sbjct: 132 YIVTIGIGTPKHDLSLVFDTGSDLTW-----TQCEPCLGSCYSQKEPK------FNPSSS 180
Query: 161 STSKKVTCNNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDLV 220
ST + V+C++ +C C + SNC Y + Y ++ T G L ++ LT N D++
Sbjct: 181 STYQNVSCSSPMCEDAESC--SASNCVYSIVY-GDKSFTQGFLAKEKFTLT----NSDVL 233
Query: 221 EANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMC---FGRDG 277
E +V FGCG+ G F VA GL GLG K+S+P+ + + FS C F +
Sbjct: 234 E-DVYFGCGENNQGLFDGVA---GLLGLGPGKLSLPAQTTTT--YNNIFSYCLPSFTSNS 287
Query: 278 IGRISFGDKGSLDQDE-TPFNVNPSHPTYNITINQVRVGTTLIDV---EFT---ALFDSG 330
G ++FG G + + TP + PS Y I I + VG + + F+ A+ DSG
Sbjct: 288 TGHLTFGSAGISESVKFTPISSFPSAFNYGIDIIGISVGDKELAITPNSFSTEGAIIDSG 347
Query: 331 TSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSMSLTMGGG 390
T FT L Y+ L F ++ + + FD CYD + +T P+++ + G
Sbjct: 348 TVFTRLPTKVYAELRSVFKEKMSSYKSTSGYGL-FDTCYDFT-GLDTVTYPTIAFSFAGS 405
Query: 391 SRFVVYDPIIIISTQSELVYCLAVVKSAEL-NIIGQNFMTGYRVVFDRGKLILGWKKSDC 449
+ + I + + V CLA + +L I G T VV+D +G+ + C
Sbjct: 406 TVVELDGSGISLPIKISQV-CLAFAGNDDLPAIFGNVQQTTLDVVYDVAGGRVGFAPNGC 464
>At1g05840 unknown protein
Length = 485
Score = 108 bits (270), Expect = 7e-24
Identities = 102/381 (26%), Positives = 165/381 (42%), Gaps = 52/381 (13%)
Query: 100 LHYTTIELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPN 158
L+Y I +GTP + V +DTGSD+ WV C C +C R S +L++YN +
Sbjct: 79 LYYAKIGIGTPAKSYYVQVDTGSDIMWVNCIQCKQC--PRRSTLG------IELTLYNID 130
Query: 159 GSSTSKKVTCNNSLCTH-----RNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHL--T 211
S + K V+C++ C + C S CPY+ Y +ST+G V+DV+
Sbjct: 131 ESDSGKLVSCDDDFCYQISGGPLSGCKANMS-CPYLEIYGDG-SSTAGYFVKDVVQYDSV 188
Query: 212 QPDDNHDLVEANVIFGCGQVQSGSF--LDVAAPNGLFGLGMEKISVPSMLSREGFTADSF 269
D +VIFGCG QSG + A +G+ G G S+ S L+ G F
Sbjct: 189 AGDLKTQTANGSVIFGCGARQSGDLDSSNEEALDGILGFGKANSSMISQLASSGRVKKIF 248
Query: 270 SMCF-GRDGIGRISFGDKGSLDQDETPFNVNPSHPTYNITINQVRVGTTLIDVEFT---- 324
+ C GR+G G + G + TP + P+ P YN+ + V+VG + +
Sbjct: 249 AHCLDGRNGGGIFAIGRVVQPKVNMTP--LVPNQPHYNVNMTAVQVGQEFLTIPADLFQP 306
Query: 325 -----ALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPF---DY-CYDMSPDS 375
A+ DSGT+ YL + Y L + SQ P ++ DY C+ S
Sbjct: 307 GDRKGAIIDSGTTLAYLPEIIYEPLVKKITSQ------EPALKVHIVDKDYKCFQYSGRV 360
Query: 376 NTSLIPSMSLTMGGGSRFVVYDPIIIISTQSELVYCLAVVKSA-------ELNIIGQNFM 428
+ P+++ VY + E ++C+ SA + ++G +
Sbjct: 361 DEG-FPNVTFHFENSVFLRVYPHDYLF--PHEGMWCIGWQNSAMQSRDRRNMTLLGDLVL 417
Query: 429 TGYRVVFDRGKLILGWKKSDC 449
+ V++D ++GW + +C
Sbjct: 418 SNKLVLYDLENQLIGWTEYNC 438
>At3g50050 unknown protein
Length = 632
Score = 105 bits (261), Expect = 7e-23
Identities = 96/373 (25%), Positives = 165/373 (43%), Gaps = 46/373 (12%)
Query: 101 HYTT-IELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPN 158
+YTT + +GTP F + +D+GS + +VPC DC +C + F P
Sbjct: 92 YYTTRLWIGTPPQMFALIVDSGSTVTYVPCSDCEQCGKHQDPKF-------------QPE 138
Query: 159 GSSTSKKVTCNNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHD 218
SST + V CN C C Y Y +S+ G+L ED++ +
Sbjct: 139 MSSTYQPVKCNMDC-----NCDDDREQCVYEREYAE-HSSSKGVLGEDLISF---GNESQ 189
Query: 219 LVEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFGRDGI 278
L +FGC V++G A +G+ GLG +S+ L +G ++SF +C+G +
Sbjct: 190 LTPQRAVFGCETVETGDLYSQRA-DGIIGLGQGDLSLVDQLVDKGLISNSFGLCYGGMDV 248
Query: 279 GRISFGDKG-SLDQDETPFNVNPSH-PTYNITINQVRVG-------TTLIDVEFTALFDS 329
G S G D + +P P YNI + +RV + + D E A+ DS
Sbjct: 249 GGGSMILGGFDYPSDMVFTDSDPDRSPYYNIDLTGIRVAGKQLSLHSRVFDGEHGAVLDS 308
Query: 330 GTSFTYLVDPTYSRLSESFHSQVEDRRR--PPDSRIPFDYCYDMSPDSNTS----LIPSM 383
GT++ YL D ++ E+ +V ++ PD D C+ ++ + S + PS+
Sbjct: 309 GTTYAYLPDAAFAAFEEAVMREVSTLKQIDGPDPNFK-DTCFQVAASNYVSELSKIFPSV 367
Query: 384 SLTMGGGSRFVVYDPIIIISTQSEL--VYCLAVVKSAE--LNIIGQNFMTGYRVVFDRGK 439
+ G +++ P + S++ YCL V + + ++G + VV+DR
Sbjct: 368 EMVFKSGQSWLL-SPENYMFRHSKVHGAYCLGVFPNGKDHTTLLGGIVVRNTLVVYDREN 426
Query: 440 LILGWKKSDCKWL 452
+G+ +++C L
Sbjct: 427 SKVGFWRTNCSEL 439
>At3g18490 chloroplast nucleoid DNA-binding protein like
Length = 500
Score = 103 bits (256), Expect = 3e-22
Identities = 95/367 (25%), Positives = 147/367 (39%), Gaps = 46/367 (12%)
Query: 101 HYTTIELGTPGVKFMVALDTGSDLFWVPCD-CTRCSATRSSAFASALASDFDLSVYNPNG 159
+++ I +GTP + + LDTGSD+ W+ C+ C C V+NP
Sbjct: 162 YFSRIGVGTPAKEMYLVLDTGSDVNWIQCEPCADCYQQSD-------------PVFNPTS 208
Query: 160 SSTSKKVTCNNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDL 219
SST K +TC+ C+ + C Y VSY + + + V N
Sbjct: 209 SSTYKSLTCSAPQCSLLETSACRSNKCLYQVSYGDGSFTVGELATDTVTFGNSGKIN--- 265
Query: 220 VEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFGRDGIG 279
NV GCG G F A GL GLG +S+ + + A SFS C G
Sbjct: 266 ---NVALGCGHDNEGLFTGAA---GLLGLGGGVLSITNQMK-----ATSFSYCLVDRDSG 314
Query: 280 RISFGDKGSLD----QDETPFNVNPSHPT-YNITINQVRVG-------TTLIDVEFT--- 324
+ S D S+ P N T Y + ++ VG + DV+ +
Sbjct: 315 KSSSLDFNSVQLGGGDATAPLLRNKKIDTFYYVGLSGFSVGGEKVVLPDAIFDVDASGSG 374
Query: 325 -ALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSM 383
+ D GT+ T L Y+ L ++F + ++ S FD CYD S +T +P++
Sbjct: 375 GVILDCGTAVTRLQTQAYNSLRDAFLKLTVNLKKGSSSISLFDTCYDFS-SLSTVKVPTV 433
Query: 384 SLTMGGGSRFVVYDPIIIISTQSELVYCLAVV-KSAELNIIGQNFMTGYRVVFDRGKLIL 442
+ GG + +I +C A S+ L+IIG G R+ +D K ++
Sbjct: 434 AFHFTGGKSLDLPAKNYLIPVDDSGTFCFAFAPTSSSLSIIGNVQQQGTRITYDLSKNVI 493
Query: 443 GWKKSDC 449
G + C
Sbjct: 494 GLSGNKC 500
>At1g77480 nucellin-like protein
Length = 466
Score = 102 bits (255), Expect = 4e-22
Identities = 101/384 (26%), Positives = 154/384 (39%), Gaps = 59/384 (15%)
Query: 97 LGFLHYTTIELGTPGVKFMVALDTGSDLFWVPCD--CTRCSATRSSAFASALASDFDLSV 154
LG+ +Y + +G P F + +DTGSDL WV CD C C+ R+
Sbjct: 64 LGY-YYVLLNIGNPPKLFDLDIDTGSDLTWVQCDAPCNGCTKPRAKQ------------- 109
Query: 155 YNPNGSSTSKKVTCNNSLCT-----HRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLH 209
Y PN ++ + C++ LC+ C C Y + Y S S+ G LV D +
Sbjct: 110 YKPNHNT----LPCSHILCSGLDLPQDRPCADPEDQCDYEIGY-SDHASSIGALVTDEVP 164
Query: 210 LTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPN-GLFGLGMEKISVPSMLSREGFTADS 268
L N ++ + FGCG Q P G+ GLG K+ + + L G T +
Sbjct: 165 LKLA--NGSIMNLRLTFGCGYDQQNPGPHPPPPTAGILGLGRGKVGLSTQLKSLGITKNV 222
Query: 269 FSMCFGRDGIGRISFGDKGSLDQDETPFNVNPSHPTYN-------ITINQVRVGTTLIDV 321
C G G +S GD+ T ++ + P+ N + N G I+V
Sbjct: 223 IVHCLSHTGKGFLSIGDELVPSSGVTWTSLATNSPSKNYMAGPAELLFNDKTTGVKGINV 282
Query: 322 EFTALFDSGTSFTYLVDPTYSRLSESFHSQVEDR---RRPPDSRIPFDYCYD-----MSP 373
+FDSG+S+TY Y + + + + D +P C+ S
Sbjct: 283 ----VFDSGSSYTYFNAEAYQAILDLIRKDLNGKPLTDTKDDKSLP--VCWKGKKPLKSL 336
Query: 374 DSNTSLIPSMSLTMG---GGSRFVVYDPIIIISTQSELVYCLAVVKSAEL-----NIIGQ 425
D +++L G G F V +I T+ V CL ++ E+ NIIG
Sbjct: 337 DEVKKYFKTITLRFGNQKNGQLFQVPPESYLIITEKGRV-CLGILNGTEIGLEGYNIIGD 395
Query: 426 NFMTGYRVVFDRGKLILGWKKSDC 449
G V++D K +GW SDC
Sbjct: 396 ISFQGIMVIYDNEKQRIGWISSDC 419
>At5g10770 nucleoid DNA-binding protein cnd41 - like protein
Length = 474
Score = 102 bits (254), Expect = 5e-22
Identities = 92/313 (29%), Positives = 139/313 (44%), Gaps = 40/313 (12%)
Query: 95 SSLGFLHY-TTIELGTPGVKFMVALDTGSDLFWVPCD-CTRCSATRSSAFASALASDFDL 152
S+LG +Y T+ LGTP + DTGSDL W C C R D
Sbjct: 125 STLGSGNYIVTVGLGTPKNDLSLIFDTGSDLTWTQCQPCVR------------TCYDQKE 172
Query: 153 SVYNPNGSSTSKKVTCNNSLCTHRNQCLG-----TFSNCPYMVSYVSAETSTSGILVEDV 207
++NP+ S++ V+C+++ C + G + SNC Y + Y ++ + G L ++
Sbjct: 173 PIFNPSKSTSYYNVSCSSAACGSLSSATGNAGSCSASNCIYGIQY-GDQSFSVGFLAKEK 231
Query: 208 LHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTAD 267
LT D V V FGCG+ G F VA GL GLG +K+S PS +
Sbjct: 232 FTLTNSD-----VFDGVYFGCGENNQGLFTGVA---GLLGLGRDKLSFPSQTATAYNKIF 283
Query: 268 SFSMCFGRDGIGRISFGDKG-SLDQDETPFN-VNPSHPTYNITINQVRVGTTLIDVEFT- 324
S+ + G ++FG G S TP + + Y + I + VG + + T
Sbjct: 284 SYCLPSSASYTGHLTFGSAGISRSVKFTPISTITDGTSFYGLNIVAITVGGQKLPIPSTV 343
Query: 325 -----ALFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIP-FDYCYDMSPDSNTS 378
AL DSGT T L Y+ L SF +++ + P S + D C+D+S T
Sbjct: 344 FSTPGALIDSGTVITRLPPKAYAALRSSFKAKMS--KYPTTSGVSILDTCFDLS-GFKTV 400
Query: 379 LIPSMSLTMGGGS 391
IP ++ + GG+
Sbjct: 401 TIPKVAFSFSGGA 413
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,268,467
Number of Sequences: 26719
Number of extensions: 488877
Number of successful extensions: 1283
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1042
Number of HSP's gapped (non-prelim): 86
length of query: 492
length of database: 11,318,596
effective HSP length: 103
effective length of query: 389
effective length of database: 8,566,539
effective search space: 3332383671
effective search space used: 3332383671
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149547.3


