

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149546.4 - phase: 0
(182 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
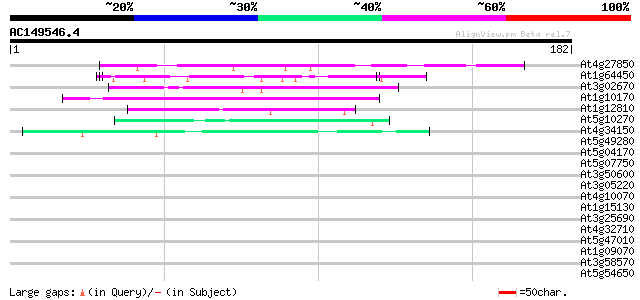
Score E
Sequences producing significant alignments: (bits) Value
At4g27850 putative proline-rich protein 51 4e-07
At1g64450 unknown protein 48 3e-06
At3g02670 unknown protein 47 7e-06
At1g10170 hypothetical protein 43 8e-05
At1g12810 unknown protein 41 3e-04
At5g10270 cdc2-like protein kinase 40 5e-04
At4g34150 unknown protein 40 5e-04
At5g49280 predicted GPI-anchored protein 39 0.002
At5g04170 EF - hand Calcium binding protein - like 39 0.002
At5g07750 putative protein 39 0.002
At3g50600 unknown protein 39 0.002
At3g05220 unknown protein 39 0.002
At4g10070 putative DNA-directed RNA polymerase 38 0.003
At1g15130 unknown protein 38 0.003
At3g25690 unknown protein 37 0.004
At4g32710 putative protein kinase 37 0.006
At5g47010 UPF1 36 0.010
At1g09070 unknown protein 36 0.013
At3g58570 ATP-dependent RNA helicase-like protein 35 0.017
At5g54650 unknown protein 35 0.022
>At4g27850 putative proline-rich protein
Length = 577
Score = 50.8 bits (120), Expect = 4e-07
Identities = 47/147 (31%), Positives = 62/147 (41%), Gaps = 28/147 (19%)
Query: 30 IPDGGPIEPIP-PRRPYSPGGQFYGIPDGGPGEPIPPRRPYSP----GGQFYGIPDGGPG 84
+P GP P+P P P SP P GP P+P P SP G P GP
Sbjct: 190 LPSPGPDSPLPLPGPPPSPS------PTPGPDSPLPSPGPDSPLPLPGPPPSSSPTPGPD 243
Query: 85 EPIP-PRRPYSPG---GQFYRIPDGGPGEPIPLRRPYSPGGQFDMPFMGRHVDDPYLYDD 140
P+P P P SP G +P GP P+P SPG +P G P+LY+
Sbjct: 244 SPLPSPGPPPSPSPTPGPDSPLPSPGPDSPLP-----SPGPDPPLPSPG-----PHLYEK 293
Query: 141 NMRGIKRPFHMTVRFCFVLYLLMCMVV 167
N I P ++++ V LL ++
Sbjct: 294 NRWLIHFP---SIKYLSVFSLLFLAIL 317
Score = 37.4 bits (85), Expect = 0.004
Identities = 29/83 (34%), Positives = 33/83 (38%), Gaps = 19/83 (22%)
Query: 38 PIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEPIP-PRRPYSPG 96
P+PP P P P P P P P SP +P GP P+P P P SP
Sbjct: 162 PLPPPPPPYPS------PLPPPPSPSPTPGPDSP------LPSPGPDSPLPLPGPPPSPS 209
Query: 97 GQFYRIPDGGPGEPIPLRRPYSP 119
P GP P+P P SP
Sbjct: 210 ------PTPGPDSPLPSPGPDSP 226
>At1g64450 unknown protein
Length = 342
Score = 48.1 bits (113), Expect = 3e-06
Identities = 44/113 (38%), Positives = 50/113 (43%), Gaps = 14/113 (12%)
Query: 29 GIPD--GGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEP 86
G PD G P P PR P SP +F G PG PI PR P SP +F P PG P
Sbjct: 182 GSPDFSGNPGPPSFPRNPGSP--EF----PGNPGAPIIPRNPGSP--EFPINPPRNPGAP 233
Query: 87 IPPRR---PYSPGGQFYRIPDGGPGEPIPLRRPYSP-GGQFDMPFMGRHVDDP 135
+ PR P PG P G PG P P +P GG P +G +P
Sbjct: 234 VIPRNPNPPVFPGNPRSMGPPGFPGIGGPPGFPGTPFGGGGTGPTLGDGYANP 286
Score = 43.5 bits (101), Expect = 6e-05
Identities = 36/93 (38%), Positives = 42/93 (44%), Gaps = 19/93 (20%)
Query: 30 IPDGGPIEPIPPR---RPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEP 86
IPD PI P PP P +P ++ PG P P P +PG + G PG P
Sbjct: 144 IPDS-PIIPGPPDFTVTPRNPDSPYF------PGYPESPDLPGNPGSPDF---SGNPGPP 193
Query: 87 IPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSP 119
PR P SP +F G PG PI R P SP
Sbjct: 194 SFPRNPGSP--EF----PGNPGAPIIPRNPGSP 220
Score = 40.0 bits (92), Expect = 7e-04
Identities = 39/101 (38%), Positives = 42/101 (40%), Gaps = 16/101 (15%)
Query: 31 PDGGPIEPIPPRRPYSPGGQFYGIPD--GGPGEPIPPRRPYSPGGQFYGIPD-------- 80
PD P P P P PG G PD G PG P PR P SP +F G P
Sbjct: 163 PDS-PYFPGYPESPDLPGNP--GSPDFSGNPGPPSFPRNPGSP--EFPGNPGAPIIPRNP 217
Query: 81 GGPGEPI-PPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPG 120
G P PI PPR P +P P PG P + P PG
Sbjct: 218 GSPEFPINPPRNPGAPVIPRNPNPPVFPGNPRSMGPPGFPG 258
>At3g02670 unknown protein
Length = 217
Score = 46.6 bits (109), Expect = 7e-06
Identities = 41/104 (39%), Positives = 46/104 (43%), Gaps = 12/104 (11%)
Query: 33 GGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQ-----FYGIPD-----GG 82
G P +P P SPGG GIP G PG +P P SPGG GIP G
Sbjct: 71 GSPGFRLPFPFPSSPGGN-PGIP-GSPGFRLPFPFPSSPGGNPGIPGIPGIPGLPGIPGS 128
Query: 83 PGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPGGQFDMP 126
PG +P P SPGG G PG +P P S GG +P
Sbjct: 129 PGFRLPFPFPSSPGGGSIPGIPGSPGFRLPFPFPPSGGGIPGLP 172
Score = 32.3 bits (72), Expect = 0.14
Identities = 28/87 (32%), Positives = 32/87 (36%), Gaps = 16/87 (18%)
Query: 51 FYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEP------IPPRRPYSPGGQ--FYRI 102
F + P PIP + F P G PG P +P P SPGG I
Sbjct: 57 FSSLQSSPPTSPIPGSPGFRLPFPFPSSPGGNPGIPGSPGFRLPFPFPSSPGGNPGIPGI 116
Query: 103 P--------DGGPGEPIPLRRPYSPGG 121
P G PG +P P SPGG
Sbjct: 117 PGIPGLPGIPGSPGFRLPFPFPSSPGG 143
>At1g10170 hypothetical protein
Length = 1188
Score = 43.1 bits (100), Expect = 8e-05
Identities = 32/103 (31%), Positives = 45/103 (43%), Gaps = 4/103 (3%)
Query: 18 PSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYG 77
PS ++H R+ + GP P RR +P Q + GP +P RR +P Q
Sbjct: 72 PSYNHHQRS----SNIGPPPPNQHRRYNAPDNQHQRSDNIGPPQPNQHRRYNAPDNQHQR 127
Query: 78 IPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPG 120
+ GP +P RR +P Q R + GP P RR + G
Sbjct: 128 SDNSGPPQPYRHRRNNAPENQHQRSDNIGPPPPNRQRRNNASG 170
Score = 35.8 bits (81), Expect = 0.013
Identities = 22/65 (33%), Positives = 29/65 (43%)
Query: 58 GPGEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPY 117
GP P RR +P Q + GP +P RR +P Q R + GP +P RR
Sbjct: 84 GPPPPNQHRRYNAPDNQHQRSDNIGPPQPNQHRRYNAPDNQHQRSDNSGPPQPYRHRRNN 143
Query: 118 SPGGQ 122
+P Q
Sbjct: 144 APENQ 148
>At1g12810 unknown protein
Length = 129
Score = 41.2 bits (95), Expect = 3e-04
Identities = 31/80 (38%), Positives = 33/80 (40%), Gaps = 7/80 (8%)
Query: 39 IPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGP--GEPIPPRRPYSPG 96
+PP PG Q + P G P P PP P SP G P P G P P RPY G
Sbjct: 6 VPPESYPPPGYQSHYPPPGYPSAPPPPGYP-SPPSHHEGYPPPQPYGGYPPPSSRPYEGG 64
Query: 97 GQFYRIPDGGP----GEPIP 112
Q Y G P G P P
Sbjct: 65 YQGYFAGGGYPHQHHGPPPP 84
Score = 37.0 bits (84), Expect = 0.006
Identities = 27/65 (41%), Positives = 27/65 (41%), Gaps = 7/65 (10%)
Query: 31 PDGGPIEPIPPRRPYSPGGQFYGIPDGGP--GEPIPPRRPYSPGGQFY----GIPDGGPG 84
P G P P PP P SP G P P G P P RPY G Q Y G P G
Sbjct: 22 PPGYPSAPPPPGYP-SPPSHHEGYPPPQPYGGYPPPSSRPYEGGYQGYFAGGGYPHQHHG 80
Query: 85 EPIPP 89
P PP
Sbjct: 81 PPPPP 85
Score = 36.2 bits (82), Expect = 0.010
Identities = 23/61 (37%), Positives = 26/61 (41%), Gaps = 1/61 (1%)
Query: 63 IPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPG-GQFYRIPDGGPGEPIPLRRPYSPGG 121
+PP PG Q + P G P P PP P P + Y P G P P RPY G
Sbjct: 6 VPPESYPPPGYQSHYPPPGYPSAPPPPGYPSPPSHHEGYPPPQPYGGYPPPSSRPYEGGY 65
Query: 122 Q 122
Q
Sbjct: 66 Q 66
>At5g10270 cdc2-like protein kinase
Length = 505
Score = 40.4 bits (93), Expect = 5e-04
Identities = 31/90 (34%), Positives = 36/90 (39%), Gaps = 5/90 (5%)
Query: 35 PIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYS 94
P PP+ P P FYG P G PG P R P S G Q G G P
Sbjct: 399 PTNNAPPQVPAGPSHNFYGKPRGPPG---PNRYPPS-GNQSGGYNQSRGGYSSGSYPPQG 454
Query: 95 PGGQFYRIPDGGPGEPIPLRRP-YSPGGQF 123
G + P G G P + P Y+ GGQ+
Sbjct: 455 RGAPYVAGPRGPSGGPYGVGPPNYTQGGQY 484
>At4g34150 unknown protein
Length = 247
Score = 40.4 bits (93), Expect = 5e-04
Identities = 39/136 (28%), Positives = 48/136 (34%), Gaps = 19/136 (13%)
Query: 5 HYAASRGGAPFAGPSRHY--HDRAWYGIPDGGPIEPIPPRRPYS--PGGQFYGIPDGGPG 60
HYA ++ + PS Y H + P P PP S P Q Y P G
Sbjct: 127 HYAGAKKHNYGSAPSAPYAPHVPQYSAPPSASPYSTAPPYSGPSLYPQVQQYPQPSG--- 183
Query: 61 EPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPG 120
PP Y P Y P PIP P P Y P +P P + Y P
Sbjct: 184 --YPPASGYPPQPSAYPPPSTSGYPPIPSAYPPPPPSSAY------PPQPYPPQPSYYPQ 235
Query: 121 GQFDMPFMGRHVDDPY 136
G P+ G++ PY
Sbjct: 236 G----PYPGQYPPPPY 247
>At5g49280 predicted GPI-anchored protein
Length = 162
Score = 38.9 bits (89), Expect = 0.002
Identities = 30/77 (38%), Positives = 31/77 (39%), Gaps = 8/77 (10%)
Query: 38 PIPPRRPYSPGGQFYGIP---DGGPGEPIPPRRPYSPGGQ--FYGIPDGGPGEPIPPRRP 92
P PP P S GG Y P G G PP PY GGQ +Y P G PP P
Sbjct: 66 PPPPSPPSSGGGSSYYYPPPSQSGGGSKYPP--PYGGGGQGYYYPPPYSGNYPTPPPPNP 123
Query: 93 YSPGGQF-YRIPDGGPG 108
P F Y P G G
Sbjct: 124 IVPYFPFYYHTPPPGSG 140
Score = 30.8 bits (68), Expect = 0.42
Identities = 32/94 (34%), Positives = 37/94 (39%), Gaps = 22/94 (23%)
Query: 35 PIEPIPPRRPYSPGGQFYGIPDGGPGE--PIPPRRPYSPGGQFYGIPDGGPGEPIPPRRP 92
P++ PP P SP P P P PP P S GG Y P PP +
Sbjct: 45 PVQSSPP--PPSPP------PPSTPTTACPPPPSPPSSGGGSSYYYP--------PPSQ- 87
Query: 93 YSPGGQFYRIPDGGPGEPIPLRRPYSPGGQFDMP 126
S GG Y P GG G+ PYS G + P
Sbjct: 88 -SGGGSKYPPPYGGGGQGYYYPPPYS--GNYPTP 118
>At5g04170 EF - hand Calcium binding protein - like
Length = 354
Score = 38.9 bits (89), Expect = 0.002
Identities = 31/102 (30%), Positives = 36/102 (34%), Gaps = 25/102 (24%)
Query: 28 YGIPDGGPIEPIPPRRPYSPGGQF-----------YGIPDGGPGEPIPPRRPYSPGGQFY 76
YG GG +P PP+ PYS GG Y +P G + G Y
Sbjct: 11 YGYGYGGGNQPPPPQPPYSSGGNNPPYGSSTTSSPYAVPYGASKPQSSSSSAPTYGSSSY 70
Query: 77 GIPDGGPGEPIPPRRPYSPGGQFYRIP------DGGPGEPIP 112
G P PP PY+P Y P GG G P P
Sbjct: 71 GAP--------PPSAPYAPSPGDYNKPPKEKPYGGGYGAPPP 104
Score = 29.6 bits (65), Expect = 0.94
Identities = 16/44 (36%), Positives = 20/44 (45%), Gaps = 1/44 (2%)
Query: 93 YSPGGQFYRIPDGGPGEPIPLRRPYSPGGQFDMPFMGRHVDDPY 136
Y P Q Y GG +P P + PYS GG + P+ PY
Sbjct: 4 YPPTSQGYGYGYGGGNQPPPPQPPYSSGGN-NPPYGSSTTSSPY 46
>At5g07750 putative protein
Length = 1289
Score = 38.5 bits (88), Expect = 0.002
Identities = 38/112 (33%), Positives = 43/112 (37%), Gaps = 21/112 (18%)
Query: 10 RGGAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPY 69
RGGAP P + G P P PP R +P + G P P PP R
Sbjct: 1082 RGGAPPPPPPP---------MRGGAPPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGG 1132
Query: 70 SPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPGG 121
+P P GG G PP P PGG+ PG P P P PGG
Sbjct: 1133 APPPP---PPPGGRGPGAPPPPP-PPGGR-------APGPP-PPPGPRPPGG 1172
Score = 33.5 bits (75), Expect = 0.065
Identities = 44/143 (30%), Positives = 47/143 (32%), Gaps = 41/143 (28%)
Query: 11 GGAPFAGPSRHYHDRAWYGIPDGGPIEPI------PPRRPYSPGGQFYGIPDGGPGEPIP 64
GGAP P H G P P P+ PP P G Q P G P P
Sbjct: 1033 GGAPPPPPPPPMHG----GAPPPPPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAPPP 1088
Query: 65 PRRPYSPGG-------QFYGIP--------DGGPGEPIPPRR------PYSPGGQFYRIP 103
P P G G P G P P PP R P PGG+
Sbjct: 1089 PPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGR----- 1143
Query: 104 DGGPGEPIPLRRPYSPGGQFDMP 126
GPG P P P PGG+ P
Sbjct: 1144 --GPGAPPP---PPPPGGRAPGP 1161
Score = 29.6 bits (65), Expect = 0.94
Identities = 25/82 (30%), Positives = 26/82 (31%), Gaps = 2/82 (2%)
Query: 38 PIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGG 97
P PP P+S P G P PP P G P P PP P P G
Sbjct: 919 PPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPG 978
Query: 98 QFYRIPDGGPGEPIPLRRPYSP 119
Y P P P P P
Sbjct: 979 --YGSPPPPPPPPPSYGSPPPP 998
Score = 28.5 bits (62), Expect = 2.1
Identities = 32/99 (32%), Positives = 33/99 (33%), Gaps = 20/99 (20%)
Query: 31 PDGGPIEPIPPRRPYSPGGQFYGIPDGGP------GEPIPPRRPYSPGGQFYGIPDGGPG 84
P G P PP P YG P P G P PP P PG YG P P
Sbjct: 938 PSYGSPPPPPPPPPS------YGSPPPPPPPPPSYGSP-PPPPPPPPG---YGSPPPPPP 987
Query: 85 EP----IPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSP 119
P PP P P IP P P+ P P
Sbjct: 988 PPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPPP 1026
Score = 27.3 bits (59), Expect = 4.6
Identities = 38/134 (28%), Positives = 42/134 (30%), Gaps = 32/134 (23%)
Query: 11 GGAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIP----DGGPGEPIPP- 65
GGAP P H G P PP P G P G P P PP
Sbjct: 1020 GGAPPPPPPPPMH----------GGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPM 1069
Query: 66 ---RRPYSPGGQFYGIP--------DGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLR 114
+P P G P G P P PP R +P + G P P P
Sbjct: 1070 FGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPM 1129
Query: 115 R------PYSPGGQ 122
R P PGG+
Sbjct: 1130 RGGAPPPPPPPGGR 1143
>At3g50600 unknown protein
Length = 852
Score = 38.5 bits (88), Expect = 0.002
Identities = 28/84 (33%), Positives = 38/84 (44%), Gaps = 16/84 (19%)
Query: 29 GIPDGGPIEPIPPRRPYSPGGQF--YGIPDGG-----PGE-PIPPRRPYSPGG---QFYG 77
G+PDGG + P + PG F G+PDGG PG+ +P + P S G
Sbjct: 545 GLPDGGVPQQYPQQTSQQPGAPFQTVGLPDGGVRQQYPGQNQVPSQVPVSTQPLDLSVLG 604
Query: 78 IPDGG-----PGEPIPPRRPYSPG 96
+P+ G PG+P P PG
Sbjct: 605 VPNTGDSGKPPGQPQSPPASVRPG 628
Score = 30.4 bits (67), Expect = 0.55
Identities = 14/38 (36%), Positives = 19/38 (49%), Gaps = 2/38 (5%)
Query: 77 GIPDGGPGEPIPPRRPYSPGGQFYRI--PDGGPGEPIP 112
G+PDGG + P + PG F + PDGG + P
Sbjct: 545 GLPDGGVPQQYPQQTSQQPGAPFQTVGLPDGGVRQQYP 582
>At3g05220 unknown protein
Length = 541
Score = 38.5 bits (88), Expect = 0.002
Identities = 32/103 (31%), Positives = 37/103 (35%), Gaps = 14/103 (13%)
Query: 9 SRGGAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRP 68
S+GG H H G GP P+ P P GG + GGP + P P
Sbjct: 366 SKGGGGGGNKGNHNHSAKGIGGGPMGPGGPMGPGGPMGQGGPMGMMGPGGPMSMMGPGGP 425
Query: 69 YSP-GGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEP 110
P GGQ G P P S GG +Y PG P
Sbjct: 426 MGPMGGQ-------GGSYPAVQGLPMSGGGGYY------PGPP 455
Score = 28.5 bits (62), Expect = 2.1
Identities = 15/40 (37%), Positives = 19/40 (47%), Gaps = 3/40 (7%)
Query: 82 GPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPGG 121
GPG P+ P P GG + GPG P+ + P P G
Sbjct: 391 GPGGPMGPGGPMGQGGPMGMM---GPGGPMSMMGPGGPMG 427
>At4g10070 putative DNA-directed RNA polymerase
Length = 748
Score = 37.7 bits (86), Expect = 0.003
Identities = 30/90 (33%), Positives = 35/90 (38%), Gaps = 21/90 (23%)
Query: 34 GPIEPIP----PRRPYSPGGQFYGIPDGG--PGEPIPPRR----------PYSPGGQFYG 77
GP P P PR PYS G +Y P G P + +PPR PYS +YG
Sbjct: 398 GPHAPHPYDYHPRGPYSSQGSYYNSPGFGGYPPQHMPPRGGYGTDWDQRPPYSGPYNYYG 457
Query: 78 IPDGGPGEPIPPRRPYSPGGQFYRIPDGGP 107
P+PP P G GGP
Sbjct: 458 RQGAQSAGPVPP-----PSGPVPSPAFGGP 482
Score = 30.0 bits (66), Expect = 0.72
Identities = 35/130 (26%), Positives = 43/130 (32%), Gaps = 22/130 (16%)
Query: 14 PFAGPSRHYHDRAWYGIPD------GGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRR 67
P++ +Y+ + G P GG R PYS +YG P+PP
Sbjct: 412 PYSSQGSYYNSPGFGGYPPQHMPPRGGYGTDWDQRPPYSGPYNYYGRQGAQSAGPVPPPS 471
Query: 68 PYSPGGQFYGIP----DGGPGEPIPPR----RPYSPG------GQFYRIP--DGGPGEPI 111
P F G P G G+ P PYS GQ Y P D P
Sbjct: 472 GPVPSPAFGGPPLSQVSYGYGQSHGPEYGHAAPYSQTGYQQTYGQTYEQPKYDSNPPMQP 531
Query: 112 PLRRPYSPGG 121
P Y P G
Sbjct: 532 PYGGSYPPAG 541
>At1g15130 unknown protein
Length = 846
Score = 37.7 bits (86), Expect = 0.003
Identities = 30/92 (32%), Positives = 35/92 (37%), Gaps = 15/92 (16%)
Query: 21 HYHDRAWYGIPDGGPIEP---IPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPG----- 72
H H A Y P P IPP P P Y P G +P PP+ P
Sbjct: 758 HPHPHAPYYRPPEQMSRPGYSIPPYGPPPP----YHTPHGQAPQPYPPQAQQQPHPSWQQ 813
Query: 73 GQFYGIPDGGPGEPIPPRRPYSP---GGQFYR 101
G +Y P P P + PY P GG +YR
Sbjct: 814 GSYYDPQGQQPRPPYPGQSPYQPPHQGGGYYR 845
Score = 31.2 bits (69), Expect = 0.32
Identities = 22/71 (30%), Positives = 27/71 (37%), Gaps = 14/71 (19%)
Query: 14 PFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPG-----GQFYGIPDGGPGEPIPPRRP 68
P GP YH P G +P PP+ P G +Y P P P + P
Sbjct: 780 PPYGPPPPYHT------PHGQAPQPYPPQAQQQPHPSWQQGSYYDPQGQQPRPPYPGQSP 833
Query: 69 YSP---GGQFY 76
Y P GG +Y
Sbjct: 834 YQPPHQGGGYY 844
>At3g25690 unknown protein
Length = 939
Score = 37.4 bits (85), Expect = 0.004
Identities = 29/80 (36%), Positives = 34/80 (42%), Gaps = 11/80 (13%)
Query: 36 IEPIPPRRPYSP-----GGQFYGIPDGGPGEPI--PPRRPYSPGGQFYGIPDGGPGEPIP 88
IE PPR P P GG+ +P P P PP P PGG P GGP P P
Sbjct: 579 IEKRPPRVPRPPPRSAGGGKSTNLPSARPPLPGGGPPPPPPPPGGGPPPPPGGGPPPPPP 638
Query: 89 P----RRPYSPGGQFYRIPD 104
P R G + +R P+
Sbjct: 639 PPGALGRGAGGGNKVHRAPE 658
Score = 31.6 bits (70), Expect = 0.25
Identities = 17/36 (47%), Positives = 18/36 (49%), Gaps = 5/36 (13%)
Query: 30 IPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPP 65
+P GGP P PP PGG P GGP P PP
Sbjct: 609 LPGGGPPPPPPP-----PGGGPPPPPGGGPPPPPPP 639
>At4g32710 putative protein kinase
Length = 731
Score = 37.0 bits (84), Expect = 0.006
Identities = 27/95 (28%), Positives = 31/95 (32%), Gaps = 13/95 (13%)
Query: 38 PIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGG 97
P PP SP P G P P P +P P P P E +PP SP
Sbjct: 88 PPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPP-------PSSPPSETVPPGNTISPPP 140
Query: 98 QFY------RIPDGGPGEPIPLRRPYSPGGQFDMP 126
+ + P P P RR P F P
Sbjct: 141 RSLPSESTPPVNTASPPPPSPPRRRSGPKPSFPPP 175
Score = 35.0 bits (79), Expect = 0.022
Identities = 25/88 (28%), Positives = 30/88 (33%), Gaps = 13/88 (14%)
Query: 31 PDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYG------IPDGGPG 84
P G P P P +P P P P E +PP SP + + P
Sbjct: 105 PSGSPPLPFLPAKPSPP-------PSSPPSETVPPGNTISPPPRSLPSESTPPVNTASPP 157
Query: 85 EPIPPRRPYSPGGQFYRIPDGGPGEPIP 112
P PPRR P F + P P P
Sbjct: 158 PPSPPRRRSGPKPSFPPPINSSPPNPSP 185
Score = 29.6 bits (65), Expect = 0.94
Identities = 28/112 (25%), Positives = 35/112 (31%), Gaps = 15/112 (13%)
Query: 12 GAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQF-----YGIPDGGPGEPIPPR 66
G + P R + + P P PPRR P F P+ P P P
Sbjct: 133 GNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNTPSLPE 192
Query: 67 RPYSPGGQFYGIPDGGPGEPIPPRRP-------YSPGGQFYRIPDGGPGEPI 111
P P P P + P + P GQ +PDG PI
Sbjct: 193 TSPPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFFGPAGQ---LPDGTVAPPI 241
>At5g47010 UPF1
Length = 1254
Score = 36.2 bits (82), Expect = 0.010
Identities = 47/163 (28%), Positives = 59/163 (35%), Gaps = 29/163 (17%)
Query: 3 GGHYAASRGGAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEP 62
GG Y S G A P H A Y IP P+ P P P P Y IP GP
Sbjct: 976 GGSYLPS-GPPNGARPGLH---PAGYPIPRV-PLSPFPGGPPSQP----YAIPTRGPVGA 1026
Query: 63 IPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPY---SP 119
+P PG +G G G + P+ Q + + GP PL P SP
Sbjct: 1027 VP--HAPQPGNHGFG---AGRGTSVGGHLPHQQATQ-HNVGTIGPSLNFPLDSPNSQPSP 1080
Query: 120 GGQFDMPFMGRH----------VDDPYLYDD-NMRGIKRPFHM 151
GG P G + +L DD +G P++M
Sbjct: 1081 GGPLSQPGYGSQAFRDGFSMGGISQDFLADDIKSQGSHDPYNM 1123
>At1g09070 unknown protein
Length = 324
Score = 35.8 bits (81), Expect = 0.013
Identities = 34/108 (31%), Positives = 41/108 (37%), Gaps = 3/108 (2%)
Query: 4 GHYAASRGGAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPI 63
GH A S AP AGPS Y + DG + P + Y G Y P P +
Sbjct: 169 GHGAPSAYPAPPAGPSSGYPPQGHDDKHDG--VYGYPQQAGYPAGTGGYPPPGAYPQQGG 226
Query: 64 PPRRPYSPGGQFYGIPDGGP-GEPIPPRRPYSPGGQFYRIPDGGPGEP 110
P P G + G P GP G P P P G + G P +P
Sbjct: 227 YPGYPPQQQGGYPGYPPQGPYGYPQQGYPPQGPYGYPQQQAHGKPQKP 274
Score = 32.0 bits (71), Expect = 0.19
Identities = 37/136 (27%), Positives = 43/136 (31%), Gaps = 26/136 (19%)
Query: 22 YHDRAWYGIPDGGPIEPIP----------PRRPYSPGG---QFYGIPDGGPGEPIPPRRP 68
+ ++ YG GP P+P P Y PG Y P GP PP+
Sbjct: 134 FGEKYTYG-SSSGPHAPVPSAMDHKTMDQPVTAYPPGHGAPSAYPAPPAGPSSGYPPQGH 192
Query: 69 YSPGGQFYGIPD------GGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRPYSPGGQ 122
YG P G G P P P G Y P G P Y P G
Sbjct: 193 DDKHDGVYGYPQQAGYPAGTGGYPPPGAYPQQGGYPGYP-PQQQGGYP-----GYPPQGP 246
Query: 123 FDMPFMGRHVDDPYLY 138
+ P G PY Y
Sbjct: 247 YGYPQQGYPPQGPYGY 262
>At3g58570 ATP-dependent RNA helicase-like protein
Length = 646
Score = 35.4 bits (80), Expect = 0.017
Identities = 26/76 (34%), Positives = 32/76 (41%), Gaps = 11/76 (14%)
Query: 45 YSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPD 104
Y GG + G+P GG G +P PGG + +P GG PY GG Y P
Sbjct: 576 YGGGGGYGGVPGGGYGA-MPGGYGPVPGGGYGNVPGGG-------YAPYGRGGGAYYGPG 627
Query: 105 GGPGEPIPLRRPYSPG 120
G P + Y PG
Sbjct: 628 GYGTVP---NQGYGPG 640
Score = 33.5 bits (75), Expect = 0.065
Identities = 30/88 (34%), Positives = 36/88 (40%), Gaps = 14/88 (15%)
Query: 9 SRGGAPFAGPSRHYHDRAWYGIPDGGPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPPRRP 68
SRGG G + +Y YG GG +P PGG + +P GG P
Sbjct: 567 SRGG----GGADYYGGGGGYGGVPGGGYGAMPGGYGPVPGGGYGNVPGGG-------YAP 615
Query: 69 YSPGGQFYGIPDGGPGEPIPPRRPYSPG 96
Y GG Y P GG G P + Y PG
Sbjct: 616 YGRGGGAYYGP-GGYG--TVPNQGYGPG 640
Score = 32.3 bits (72), Expect = 0.14
Identities = 23/65 (35%), Positives = 29/65 (44%), Gaps = 8/65 (12%)
Query: 69 YSPGGQFYGIPDGGPGEPIPPRRPYSPGGQFYRIPDGGPGEPIPLRRP----YSPGGQFD 124
Y GG + G+P GG G +P PGG + +P GG P R Y PGG
Sbjct: 576 YGGGGGYGGVPGGGYGA-MPGGYGPVPGGGYGNVPGGGYA---PYGRGGGAYYGPGGYGT 631
Query: 125 MPFMG 129
+P G
Sbjct: 632 VPNQG 636
>At5g54650 unknown protein
Length = 900
Score = 35.0 bits (79), Expect = 0.022
Identities = 30/98 (30%), Positives = 36/98 (36%), Gaps = 19/98 (19%)
Query: 34 GPIEPIPPRRPYSPGGQFYGIPDGGPGEPIPP-RRPYSPGGQFYGIPDGGPGEPIPPRRP 92
G +EP+PP P +F + P PP P P GP P PP P
Sbjct: 347 GKVEPLPPEPP-----KFLKVSSKKASAPPPPVPAPQMPSS-------AGPPRPPPPAPP 394
Query: 93 YSPGGQFYRIPDG--GPGEPIPL----RRPYSPGGQFD 124
GG P G GP P P+ + P P G D
Sbjct: 395 PGSGGPKPPPPPGPKGPRPPPPMSLGPKAPRPPSGPAD 432
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.153 0.530
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,799,115
Number of Sequences: 26719
Number of extensions: 356183
Number of successful extensions: 1099
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 679
Number of HSP's gapped (non-prelim): 276
length of query: 182
length of database: 11,318,596
effective HSP length: 93
effective length of query: 89
effective length of database: 8,833,729
effective search space: 786201881
effective search space used: 786201881
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149546.4


