

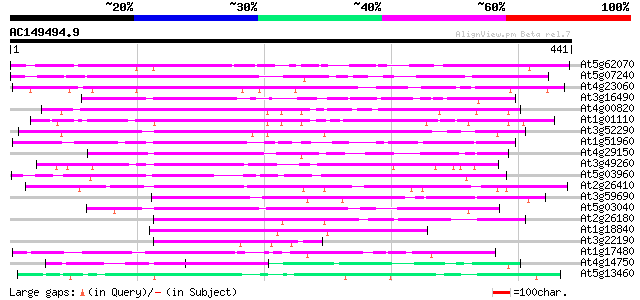
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.9 + phase: 0
(441 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g62070 unknown protein 271 5e-73
At5g07240 unknown protein 263 2e-70
At4g23060 unknown protein 197 1e-50
At3g16490 putative calmodulin-binding protein 169 4e-42
At4g00820 unknown protein 142 4e-34
At1g01110 unknown protein 133 2e-31
At3g52290 unknown protein (At3g52290) 127 2e-29
At1g51960 hypothetical protein 126 3e-29
At4g29150 unknown protein (At4g29150) 124 1e-28
At3g49260 SF16 -like protein 115 5e-26
At5g03960 unknown protein 108 8e-24
At2g26410 putative SF16 protein {Helianthus annuus} 107 1e-23
At3g59690 unknown protein 99 6e-21
At5g03040 unknown protein 95 7e-20
At2g26180 putative SF16 protein {Helianthus annuus} 94 1e-19
At1g18840 hypothetical protein 89 5e-18
At3g22190 unknown protein 86 3e-17
At1g17480 hypothetical protein 86 4e-17
At4g14750 unknown protein 84 2e-16
At5g13460 unknown protein 83 3e-16
>At5g62070 unknown protein
Length = 403
Score = 271 bits (693), Expect = 5e-73
Identities = 213/465 (45%), Positives = 252/465 (53%), Gaps = 88/465 (18%)
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
MGF RLFG+KK S K + +KRRWSF +SS + SA
Sbjct: 1 MGFFGRLFGSKKK------SDKAASSRDKRRWSFTTRSSNSSKRAPAVTSA--SVVEQNG 52
Query: 61 LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSG----VAGSSNKTRGQLR----- 111
L+ +KHAIAVAAATAAVAEAAL AAHAAAEVVRLTS V G N + Q+
Sbjct: 53 LDADKHAIAVAAATAAVAEAALTAAHAAAEVVRLTSGNGGRNVGGGGNSSVFQIGRSNRR 112
Query: 112 -LPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQ 170
E AA+KIQSAFRGYLARRALRALKALVKLQALVRGHIVRK+TADMLRRMQTLVRLQ
Sbjct: 113 WAQENIAAMKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKQTADMLRRMQTLVRLQ 172
Query: 171 TKARASRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI 230
++ARA RA SS + SF SS + P P +++ RC SN+ ++
Sbjct: 173 SQARA-RASRSSHSSASFHSSTA-LLFPSSSSSPRSLHT----------RCVSNA---EV 217
Query: 231 ESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVN 290
S R GS LD W E S DKILEVDTWKP + +
Sbjct: 218 SSLDHRGGSKRLD-WQAEES----------------ENGDKILEVDTWKPHYHPKPLR-S 259
Query: 291 SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT 350
+NESP K ++ + R+ +NSPQ S SG RR TPFT
Sbjct: 260 ERNNESP----------------RKRQQSLLGPRSTENSPQVGS------SGSRRRTPFT 297
Query: 351 PT-RSECSWSFLG-GYSGY-PNYMANTESSRAKVRSQSAPRQRHEF-EEYSSTRRPFQGL 406
PT RSE SW YSGY PNYMANTES +AKVRSQSAP+QR E E S +R QG
Sbjct: 298 PTSRSEYSWGCNNYYYSGYHPNYMANTESYKAKVRSQSAPKQRVEVSNETSGYKRSVQGQ 357
Query: 407 W-----------DVGSTNSDNDSDSRSNKVSPALSRFNRIGSSNL 440
+ DVGS S++++ S +R+ SS L
Sbjct: 358 YYYYTAVEEESLDVGSAGYYGGGGGDSDRLNRNQSAKSRMHSSFL 402
>At5g07240 unknown protein
Length = 401
Score = 263 bits (671), Expect = 2e-70
Identities = 192/435 (44%), Positives = 235/435 (53%), Gaps = 82/435 (18%)
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
MGF RLFG+KK ++ N+RRWSF +SS ++ S+ + D
Sbjct: 1 MGFFGRLFGSKK---------QEKATPNRRRWSFATRSSHPEN-DSSSHSSKRRGDEDV- 49
Query: 61 LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNK-TRGQLRLPEE-TAA 118
L +KHAIAVAAATAAVAEAALAAA AAAEVVRLT+ G S + +R R +E AA
Sbjct: 50 LNGDKHAIAVAAATAAVAEAALAAARAAAEVVRLTNGGRNSSVKQISRSNRRWSQEYKAA 109
Query: 119 VKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRA 178
+KIQSAFRGYLARRALRALKALVKLQALV+GHIVRK+TADMLRRMQTLVRLQ +ARASR+
Sbjct: 110 MKIQSAFRGYLARRALRALKALVKLQALVKGHIVRKQTADMLRRMQTLVRLQARARASRS 169
Query: 179 HLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI-------- 230
SD+ H + P S RC S + + K+
Sbjct: 170 SHVSDSSHPPTLMIPSSP------------------QSFHARCVSEAEYSKVIAMDHHHN 211
Query: 231 ESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVN 290
P S LD W E S+ S+ + D+ DKILEVDTWKP
Sbjct: 212 NHRSPMGSSRLLDQWRTEESL-----WSAPKYNEDD---DKILEVDTWKPHF-------- 255
Query: 291 SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT 350
ESP K + + + +NSPQ S + + G RR TPFT
Sbjct: 256 ---RESPRKRGS-----------------LVVPTSVENSPQLRSRTGSSSGGSRRKTPFT 295
Query: 351 PTRSECSWSFLGGYSGY-PNYMANTESSRAKVRSQSAPRQR-HEFEEYSSTRRPFQGLWD 408
P RSE + YSGY PNYMANTES +AKVRSQSAPRQR + S +R QG +
Sbjct: 296 PARSEYEY-----YSGYHPNYMANTESYKAKVRSQSAPRQRLQDLPSESGYKRSIQGQYY 350
Query: 409 VGSTNSDNDSDSRSN 423
+ ++ D RS+
Sbjct: 351 YYTPAAERSFDQRSD 365
>At4g23060 unknown protein
Length = 484
Score = 197 bits (500), Expect = 1e-50
Identities = 172/501 (34%), Positives = 236/501 (46%), Gaps = 97/501 (19%)
Query: 3 FLRRLFGAKKPIP-----PSDGSGKKSDKDNKRRWSFGKQSSKTKSLP------------ 45
+ R LFG KKP P + + + + KRRWSF K + +S P
Sbjct: 7 WFRSLFGVKKPDPGYPDLSVETPSRSTSSNLKRRWSFVKSKREKESTPINQVPHTPSLPN 66
Query: 46 -QPPPSAFNQFDSSTPLER---------------NKHAIAVAAATAAVAEAALAAAHAAA 89
PPP + +Q S+P R +KHAIAVAAATAAVAEAA+AAA+AAA
Sbjct: 67 STPPPPSHHQ---SSPRRRRKQKPMWEDEGSEDSDKHAIAVAAATAAVAEAAVAAANAAA 123
Query: 90 EVVRLTSSG------------------VAGSSNKTRGQLRLPEETAAVKIQSAFRGYLAR 131
VVRLTS+ V +K G R E A +KIQS FRGYLA+
Sbjct: 124 AVVRLTSTSGRSTRSPVKARFSDGFDDVVAHGSKFYGHGRDSCELAVIKIQSIFRGYLAK 183
Query: 132 RALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLS--SDNLHSFK 189
RALRALK LV+LQA+VRGHI RK+ + LRRM LVR Q + RA+R ++ S + S
Sbjct: 184 RALRALKGLVRLQAIVRGHIERKRMSVHLRRMHALVRAQARVRATRVIVTPESSSSQSNN 243
Query: 190 SSLSHY-----PVPEEYEQPHHVYSTKFGGSSILKRCSS--NSNFRKIESEKPRFGSNWL 242
+ SH+ P PE+ E S+K S + KR S + N R + + F +
Sbjct: 244 TKSSHFQNPGPPTPEKLEHSISSRSSKLAHSHLFKRNGSKASDNNRLYPAHRETFSATDE 303
Query: 243 DHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSKHST 302
+ + + + + +NR + S IL+ N + S
Sbjct: 304 EEKILQIDRKHISSYTRRNRPDMFYSSHLILD-------------NAGLSEPVFATPFSP 350
Query: 303 KAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLG 362
+ ++ ++ +F TA+NSPQ +SA+SR+ + P S+C+ S
Sbjct: 351 SSSHEEITSQF----------CTAENSPQLYSATSRSKRSAFTASSIAP--SDCTKSCCD 398
Query: 363 GYSGYPNYMANTESSRAKVRSQSAPRQRHE--FEEYSSTRRPFQGLWDVGSTNSDNDSDS 420
G +P+YMA TESSRAK RS SAP+ R + +E SS R F L G T S S
Sbjct: 399 G--DHPSYMACTESSRAKARSASAPKSRPQLFYERPSSKRFGFVDLPYCGDTKSGPQKGS 456
Query: 421 R-----SNKVSPALSRFNRIG 436
NK P R +R+G
Sbjct: 457 ALHTSFMNKAYPGSGRLDRLG 477
>At3g16490 putative calmodulin-binding protein
Length = 389
Score = 169 bits (427), Expect = 4e-42
Identities = 134/344 (38%), Positives = 181/344 (51%), Gaps = 63/344 (18%)
Query: 57 SSTPLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEET 116
+ T E+NKHAIAVAAATAA A+AA+AAA AA VVRLTS+G +G + E
Sbjct: 54 AETDKEQNKHAIAVAAATAAAADAAVAAAQAAVAVVRLTSNGRSGGYSGNA-----MERW 108
Query: 117 AAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARAS 176
AAVKIQS F+GYLAR+ALRALK LVKLQALVRG++VRK+ A+ L MQ L+R QT R+
Sbjct: 109 AAVKIQSVFKGYLARKALRALKGLVKLQALVRGYLVRKRAAETLHSMQALIRAQTSVRSQ 168
Query: 177 RAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPR 236
R + ++++ H P H + +++ +
Sbjct: 169 RIN---------RNNMFH---------PRH-------------------SLERLDDSRSE 191
Query: 237 FGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNES 296
S + SIS K ++ N DE S KI+E+DT+K + NV +E
Sbjct: 192 IHSKRI-------SISVEKQSNHNNNAYDE-TSPKIVEIDTYKTKSRSKRMNV--AVSEC 241
Query: 297 PSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSEC 356
+A++ S K K TA N+P+ FS+S N + TP +P +S C
Sbjct: 242 GDDFIYQAKDFEWSFPGEKCK-----FPTAQNTPR-FSSSMANNN--YYYTPPSPAKSVC 293
Query: 357 -SWSFLGGYSGY--PNYMANTESSRAKVRSQSAPRQRHEFEEYS 397
F Y G P+YMANT+S +AKVRS SAPRQR + + S
Sbjct: 294 RDACFRPSYPGLMTPSYMANTQSFKAKVRSHSAPRQRPDRKRLS 337
>At4g00820 unknown protein
Length = 534
Score = 142 bits (358), Expect = 4e-34
Identities = 142/443 (32%), Positives = 194/443 (43%), Gaps = 85/443 (19%)
Query: 26 KDNKRRWSFGKQSS-----KTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAAVAEA 80
K KRRW F K ++ KT + + P+ ST + +++ T +A
Sbjct: 43 KKEKRRWLFRKSTNHDSPVKTSGVGKDAPA-----QKSTETTTIINPTVLSSVTEQRYDA 97
Query: 81 ALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALRALKAL 140
+ A +A S N TR E+ AAV IQ+ FRGYLARRALRALK L
Sbjct: 98 STPPATVSAASETHPPSTTKELPNLTRRTYTAREDYAAVVIQTGFRGYLARRALRALKGL 157
Query: 141 VKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKSSLSHYPVPEE 200
VKLQALVRGH VRK+ LR MQ LVR+Q++ R LS D S + +
Sbjct: 158 VKLQALVRGHNVRKQAKMTLRCMQALVRVQSRVLDQRKRLSHDGSRKSAFSDTQSVLESR 217
Query: 201 Y----------------------EQPHHVYSTKF----GGSSILKRCSSNSNFR------ 228
Y ++PH + K + L+R S+NS +
Sbjct: 218 YLQEISDRRSMSREGSSIAEDWDDRPHTIEEVKAMLQQRRDNALRRESNNSISQAFSHQV 277
Query: 229 -----------KIESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDT 277
+ E E+P+ WLD WM S K AS+ R P +K+ +E+DT
Sbjct: 278 RRTRGSYSTGDEYEEERPK----WLDRWMA--SKPWDKRASTDQRVPPVYKT---VEIDT 328
Query: 278 WKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSP-QTFSAS 336
+P L + NS + SPS+ + S + + A A + P Q SAS
Sbjct: 329 SQPYLTRG----NSRTGASPSRSQRPSSPSRTSHHYQQHNFSSATPSPAKSRPIQIRSAS 384
Query: 337 ---SRNGSGVRRNTPFTPT-RSECSWSFLGGYS-----------GYPNYMANTESSRAKV 381
R+ T TP+ RS S++ GYS PNYMA TES++A++
Sbjct: 385 PRIQRDDRSAYNYTSNTPSLRSNYSFTARSGYSVCTTTTTATNAALPNYMAITESAKARI 444
Query: 382 RSQSAPRQR---HEFEEYSSTRR 401
RSQSAPRQR E E SS R+
Sbjct: 445 RSQSAPRQRPSTPEKERISSARK 467
>At1g01110 unknown protein
Length = 527
Score = 133 bits (335), Expect = 2e-31
Identities = 158/510 (30%), Positives = 214/510 (40%), Gaps = 118/510 (23%)
Query: 17 SDGSGKKSDKDNKRRWSFGK---QSSKTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAA 73
+D + K KRRW F K Q S KS PP+ Q DS L N A
Sbjct: 28 NDVEEDEEKKREKRRW-FRKPATQESPVKSSGISPPAP--QEDS---LNVNSKPSPETAP 81
Query: 74 TAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRL---PEETAAVKIQSAFRGYLA 130
+ A A + VV +A S++KT R+ E AAV IQ++FRGYLA
Sbjct: 82 SYATTTPPSNAGKPPSAVVP-----IATSASKTLAPRRIYYARENYAAVVIQTSFRGYLA 136
Query: 131 RRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKS 190
RRALRALK LVKLQALVRGH VRK+ LR MQ LVR+Q++ R LS D
Sbjct: 137 RRALRALKGLVKLQALVRGHNVRKQAKMTLRCMQALVRVQSRVLDQRKRLSHDGSRKSAF 196
Query: 191 SLSHYPVPEEY----------------------EQPHHVYSTKF-----------GGSSI 217
S SH Y ++PH + + K +
Sbjct: 197 SDSHAVFESRYLQDLSDRQSMSREGSSAAEDWDDRPHTIDAVKVMLQRRRDTALRHDKTN 256
Query: 218 LKRCSSNSNFR-------------KIESEKPRFGSNWLDHWMQENSISQTKNASSKNRHP 264
L + S +R ++E E+P+ WLD WM K ASS+
Sbjct: 257 LSQAFSQKMWRTVGNQSTEGHHEVELEEERPK----WLDRWMATR--PWDKRASSR-ASV 309
Query: 265 DEHKSDKILEVDTWKPQLNKNENNVN-SMSNESPSKHSTKAQNQ-SLSVKFHKAKEEVAA 322
D+ S K +E+DT +P + + SPS+ S Q++ + S AK
Sbjct: 310 DQRVSVKTVEIDTSQPYSRTGAGSPSRGQRPSSPSRTSHHYQSRNNFSATPSPAKSRPIL 369
Query: 323 SRTA----------DNSPQTFSASSRNGSGVRRNTPFTPTRSECSWS--FLGGYSGYPNY 370
R+A D +S +S N +R N FT RS CS S + S PNY
Sbjct: 370 IRSASPRCQRDPREDRDRAAYSYTS-NTPSLRSNYSFT-ARSGCSISTTMVNNASLLPNY 427
Query: 371 MANTESSRAKVRSQSAPRQR-----------------------HEFEEYSSTRRP----- 402
MA+TES++A++RS SAPRQR E+E+ +S R P
Sbjct: 428 MASTESAKARIRSHSAPRQRPSTPERDRAGLVKKRLSYPVPPPAEYEDNNSLRSPSFKSV 487
Query: 403 ----FQGLWDVGSTNSDNDSDSRSNKVSPA 428
F G+ + S S ++S ++SPA
Sbjct: 488 AGSHFGGMLEQQSNYSSCCTESNGVEISPA 517
>At3g52290 unknown protein (At3g52290)
Length = 430
Score = 127 bits (318), Expect = 2e-29
Identities = 122/439 (27%), Positives = 184/439 (41%), Gaps = 71/439 (16%)
Query: 8 FGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSS---KTKSLPQPPPSAFNQFDSSTPLERN 64
F A K + KK K +K + FGK P + + +++
Sbjct: 6 FSAVKKALSPEPKQKKEQKPHKSKKWFGKSKKLDVTNSGAAYSPRTVKDAKLKEIEEQQS 65
Query: 65 KHAIAVAAATAAVAEAALAAAHAAAEVVRLTS-SGVAGSSNKTRGQLRLPEETAAVKIQS 123
+HA +VA ATAA AEAA+AAA AAAEVVRL++ S G S EE AA+KIQ+
Sbjct: 66 RHAYSVAIATAAAAEAAVAAAQAAAEVVRLSALSRFPGKSM---------EEIAAIKIQT 116
Query: 124 AFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSD 183
AFRGY+ARRALRAL+ LV+L++LV+G VR++ L+ MQTL R+Q + R R LS D
Sbjct: 117 AFRGYMARRALRALRGLVRLKSLVQGKCVRRQATSTLQSMQTLARVQYQIRERRLRLSED 176
Query: 184 NLHSFK-------------------SSLSHYPVPEEY-------EQPHHVYSTKFGGSSI 217
+ S+LS V + + F +
Sbjct: 177 KQALTRQLQQKHNKDFDKTGENWNDSTLSREKVEANMLNKQVATMRREKALAYAFSHQNT 236
Query: 218 LKRCSSNSNFRKIESEKPRFGSNWLDHWM---QENSISQTKNASSKNRHPDEHKSDKILE 274
K + + ++ P +G +WL+ WM + S T + + K+ S + E
Sbjct: 237 WKNSTKMGSQTFMDPNNPHWGWSWLERWMAARPNENHSLTPDNAEKDSSARSVASRAMSE 296
Query: 275 VDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFS 334
+ L+ NS SP +++ + V F + T + P T
Sbjct: 297 MIPRGKNLSPRGKTPNSRRGSSPRVRQVPSEDSNSIVSFQSEQPCNRRHSTCGSIPST-- 354
Query: 335 ASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSRAKVR--------SQSA 386
R + FT + S+ P YMA T++++A+ R S+
Sbjct: 355 ---------RDDESFTSSFSQ----------SVPGYMAPTQAAKARARFSNLSPLSSEKT 395
Query: 387 PRQRHEFEEYSSTRRPFQG 405
++R F T R F G
Sbjct: 396 AKKRLSFSGSPKTVRRFSG 414
>At1g51960 hypothetical protein
Length = 351
Score = 126 bits (316), Expect = 3e-29
Identities = 126/395 (31%), Positives = 174/395 (43%), Gaps = 81/395 (20%)
Query: 3 FLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLE 62
+ + +FG KK S SG S K F + T +
Sbjct: 7 WFKGMFGTKKSKDRSHVSGGDSVKGGDHSGDFNVPRDSV---------LLGTILTDTEKD 57
Query: 63 RNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQ 122
+NK+AIAVA ATA A+AA++AA VVRLTS G AG T+ E AAVKIQ
Sbjct: 58 QNKNAIAVATATATAADAAVSAA-----VVRLTSEGRAGDIIITK-----EERWAAVKIQ 107
Query: 123 SAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSS 182
FRG LAR+ALRALK +VKLQALVRG++VRK+ A ML+ +QTL+R+QT R+ R + S
Sbjct: 108 KVFRGSLARKALRALKGIVKLQALVRGYLVRKRAAAMLQSIQTLIRVQTAMRSKRINRSL 167
Query: 183 DNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWL 242
+ ++ QP + KF ++ R + KI + R+
Sbjct: 168 NKEYN------------NMFQPRQSFD-KFDEATFDDRRT------KIVEKDDRY----- 203
Query: 243 DHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSKHST 302
+ +SS++R H NV SMS+ +
Sbjct: 204 -----------MRRSSSRSRSRQVH--------------------NVVSMSD---YEGDF 229
Query: 303 KAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLG 362
+ L + F K + A TA N+P+ S N +P + +
Sbjct: 230 VYKGNDLELCFSDEKWKFA---TAQNTPRLLHHHSANNRYYVMQSPAKSVGGKALCDYES 286
Query: 363 GYSGYPNYMANTESSRAKVRSQSAPRQRHEFEEYS 397
S P YM T+S +AKVRS SAPRQR E + S
Sbjct: 287 SVS-TPGYMEKTKSFKAKVRSHSAPRQRSERQRLS 320
>At4g29150 unknown protein (At4g29150)
Length = 383
Score = 124 bits (310), Expect = 1e-28
Identities = 114/334 (34%), Positives = 151/334 (45%), Gaps = 61/334 (18%)
Query: 62 ERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKI 121
ER HAIAVAAATAA A+AA+AAA AAA VVRL G +G + + E AA++I
Sbjct: 66 ERRTHAIAVAAATAAAADAAVAAAKAAAAVVRLQGQGKSGPLGGGKSR----EHRAAMQI 121
Query: 122 QSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLS 181
Q AFRGYLAR+ALRAL+ +VK+QALVRG +VR + A LR M+ LVR Q + RA
Sbjct: 122 QCAFRGYLARKALRALRGVVKIQALVRGFLVRNQAAATLRSMEALVRAQKTVKIQRALRR 181
Query: 182 SDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFR--KIESEKPRF-G 238
+ N + S + E + G K ++ R P G
Sbjct: 182 NGNAAPARKSTERFSGSLE---------NRNNGEETAKIVEVDTGTRPGTYRIRAPVLSG 232
Query: 239 SNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPS 298
S++LD N +T ++ R P P+L+ + S++ P+
Sbjct: 233 SDFLD-----NPFRRTLSSPLSGRVP---------------PRLSMPKPEWEECSSKFPT 272
Query: 299 KHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSW 358
ST R + SP S G +T R +
Sbjct: 273 AQST--------------------PRFSGGSPARSVCCSGGGVEAEVDTEADANR----F 308
Query: 359 SFLGGYSGYPNYMANTESSRAKVRSQSAPRQRHE 392
FL G YMA+T S RAK+RS SAPRQR E
Sbjct: 309 CFLSGEFN-SGYMADTTSFRAKLRSHSAPRQRPE 341
>At3g49260 SF16 -like protein
Length = 471
Score = 115 bits (288), Expect = 5e-26
Identities = 123/412 (29%), Positives = 184/412 (43%), Gaps = 85/412 (20%)
Query: 22 KKSDKDNKRRWSFG-------KQSSKTKSL---------PQPPPSAFNQFDSSTPLER-- 63
K S KD+KR + G +Q T+ + P S + +STP
Sbjct: 19 KSSPKDSKRENNIGSNNADIWQQQHDTQEVVSFEHFPAESSPEISHDVESTASTPATNVG 78
Query: 64 -NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQ 122
KHA+AVA ATAA AEAA+AAA AAA+VVRL AG + +T E++AAV IQ
Sbjct: 79 DRKHAMAVAIATAAAAEAAVAAAQAAAKVVRL-----AGYNRQTE------EDSAAVLIQ 127
Query: 123 SAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSS 182
S +RGYLARRALRALK LV+LQALVRG+ VRK+ ++ MQ LVR+Q + RA R ++
Sbjct: 128 SHYRGYLARRALRALKGLVRLQALVRGNHVRKQAQMTMKCMQALVRVQGRVRARRLQVAH 187
Query: 183 DNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWL 242
D FK E+P+ ++ K E EKP+
Sbjct: 188 DR---FKKQFEEEEKRSGMEKPNKGFAN-----------------LKTEREKPK-----K 222
Query: 243 DHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQL-NKNENNVNSMSNESPSKH- 300
H + S+ QT+ + + + T++ Q+ + N +S+ P ++
Sbjct: 223 LHEVNRTSLYQTQGKEKERSEGMMKRERALAYAYTYQRQMQHTNSEEGIGLSSNGPDRNQ 282
Query: 301 -STKAQNQSLSVKFHKAKEEVAASRTADNSPQTF-----SASSRNGSGVRRNT----PFT 350
+ + +S + + ++ +P + +A++ V T T
Sbjct: 283 WAWNWLDHWMSSQPYTGRQTGPGPGPGQYNPPPYPPFPTAAATTTSDDVSEKTVEMDVTT 342
Query: 351 PTR--------SECSWSFLGGY----------SGYPNYMANTESSRAKVRSQ 384
PT + + LG Y + P+YMA T S++AKVR Q
Sbjct: 343 PTSLKGNIIGLIDREYIDLGSYRQGHKQRKSPTHIPSYMAPTASAKAKVRDQ 394
>At5g03960 unknown protein
Length = 403
Score = 108 bits (269), Expect = 8e-24
Identities = 116/402 (28%), Positives = 175/402 (42%), Gaps = 66/402 (16%)
Query: 2 GFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPL 61
G+++RLF + + +++K + RW F K L P A ++ T
Sbjct: 9 GWMKRLFICE--------AKARAEKPRRLRWVF-------KRLKLRPQLATCGQETRTLN 53
Query: 62 E----RNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETA 117
E + KHA+ VA ATAA AEAA+AAA AAAEVVR+ + + L A
Sbjct: 54 EATQDQRKHAMNVAIATAAAAEAAVAAAKAAAEVVRMAGNAFTSQHFVKK----LAPNVA 109
Query: 118 AVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASR 177
A+KIQSAFR LAR+ALRALKALV+LQA+VRG VR+K + +L+
Sbjct: 110 AIKIQSAFRASLARKALRALKALVRLQAIVRGRAVRRKVSALLK---------------- 153
Query: 178 AHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRF 237
+ HS K+S S+ + + + H +TK S +K SN S+
Sbjct: 154 ------SSHSNKASTSN--IIQRQTERKHWSNTK----SEIKEELQVSNHSLCNSKVKCN 201
Query: 238 GSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESP 297
G W + A + K D++L+ + + V S+ +
Sbjct: 202 G------WDSSALTKEDIKAIWLRKQEGVIKRDRMLKYSRSQRERRSPHMLVESLYAKDM 255
Query: 298 SKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECS 357
S + ++ S + S + + R SG +++PF+ R S
Sbjct: 256 GMRSCRLEHWGESKSAKSINSFLIPSEMLVPTKVKLRSLQRQDSGDGQDSPFSFPRRSFS 315
Query: 358 ---------WSFLGGYSGYPNYMANTESSRAKVRSQSAPRQR 390
S+ +G+ YM+ TES+R K+RS S PRQR
Sbjct: 316 RLEQSILEDESWFQRSNGFQPYMSVTESAREKMRSLSTPRQR 357
>At2g26410 putative SF16 protein {Helianthus annuus}
Length = 516
Score = 107 bits (267), Expect = 1e-23
Identities = 124/503 (24%), Positives = 205/503 (40%), Gaps = 104/503 (20%)
Query: 13 PIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFN---QFDSSTPLERNKHAIA 69
P+ PS S + F Q P PPP A+ + + +N+ A+A
Sbjct: 41 PVDPSPSSVHRPYPPPPPLPDFAPQPLLPPPSPPPPPPAYTINTRIYGESKESKNRQALA 100
Query: 70 VAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYL 129
+A+A AA EAA+ AAHAAAEV+RLT+ EETAA+KIQ+A+R Y
Sbjct: 101 LASAVAA--EAAVVAAHAAAEVIRLTTPSTPQIEESK-------EETAAIKIQNAYRCYT 151
Query: 130 ARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFK 189
ARR LRAL+ + +L++L++G V+++ ML MQTL RLQT+ + R LS++N +
Sbjct: 152 ARRTLRALRGMARLKSLLQGKYVKRQMNAMLSSMQTLTRLQTQIQERRNRLSAEN--KTR 209
Query: 190 SSLSHYPVPEEYEQPHHVYSTKFGGSSILK-RCSSNSNFRK------------------- 229
L ++ + V + F S+ K + + S RK
Sbjct: 210 HRLIQQKGHQKENHQNLVTAGNFDSSNKSKEQIVARSVNRKEASVRRERALAYAYSHQQT 269
Query: 230 ------------IESEKPRFGSNWLDHWM-----QENSISQTKNASSKNRHPDEHKSDKI 272
+++ +G +WL+ WM SI + S + + KS
Sbjct: 270 WRNSSKLPHQTLMDTNTTDWGWSWLERWMASRPWDAESIDDQVSVKSSLKRENSIKS--- 326
Query: 273 LEVDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFH----------KAKEEVAA 322
P +K + + + S + P + TK++ ++ + H K E V +
Sbjct: 327 ------SPARSKTQKSASQSSIQWPVNNDTKSRKIEVTNRRHSIGGGSSENAKDDESVGS 380
Query: 323 S-------------------RTADNSPQTFSASSRNGSGVRRNTPFTPT-RSECSWSFLG 362
S T N + + G +RN T T +S+ S G
Sbjct: 381 SSSRRNSLDNTQTVKSKVSVETTSNVSNAQTVKPKANVGAKRNLDNTKTLKSKSSVGTTG 440
Query: 363 GYSGYPNYMANTESSRAKVR--SQSAPRQ-----RHEFEEYSSTRRPFQGLWDVGSTNSD 415
+ANTE+ ++KV + S P++ + + + ++ +G T
Sbjct: 441 N-------LANTEAVKSKVNVGTTSMPKKEVVADKKKPPQMVLPKKRLSSSTSLGKTKKL 493
Query: 416 NDSDSRSNKVSPALSRFNRIGSS 438
+DSD + V+ + GSS
Sbjct: 494 SDSDKATTGVANGEKKRRNGGSS 516
>At3g59690 unknown protein
Length = 517
Score = 98.6 bits (244), Expect = 6e-21
Identities = 87/325 (26%), Positives = 154/325 (46%), Gaps = 33/325 (10%)
Query: 112 LPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQT 171
L + A+KIQ+AFRGY+ARR+ RALK LV+LQ +VRGH V+++T + ++ MQ LVR+QT
Sbjct: 166 LVKNAYAIKIQAAFRGYMARRSFRALKGLVRLQGVVRGHSVKRQTMNAMKYMQLLVRVQT 225
Query: 172 KARASRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIE 231
+ ++ R + + + K ++ + S + S + K RKI+
Sbjct: 226 QVQSRRIQMLENRARNDK---------DDTKLVSSRMSDDWDDSVLTKEEKDVRLHRKID 276
Query: 232 S--EKPRFGSNWLDHWMQENSISQTKNASSK------NRHPDEHKSDKILEVDTWKPQLN 283
+ ++ R + H + +NS ++ + N + ++ + +P L+
Sbjct: 277 AMIKRERSMAYAYSHQLWKNSPKSAQDIRTSGFPLWWNWVDRQKNQNQPFRLTPTRPSLS 336
Query: 284 KNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSAS-SRNGSG 342
+ N + T N S S V SR ++PQ +S+S SR G
Sbjct: 337 PQPQSSNQNHFRLNNSFDTSTPNSSKST-------FVTPSRPI-HTPQPYSSSVSRYSRG 388
Query: 343 VRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSRAKVRSQSAPRQRHEFEEYSSTRRP 402
R T +P + + S + +S P+YMA T S++AK+R+ S P++R + S+ +
Sbjct: 389 GGRATQDSPFKDDDSLTSCPPFSA-PSYMAPTVSAKAKLRANSNPKERMDRTPVSTNEKR 447
Query: 403 FQGL------WDVGSTNSDNDSDSR 421
W+ GS N+S+++
Sbjct: 448 RSSFPLGSFKWNKGSLFMSNNSNNK 472
>At5g03040 unknown protein
Length = 446
Score = 95.1 bits (235), Expect = 7e-20
Identities = 98/365 (26%), Positives = 157/365 (42%), Gaps = 79/365 (21%)
Query: 61 LERNKHAIAVAAATAAVAEAA----LAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEET 116
+ERN+ + A A A + ++ A V R T + AG SN EE
Sbjct: 66 VERNRDLSPPSTADAVNVTATDVPVVPSSSAPGVVRRATPTRFAGKSN---------EEA 116
Query: 117 AAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARAS 176
AA+ IQ+ FRGYLARRALRA++ LV+L+ L+ G +V+++ A+ L+ MQTL R+Q++ RA
Sbjct: 117 AAILIQTIFRGYLARRALRAMRGLVRLKLLMEGSVVKRQAANTLKCMQTLSRVQSQIRAR 176
Query: 177 RAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSN-FRKIESEKP 235
R +S +N K L + ++ + + SI + +N K E+
Sbjct: 177 RIRMSEENQARQKQLLQKHA-----KELAGLKNGDNWNDSIQSKEKVEANLLSKYEATMR 231
Query: 236 RFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNE 295
R + + Q+N + +K+ + P TW P+ NK+ +N
Sbjct: 232 RERALAYSYSHQQNWKNNSKSGNPMFMDPSN---------PTWVPRKNKSNSN------- 275
Query: 296 SPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT-PTRS 354
+ + SVK + E A S T + S Q + SS G+ +N+ F+ PT S
Sbjct: 276 ---------NDNAASVKGSINRNEAAKSLTRNGSTQPNTPSSARGTPRNKNSFFSPPTPS 326
Query: 355 ECSWS----------------------------------FLGGYSGYPNYMANTESSRAK 380
+ S L G P+YM T+S+RA+
Sbjct: 327 RLNQSSRKSNDDDSKSTISVLSERNRRHSIAGSSVRDDESLAGSPALPSYMVPTKSARAR 386
Query: 381 VRSQS 385
++ QS
Sbjct: 387 LKPQS 391
>At2g26180 putative SF16 protein {Helianthus annuus}
Length = 416
Score = 94.4 bits (233), Expect = 1e-19
Identities = 91/334 (27%), Positives = 142/334 (42%), Gaps = 71/334 (21%)
Query: 114 EETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKA 173
EE AA++IQ+AFRG+LARRALRALK +V+LQALVRG VRK+ A LR MQ LVR+Q +
Sbjct: 83 EEWAAIRIQTAFRGFLARRALRALKGIVRLQALVRGRQVRKQAAVTLRCMQALVRVQARV 142
Query: 174 RASRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFG-------------------- 213
RA R ++ + K H + ++ + + G
Sbjct: 143 RARRVRMTVEGQAVQKLLDEHRTKSDLLKEVEEGWCDRKGTVDDIKSKLQQRQEGAFKRE 202
Query: 214 ---------------GSSILKRCSSNSNFRKIESEKPRFGSNWLDHWM-----QENSISQ 253
SS LK SS S + E +K +G +WL+ WM + +
Sbjct: 203 RALAYALAQKQWRSTTSSNLKTNSSISYLKSQEFDKNSWGWSWLERWMAARPWETRLMDT 262
Query: 254 TKNASSKNRHPDEH-KSDKILEVDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVK 312
A++ P +H KS + +V + NNV + + P H + S +
Sbjct: 263 VDTAATPPPLPHKHLKSPETADV------VQVRRNNVTTRVSAKPPPHML---SSSPGYE 313
Query: 313 FHKAK-EEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYM 371
F+++ + T S +T S + S +++ P+YM
Sbjct: 314 FNESSGSSSICTSTTPVSGKTGLVSDNSSSQAKKHK--------------------PSYM 353
Query: 372 ANTESSRAKVRSQSAPRQRHEFEEYSSTRRPFQG 405
+ TES++AK R+ RQ + ++ F G
Sbjct: 354 SLTESTKAKRRTNRGLRQSMDEFQFMKNSGMFTG 387
>At1g18840 hypothetical protein
Length = 563
Score = 89.0 bits (219), Expect = 5e-18
Identities = 61/227 (26%), Positives = 110/227 (47%), Gaps = 9/227 (3%)
Query: 111 RLPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQ 170
++ +E AAV +Q+A+RGYLARRA + LK +++LQAL+RGH+VR++ L + +VRLQ
Sbjct: 105 KIQQEIAAVTVQAAYRGYLARRAFKILKGIIRLQALIRGHMVRRQAVSTLCCVMGIVRLQ 164
Query: 171 TKARASRAHLSSDNLH-SFKSSLSHYPVPEEYEQPHHVYS----TKFGGSSIL-KRCSSN 224
AR S + K L H P+ + +S K G++ K +S+
Sbjct: 165 ALARGREIRHSDIGVEVQRKCHLHHQPLENKANSVVDTHSYLGINKLTGNAFAQKLLASS 224
Query: 225 SNFRKIESEKPRFGSNWLDHWMQE---NSISQTKNASSKNRHPDEHKSDKILEVDTWKPQ 281
N + + S WL++W + Q K AS + + +I+E + +P+
Sbjct: 225 PNVLPLSLDNDSSNSIWLENWSASCFWKPVPQPKKASLRKSQKKFASNPQIVEAEFARPK 284
Query: 282 LNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADN 328
+ + +++ N S ++ S++ + S + + V + DN
Sbjct: 285 KSVRKVPSSNLDNSSVAQTSSELEKPKRSFRKVSTSQSVEPLPSMDN 331
Score = 41.2 bits (95), Expect = 0.001
Identities = 39/139 (28%), Positives = 64/139 (45%), Gaps = 7/139 (5%)
Query: 265 DEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASR 324
DE K D++ E +P+ + + +NES +S + KE+V R
Sbjct: 380 DEEKEDEVAETVVQQPE-ELIQTHTPLGTNESLDSTLVNQIEESEENVMAEEKEDVKEER 438
Query: 325 TADNSPQTFSASSRNGSGVRRNTPFTPTRS-ECSWSFLGGYS---GYPNYMANTESSRAK 380
T + + SA N ++ + T T++ E S G + G P+YM T+S++AK
Sbjct: 439 TPKQNHKENSAGKENQKSGKKASSVTATQTAEFQESGNGNQTSSPGIPSYMQATKSAKAK 498
Query: 381 VRSQ--SAPRQRHEFEEYS 397
+R Q S+PRQ E+ S
Sbjct: 499 LRLQGSSSPRQLGTTEKAS 517
>At3g22190 unknown protein
Length = 400
Score = 86.3 bits (212), Expect = 3e-17
Identities = 62/165 (37%), Positives = 85/165 (50%), Gaps = 35/165 (21%)
Query: 114 EETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKA 173
E AA +IQ+A+RG+LARRALRALK LV+LQALVRGH VRK+ A LR MQ LVR+Q +
Sbjct: 87 ENRAATRIQTAYRGFLARRALRALKGLVRLQALVRGHAVRKQAAVTLRCMQALVRVQARV 146
Query: 174 RASRAHL-----SSDNLHSFKSSLSHYPVPEEYEQP--HHVYSTKFGGSSILKR------ 220
RA R L S + + + L+ E E+ + S + + +LKR
Sbjct: 147 RARRVRLALELESETSQQTLQQQLADEARVREIEEGWCDSIGSVEQIQAKLLKRQEAAAK 206
Query: 221 -------------------CSSNSNFRKIESEKPRFGSNWLDHWM 246
S++S F + +K +G NWL+ WM
Sbjct: 207 RERAMAYALTHQWQAGTRLLSAHSGF---QPDKNNWGWNWLERWM 248
>At1g17480 hypothetical protein
Length = 371
Score = 85.9 bits (211), Expect = 4e-17
Identities = 103/390 (26%), Positives = 174/390 (44%), Gaps = 70/390 (17%)
Query: 3 FLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLE 62
++R L +KP+ +D K SDK +K++W + SS++ + SS+
Sbjct: 7 WIRSLISNRKPV--NDQQEKLSDKSSKKKWKLWRISSESLA-------------SSSFKS 51
Query: 63 RNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQ 122
R +A + + A A A A ++R R L + E A+ +IQ
Sbjct: 52 RGSYAASSLGSELPSFSADEAFTTAMAALIRAPP----------RDFLMVKREWASTRIQ 101
Query: 123 SAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSS 182
+AFR +LAR+A RALKA+V++QA+ RG VRK+ A LR MQ LVR+Q++ RA R S
Sbjct: 102 AAFRAFLARQAFRALKAVVRIQAIFRGRQVRKQAAVTLRCMQALVRVQSRVRAHR-RAPS 160
Query: 183 DNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIES--EKPRFGSN 240
D+L E + P V T+ G C S + +++++ + + G+
Sbjct: 161 DSL--------------ELKDP--VKQTEKGW------CGSPRSIKEVKTKLQMKQEGAI 198
Query: 241 WLDHWMQENSISQTKNA-SSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSK 299
+ M Q++ S R H K W ++V + S +S S+
Sbjct: 199 KRERAMVYALTHQSRTCPSPSGRAITHHGLRKSSPGWNW-------YDDVGTFSRKS-SE 250
Query: 300 HSTKAQNQSLSVKFHKAKEEVAASRTADNSP-------QTFSASSRNGSGVRRNTPFTPT 352
S ++ ++++V+ K ++++R P S S + +T +P
Sbjct: 251 SSVLSEYETVTVR----KNNLSSTRVLARPPLLLPPVSSGMSYDSLHDETSTSSTSQSPV 306
Query: 353 RSECSWSFLGGYSGYPNYMANTESSRAKVR 382
S GGY P+YM+ T+S++AK R
Sbjct: 307 AFSSSVLDGGGYYRKPSYMSLTQSTQAKQR 336
>At4g14750 unknown protein
Length = 387
Score = 83.6 bits (205), Expect = 2e-16
Identities = 61/175 (34%), Positives = 85/175 (47%), Gaps = 22/175 (12%)
Query: 29 KRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAAVAEAALAAAHAA 88
KRRWSF + S+ P PP A DS P +
Sbjct: 41 KRRWSFRRSSATG---PPPPACAITLKDSPPPPPPPPPPPPLQQPF-------------- 83
Query: 89 AEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALRALKALVKLQALVR 148
VV + + N + ++ EE AA+KIQ+ +R +LAR+ALRALK LVKLQALVR
Sbjct: 84 --VVEIVDNEDEQIKNVSAEEI---EEFAAIKIQACYRSHLARKALRALKGLVKLQALVR 138
Query: 149 GHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKSSLSHYPVPEEYEQ 203
GH+VRK+ LR MQ L+ LQ KAR R + + + ++S+ + Y +
Sbjct: 139 GHLVRKQATATLRCMQALITLQAKAREQRIRMIGGDSTNPRTSIHKTRINNFYHE 193
Score = 53.9 bits (128), Expect = 2e-07
Identities = 68/265 (25%), Positives = 102/265 (37%), Gaps = 72/265 (27%)
Query: 139 ALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKSSLSHYPVP 198
A +K+QA R H+ RK LR ++ LV+LQ R HL
Sbjct: 107 AAIKIQACYRSHLARKA----LRALKGLVKLQALVRG---HLVRKQ-------------- 145
Query: 199 EEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWLDHWMQENSISQTKNAS 258
++ RC + ++ + R D SI +T+
Sbjct: 146 ----------------ATATLRCMQALITLQAKAREQRIRMIGGDSTNPRTSIHKTR--- 186
Query: 259 SKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKE 318
N + + ++ KI+E+D + + + MS + S H E
Sbjct: 187 INNFYHENEENIKIVEMDIQSKMYSPAPSALTEMSPRAYSSHF----------------E 230
Query: 319 EVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSR 378
+ + TA +SPQ FS +G T S Y +PNYMANT+SS+
Sbjct: 231 DCNSFNTAQSSPQCFSRFKEYYNG--------DTLSSYD------YPLFPNYMANTQSSK 276
Query: 379 AKVRSQSAPRQR--HEFEEYSSTRR 401
AK RSQSAP+QR +E+ S RR
Sbjct: 277 AKARSQSAPKQRPPEIYEKQMSGRR 301
>At5g13460 unknown protein
Length = 443
Score = 83.2 bits (204), Expect = 3e-16
Identities = 113/460 (24%), Positives = 186/460 (39%), Gaps = 91/460 (19%)
Query: 7 LFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQ--PPPSAFNQFDSSTPLERN 64
LF K I S+ + +K +K +R+W+F K K K LP PP +S E
Sbjct: 7 LFTVLKRIFISEVNSEKKEK--RRKWTFWKLRIK-KRLPSITAPPEHRTSHESH---EEQ 60
Query: 65 KHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRL-----PEETAAV 119
K I + V E + + + + + + + Q ++ E AA
Sbjct: 61 KEEIV-----SDVGEISQVSCSRQLDSIEESKGSTSPETADLVVQYQMFLNRQEEVLAAT 115
Query: 120 KIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAH 179
+IQ+AFRG+LAR+ALRALK +VKLQA +RG VR++ L+ +Q++V +Q++ R
Sbjct: 116 RIQTAFRGHLARKALRALKGIVKLQAYIRGRAVRRQAMTTLKCLQSVVNIQSQVCGKRTQ 175
Query: 180 LSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESE-KPRFG 238
+ V +YE+ S+I N N K+++ + R+
Sbjct: 176 IPGG-------------VHRDYEE-----------SNIF-----NDNILKVDTNGQKRWD 206
Query: 239 SNWLDHWMQENSISQTKNASSKNR--------HPDEHKSDKILEVDTWKPQLNKNENNVN 290
+ L +E + K AS + H +S + WK L++ +
Sbjct: 207 DSLLTKEEKEAVVMSKKEASLRRERIKEYAVTHRKSAESYQKRSNTKWKYWLDEWVDTQL 266
Query: 291 SMSNESPS---KHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNT 347
+ S E TK ++++L+ K K + R +N + S G +
Sbjct: 267 TKSKELEDLDFSSKTKPKDETLNEKQLKTPRNSSPRRLVNNHRRQVSI------GEDEQS 320
Query: 348 PFTPTRSECSWSFLGGYSGYPNYMANTESSRAKVRSQSAPRQR-HEFEEYSSTRRPFQGL 406
P T + P YM TES++AK RS S+PR R F+ S + P++
Sbjct: 321 PAAVTITT------------PTYMVATESAKAKSRSLSSPRIRPRSFDTQSESYSPYKNK 368
Query: 407 WDVGST-------------NSDNDSDSRSNKVSPALSRFN 433
+ ++ N N S + SP L FN
Sbjct: 369 LCLTTSMMSEAPSKVRIANNGSNTRPSAYQQRSPGLRGFN 408
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.123 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,823,541
Number of Sequences: 26719
Number of extensions: 415460
Number of successful extensions: 1728
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 1552
Number of HSP's gapped (non-prelim): 178
length of query: 441
length of database: 11,318,596
effective HSP length: 102
effective length of query: 339
effective length of database: 8,593,258
effective search space: 2913114462
effective search space used: 2913114462
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149494.9


