

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149546.1 - phase: 0
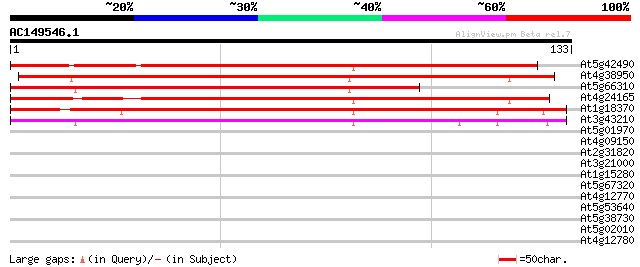
(133 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g42490 kinesin heavy chain-like protein 117 2e-27
At4g38950 kinesin like protein 112 4e-26
At5g66310 kinesin heavy chain DNA binding protein-like 106 3e-24
At4g24165 kinesin heavy chain like protein 104 1e-23
At1g18370 AtNACK1 kinesin-like protein (AtNACK1) 101 1e-22
At3g43210 kinesin -like protein 91 1e-19
At5g01970 unknown protein 31 0.17
At4g09150 unknown protein 27 2.4
At2g31820 ankyrin-like protein 27 3.1
At3g21000 unknown protein 27 4.1
At1g15280 unknown protein 26 5.3
At5g67320 unknown protein 26 6.9
At4g12770 auxilin-like protein 26 6.9
At5g53640 heat shock transcription factor HSF30-like protein 25 9.1
At5g38730 putative protein 25 9.1
At5g02010 unknown protein 25 9.1
At4g12780 auxilin-like protein 25 9.1
>At5g42490 kinesin heavy chain-like protein
Length = 1087
Score = 117 bits (292), Expect = 2e-27
Identities = 61/126 (48%), Positives = 88/126 (69%), Gaps = 3/126 (2%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
MEVELRRLS+LK I D T ++ K L RE++ +S+Q+ +K ++R +Y KWG
Sbjct: 951 MEVELRRLSFLKQT-ISNDMETSRMQTVKA-LTREKEWISKQLPKKFPWNQRIGLYQKWG 1008
Query: 61 ISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRR 119
+ ++SK R LQ+AH+LW+ T D++H++ESA++VAKL+G VEP + KEMFGL+ PR
Sbjct: 1009 VEVNSKQRSLQVAHKLWTNTQDMDHIKESASLVAKLLGFVEPSRMPKEMFGLSLLPRTEN 1068
Query: 120 KKSFGW 125
KS GW
Sbjct: 1069 VKSSGW 1074
>At4g38950 kinesin like protein
Length = 834
Score = 112 bits (281), Expect = 4e-26
Identities = 66/135 (48%), Positives = 86/135 (62%), Gaps = 8/135 (5%)
Query: 3 VELRRLSYLKD-----NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYL 57
VELRRL ++KD NQ LE G TLT SS++ L RER+MLS+ + ++ S ER +Y
Sbjct: 696 VELRRLLFMKDSFSQGNQALEGGETLTLASSRKELHRERKMLSKLVGKRFSGEERKRIYH 755
Query: 58 KWGISMSSKHRRLQLAHRLWSE-TDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPR 116
K+GI+++SK RRLQL + LWS D+ V ESA +VAKLV E +A KEMFGL F P
Sbjct: 756 KFGIAINSKRRRLQLVNELWSNPKDMTQVMESADVVAKLVRFAEQGRAMKEMFGLTFTPP 815
Query: 117 R--RRKKSFGWTSSM 129
++S W SM
Sbjct: 816 SFLTTRRSHSWRKSM 830
>At5g66310 kinesin heavy chain DNA binding protein-like
Length = 1037
Score = 106 bits (265), Expect = 3e-24
Identities = 55/100 (55%), Positives = 74/100 (74%), Gaps = 3/100 (3%)
Query: 1 MEVELRRLSYLKDN--QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLK 58
+EVELRRL Y++++ Q DG +T S R L RER LS+ MQRKLSK ER+N++L+
Sbjct: 925 LEVELRRLKYIRESFAQNSNDGNNMTLISCTRALTRERYKLSKLMQRKLSKEERENLFLR 984
Query: 59 WGISMSSKHRRLQLAHRLWSE-TDINHVRESATIVAKLVG 97
WGI +++ HRR+QLA RLWS+ D+ HVRESA++V KL G
Sbjct: 985 WGIGLNTNHRRVQLARRLWSDYKDMGHVRESASLVGKLNG 1024
>At4g24165 kinesin heavy chain like protein
Length = 1004
Score = 104 bits (260), Expect = 1e-23
Identities = 59/131 (45%), Positives = 82/131 (62%), Gaps = 9/131 (6%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
MEVELRRLS+LKD+ E R T ++ + RER+ L++Q+ K K E++ +Y KWG
Sbjct: 870 MEVELRRLSFLKDST--ETSRKQTAKA----VTREREWLAKQIPNKFGKKEKEEVYKKWG 923
Query: 61 ISMSSKHRRLQLAHRLWSET--DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRR- 117
+ +SSK R LQ+ H+LW+ DI H +ESA+++A LVG V+ KEMFGL+ P
Sbjct: 924 VELSSKRRSLQVTHKLWNNNTKDIEHCKESASLIATLVGFVDSTLTPKEMFGLSLTPTTF 983
Query: 118 RRKKSFGWTSS 128
K S GW S
Sbjct: 984 NIKPSSGWKFS 994
>At1g18370 AtNACK1 kinesin-like protein (AtNACK1)
Length = 974
Score = 101 bits (251), Expect = 1e-22
Identities = 61/147 (41%), Positives = 90/147 (60%), Gaps = 17/147 (11%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTP-----------ESSKRYLRRERQMLSRQMQRKLSK 49
MEVELRRL++L+ Q L + TP SS + LRRER+ L++++ +L+
Sbjct: 830 MEVELRRLTWLE--QHLAEVGNATPARNCDESVVSLSSSIKALRREREFLAKRVNSRLTP 887
Query: 50 SERDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEM 108
ER+ +Y+KW + + K R+LQ ++LW++ D HV+ESA IVAKLVG E KEM
Sbjct: 888 EEREELYMKWDVPLEGKQRKLQFVNKLWTDPYDSRHVQESAEIVAKLVGFCESGNISKEM 947
Query: 109 FGLNFA-PRRRRKKSFGW--TSSMKHI 132
F LNFA P +R+ + GW S++ H+
Sbjct: 948 FELNFAVPSDKRQWNIGWDNISNLLHL 974
>At3g43210 kinesin -like protein
Length = 932
Score = 91.3 bits (225), Expect = 1e-19
Identities = 58/146 (39%), Positives = 86/146 (58%), Gaps = 14/146 (9%)
Query: 1 MEVELRRLSYLKDN---------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSE 51
MEVELRRL++L+ + +L D SS R L++ER+ L++++ KL E
Sbjct: 787 MEVELRRLTWLEQHLAELGNASPALLGDEPASYVASSIRALKQEREYLAKRVNTKLGAEE 846
Query: 52 RDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAF-KEMF 109
R+ +YLKW + K RR Q ++LW++ ++ HVRESA IVAKLVG + + KEMF
Sbjct: 847 REMLYLKWDVPPVGKQRRQQFINKLWTDPHNMQHVRESAEIVAKLVGFCDSGETIRKEMF 906
Query: 110 GLNFA-PRRRRKKSFGWT--SSMKHI 132
LNFA P ++ GW S++ H+
Sbjct: 907 ELNFASPSDKKTWMMGWNFISNLLHL 932
>At5g01970 unknown protein
Length = 351
Score = 31.2 bits (69), Expect = 0.17
Identities = 27/98 (27%), Positives = 48/98 (48%), Gaps = 14/98 (14%)
Query: 18 EDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMS---SKHRRLQLAH 74
EDGRTL + ++ R++ Q +R+ + E +N +G+S S S + +QL H
Sbjct: 126 EDGRTLVENKTADIIQETRKL---QTRRRGTGGEDENQNQSYGVSSSWKKSPEQPMQLNH 182
Query: 75 RLWSETDINHVRE-------SATIVAKLVGTVEPDQAF 105
+ ET + R+ A ++ + + TV+ D AF
Sbjct: 183 -IEHETQLKASRDVAMATAAKAKLLLRELKTVKADLAF 219
>At4g09150 unknown protein
Length = 1097
Score = 27.3 bits (59), Expect = 2.4
Identities = 29/134 (21%), Positives = 60/134 (44%), Gaps = 13/134 (9%)
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
+E EL RL+ + ++ R ++ + +ER L +++ ++ K+E++ M L
Sbjct: 106 LEKELARLAKM------DEARQAAKNGLEQRVEKERDELESKVEERVLKAEKNRMLLFKA 159
Query: 61 ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRK 120
++ +R + A L + I R ++ A + + A + G+ A RRR
Sbjct: 160 MAQRRAAKRQRAAQSLMKKA-IQETRYKESVRAAIY--QKRAAAESKRMGILEAERRRAN 216
Query: 121 ----KSFGWTSSMK 130
+ FG SS++
Sbjct: 217 ARLTRVFGAASSVR 230
>At2g31820 ankyrin-like protein
Length = 662
Score = 26.9 bits (58), Expect = 3.1
Identities = 18/75 (24%), Positives = 38/75 (50%), Gaps = 10/75 (13%)
Query: 25 PESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINH 84
P++ + L++ + ++Q +L +S + G+ + +RL+ H + +N+
Sbjct: 427 PQNPAKQLKQTVSDIKHEVQSQLQQSRQT------GVRVQKIAKRLKKLHI----SGLNN 476
Query: 85 VRESATIVAKLVGTV 99
SAT+VA L+ TV
Sbjct: 477 AINSATVVAVLIATV 491
>At3g21000 unknown protein
Length = 405
Score = 26.6 bits (57), Expect = 4.1
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query: 2 EVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSE 51
+V +RRL + LED + + ES YL + ++L R + KL KS+
Sbjct: 116 QVTIRRLE-----KQLEDLKMVDKESGSSYLDKALEILERLGRAKLEKSD 160
>At1g15280 unknown protein
Length = 584
Score = 26.2 bits (56), Expect = 5.3
Identities = 13/48 (27%), Positives = 25/48 (52%)
Query: 62 SMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMF 109
S+SS ++ L+ + +I E+A ++AK GT+ P + M+
Sbjct: 393 SLSSSPQKTSLSRNRYPPDEIESSSETAALIAKGKGTLRPGGSSSFMY 440
>At5g67320 unknown protein
Length = 613
Score = 25.8 bits (55), Expect = 6.9
Identities = 13/42 (30%), Positives = 21/42 (49%)
Query: 12 KDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERD 53
KD + R E +R RER+ + R+ +R+ K ER+
Sbjct: 136 KDRHEKQKEREREREKLEREKEREREKIEREKEREREKMERE 177
>At4g12770 auxilin-like protein
Length = 909
Score = 25.8 bits (55), Expect = 6.9
Identities = 11/52 (21%), Positives = 28/52 (53%)
Query: 2 EVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERD 53
E + R ++ + L+ R+ + ++ Y RER++ +Q++ ++ER+
Sbjct: 466 EAKFRHAKERREKESLKASRSREGDHTENYDSRERELREKQVRLDRERAERE 517
>At5g53640 heat shock transcription factor HSF30-like protein
Length = 454
Score = 25.4 bits (54), Expect = 9.1
Identities = 11/39 (28%), Positives = 21/39 (53%)
Query: 30 RYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHR 68
RY R + +++ +S++E D YLK I ++K +
Sbjct: 83 RYFDSNRVLCINKLKLTISENEEDGFYLKSWIDAAAKRK 121
>At5g38730 putative protein
Length = 596
Score = 25.4 bits (54), Expect = 9.1
Identities = 14/54 (25%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Query: 50 SERDNMYLKWG-ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPD 102
S + ++ W I + +KH+ + AH+L + + S ++ LVG V D
Sbjct: 75 SSKHSLQSSWKMILILTKHKHFKTAHQLLDKLAQRELLSSPLVLRSLVGGVSED 128
>At5g02010 unknown protein
Length = 546
Score = 25.4 bits (54), Expect = 9.1
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 12/96 (12%)
Query: 6 RRLSYLKD--NQILEDGRTLT---------PESSKRYL-RRERQMLSRQMQRKLSKSERD 53
++L + +D NQIL+ + PES L R+ R L + R +S +
Sbjct: 245 KQLQHKRDCTNQILKAAMAINSITLADMEIPESYLESLPRKGRSCLGDLIYRYISSDQFS 304
Query: 54 NMYLKWGISMSSKHRRLQLAHRLWSETDINHVRESA 89
L + +SS+H+ +++A+R+ S + H R ++
Sbjct: 305 PECLLDCLDLSSEHQAIEIANRVESSIYLWHKRTNS 340
>At4g12780 auxilin-like protein
Length = 904
Score = 25.4 bits (54), Expect = 9.1
Identities = 11/52 (21%), Positives = 28/52 (53%)
Query: 2 EVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERD 53
E + R ++ + L+ R+ + ++ Y RER++ +Q++ ++ER+
Sbjct: 468 EAKFRHAKERREKENLKASRSREGDHTENYDSRERELREKQVRLDRERAERE 519
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,719,119
Number of Sequences: 26719
Number of extensions: 90927
Number of successful extensions: 369
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 342
Number of HSP's gapped (non-prelim): 19
length of query: 133
length of database: 11,318,596
effective HSP length: 88
effective length of query: 45
effective length of database: 8,967,324
effective search space: 403529580
effective search space used: 403529580
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149546.1


