

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.6 + phase: 0 /pseudo
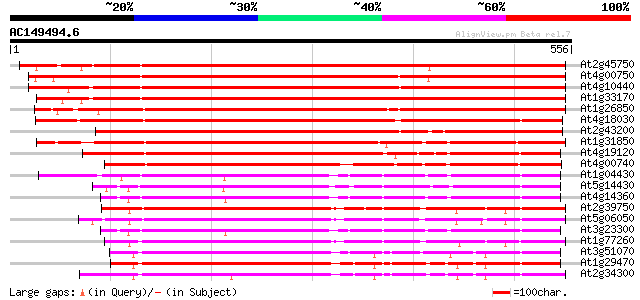
(556 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45750 hypothetical protein 716 0.0
At4g00750 unknown protein 712 0.0
At4g10440 putative protein 665 0.0
At1g33170 unknown protein 651 0.0
At1g26850 unknown protein 615 e-176
At4g18030 unknown protein 600 e-172
At2g43200 hypothetical protein 532 e-151
At1g31850 unknown protein 515 e-146
At4g19120 ERD3 protein (ERD3) 491 e-139
At4g00740 unknown protein 414 e-116
At1g04430 unknown protein 399 e-111
At5g14430 unknown protein 394 e-110
At4g14360 390 e-108
At2g39750 unknown protein 387 e-108
At5g06050 ankyrin-like protein 384 e-107
At3g23300 ankyrin repeat protein 381 e-106
At1g77260 unknown protein 374 e-103
At3g51070 putative protein 364 e-101
At1g29470 unknown protein 358 6e-99
At2g34300 unknown protein 355 5e-98
>At2g45750 hypothetical protein
Length = 631
Score = 716 bits (1849), Expect = 0.0
Identities = 338/553 (61%), Positives = 426/553 (76%), Gaps = 16/553 (2%)
Query: 10 NKFPRPSSM-RKPHLYF--LIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVN 66
N F R SS +K +LY+ L+A LC A YLLG +Q A + A S +
Sbjct: 2 NLFTRISSRTKKANLYYVTLVALLCIASYLLGIWQNTAVNP---RAAFDDSDGTPCEGFT 58
Query: 67 KPT----LDFQSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYR 122
+P LDF +HHN D ++ +FP C +E+TPCED RSL++ R R+ YR
Sbjct: 59 RPNSTKDLDFDAHHNIQDPP-PVTETAVSFPSCAAALSEHTPCEDAKRSLKFSRERLEYR 117
Query: 123 ERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGD 182
+RHCP + EE LKCR+P P+GYKTPF WPASRDVAW+ANVPH ELTVEK QNW+RY+ D
Sbjct: 118 QRHCPER-EEILKCRIPAPYGYKTPFRWPASRDVAWFANVPHTELTVEKKNQNWVRYEND 176
Query: 183 RFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSL 242
RF+FPGGGTMFP GA AYIDDIG+LI+L DGSIRTA+DTGCGVAS+GAYL SRNI T+S
Sbjct: 177 RFWFPGGGTMFPRGADAYIDDIGRLIDLSDGSIRTAIDTGCGVASFGAYLLSRNITTMSF 236
Query: 243 APRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEV 302
APRDTHEAQVQFALERGVPA+IG++A+ RLP+PSRAFD++HCSRCLIPW + DG +L EV
Sbjct: 237 APRDTHEAQVQFALERGVPAMIGIMATIRLPYPSRAFDLAHCSRCLIPWGQNDGAYLMEV 296
Query: 303 DRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAI 362
DRVLRPGGYWILSGPPINW K +GW+RT DLN EQT+IE+VA+SLCW K++++DD+AI
Sbjct: 297 DRVLRPGGYWILSGPPINWQKRWKGWERTMDDLNAEQTQIEQVARSLCWKKVVQRDDLAI 356
Query: 363 WQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEE---TAGGV 419
WQKP NH+DC+ R++ + FC ++PD AWYT + +CL P+P+V + E+ AGG
Sbjct: 357 WQKPFNHIDCKKTREVLKNPEFCRHDQDPDMAWYTKMDSCLTPLPEVDDAEDLKTVAGGK 416
Query: 420 LKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLG-TKRYRNLVD 478
++ WP RL ++PPR++ G +E +T E + ++ +LWK+R+ +YKK++ QLG T RYRNLVD
Sbjct: 417 VEKWPARLNAIPPRVNKGALEEITPEAFLENTKLWKQRVSYYKKLDYQLGETGRYRNLVD 476
Query: 479 MNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDL 538
MNA LGGFA+AL +PVWVMNVVPV+AK++TLG IYERGLIGTY +WCEAMSTYPRTYD
Sbjct: 477 MNAYLGGFAAALADDPVWVMNVVPVEAKLNTLGVIYERGLIGTYQNWCEAMSTYPRTYDF 536
Query: 539 IHADSLFSLYNGR 551
IHADS+F+LY G+
Sbjct: 537 IHADSVFTLYQGQ 549
>At4g00750 unknown protein
Length = 633
Score = 712 bits (1837), Expect = 0.0
Identities = 336/541 (62%), Positives = 413/541 (76%), Gaps = 10/541 (1%)
Query: 19 RKPHLY--FLIAFLCAAFYLLGAYQQ--RASFTSLTKKAIITSPSCTIQQVNKPTLDFQS 74
++ +LY LIA LC FY +G +Q R S +TS CT P L+F S
Sbjct: 18 KQTNLYRVILIAILCVTFYFVGVWQHSGRGISRSSISNHELTSVPCTFPHQTTPILNFAS 77
Query: 75 HHNSSDTIIALS-SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEED 133
H + D ++ + P CGV F+EYTPCE RSL + R R+IYRERHCP K E
Sbjct: 78 RHTAPDLPPTITDARVVQIPSCGVEFSEYTPCEFVNRSLNFPRERLIYRERHCPEK-HEI 136
Query: 134 LKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMF 193
++CR+P P+GY PF WP SRDVAW+ANVPH ELTVEK QNW+RY+ DRF FPGGGTMF
Sbjct: 137 VRCRIPAPYGYSLPFRWPESRDVAWFANVPHTELTVEKKNQNWVRYEKDRFLFPGGGTMF 196
Query: 194 PNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQ 253
P GA AYID+IG+LINLKDGSIRTA+DTGCGVAS+GAYL SRNI+T+S APRDTHEAQVQ
Sbjct: 197 PRGADAYIDEIGRLINLKDGSIRTAIDTGCGVASFGAYLMSRNIVTMSFAPRDTHEAQVQ 256
Query: 254 FALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWI 313
FALERGVPA+IGVLAS RLPFP+RAFDI+HCSRCLIPW +Y+G +L EVDRVLRPGGYWI
Sbjct: 257 FALERGVPAIIGVLASIRLPFPARAFDIAHCSRCLIPWGQYNGTYLIEVDRVLRPGGYWI 316
Query: 314 LSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCR 373
LSGPPINW +H +GW+RT+ DLN EQ++IE+VA+SLCW KL++++D+A+WQKP NH+ C+
Sbjct: 317 LSGPPINWQRHWKGWERTRDDLNSEQSQIERVARSLCWRKLVQREDLAVWQKPTNHVHCK 376
Query: 374 SARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKE--ETAGGVLKNWPQRLESVP 431
R PFC + P++ WYT L+TCL P+P+V+ E E AGG L WP+RL ++P
Sbjct: 377 RNRIALGRPPFC-HRTLPNQGWYTKLETCLTPLPEVTGSEIKEVAGGQLARWPERLNALP 435
Query: 432 PRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLG-TKRYRNLVDMNANLGGFASAL 490
PRI G++EG+T + + + E W++R+ +YKK + QL T RYRN +DMNA+LGGFASAL
Sbjct: 436 PRIKSGSLEGITEDEFVSNTEKWQRRVSYYKKYDQQLAETGRYRNFLDMNAHLGGFASAL 495
Query: 491 VKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNG 550
V +PVWVMNVVPV+A V+TLG IYERGLIGTY +WCEAMSTYPRTYD IHADS+FSLY
Sbjct: 496 VDDPVWVMNVVPVEASVNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFSLYKD 555
Query: 551 R 551
R
Sbjct: 556 R 556
>At4g10440 putative protein
Length = 633
Score = 665 bits (1717), Expect = 0.0
Identities = 313/539 (58%), Positives = 401/539 (74%), Gaps = 11/539 (2%)
Query: 19 RKPHLYFLIAFLCAAFYLLGAYQQRASFTSLTKKAIIT-----SPSCTIQQVNKPTLDFQ 73
+K L ++ LC FY+LGA+Q +S++K T S S + LDF+
Sbjct: 17 KKLTLILGVSGLCILFYVLGAWQANTVPSSISKLGCETQSNPSSSSSSSSSSESAELDFK 76
Query: 74 SHHNSSDTIIALSSETFN-FPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEE 132
SH+ + +++T F C ++ +EYTPCED R R+ R+ M YRERHCPVK +E
Sbjct: 77 SHNQIE---LKETNQTIKYFEPCELSLSEYTPCEDRQRGRRFDRNMMKYRERHCPVK-DE 132
Query: 133 DLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTM 192
L C +PPP YK PF WP SRD AWY N+PH+EL+VEKAVQNWI+ +GDRF FPGGGTM
Sbjct: 133 LLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSVEKAVQNWIQVEGDRFRFPGGGTM 192
Query: 193 FPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQV 252
FP GA AYIDDI +LI L DG IRTA+DTGCGVAS+GAYL R+I+ +S APRDTHEAQV
Sbjct: 193 FPRGADAYIDDIARLIPLTDGGIRTAIDTGCGVASFGAYLLKRDIMAVSFAPRDTHEAQV 252
Query: 253 QFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYW 312
QFALERGVPA+IG++ S+RLP+P+RAFD++HCSRCLIPW + DG++L EVDRVLRPGGYW
Sbjct: 253 QFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFKNDGLYLMEVDRVLRPGGYW 312
Query: 313 ILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDC 372
ILSGPPINW ++ RGW+RT++DL +EQ IE VAKSLCW K+ EK D++IWQKP+NH++C
Sbjct: 313 ILSGPPINWKQYWRGWERTEEDLKKEQDSIEDVAKSLCWKKVTEKGDLSIWQKPLNHIEC 372
Query: 373 RSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPP 432
+ ++ P C +N D AWY DL+TC+ P+P+ +N +++AGG L++WP R +VPP
Sbjct: 373 KKLKQNNKSPPICS-SDNADSAWYKDLETCITPLPETNNPDDSAGGALEDWPDRAFAVPP 431
Query: 433 RIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVK 492
RI GTI + +E + +DNE+WK+RI HYKK+ +L R+RN++DMNA LGGFA++++K
Sbjct: 432 RIIRGTIPEMNAEKFREDNEVWKERIAHYKKIVPELSHGRFRNIMDMNAFLGGFAASMLK 491
Query: 493 NPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
P WVMNVVPV A+ TLG IYERGLIGTY DWCE STYPRTYD+IHA LFSLY R
Sbjct: 492 YPSWVMNVVPVDAEKQTLGVIYERGLIGTYQDWCEGFSTYPRTYDMIHAGGLFSLYEHR 550
Score = 30.4 bits (67), Expect = 2.7
Identities = 35/156 (22%), Positives = 57/156 (36%), Gaps = 37/156 (23%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D + + A + +++ P D + + ERG LIG
Sbjct: 467 LSHGRFRNIMDMNAFLGGFAASMLKYPSWVMNVVPVDAEKQTLGVIYERG---LIGTYQD 523
Query: 270 KRLPFPS--RAFDISHCS--------RCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPI 319
F + R +D+ H RC + + L E+DR+LRP G +L
Sbjct: 524 WCEGFSTYPRTYDMIHAGGLFSLYEHRCDLT------LILLEMDRILRPEGTVVL----- 572
Query: 320 NWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLI 355
+D + K+EK+ K + W I
Sbjct: 573 -------------RDNVETLNKVEKIVKGMKWKSQI 595
>At1g33170 unknown protein
Length = 709
Score = 651 bits (1680), Expect = 0.0
Identities = 302/549 (55%), Positives = 398/549 (72%), Gaps = 28/549 (5%)
Query: 27 IAFLCAAFYLLGAYQQRASFTSLTK--------KAIITSPSCTIQQVNKPT--------- 69
++ LC Y+LG++Q TS ++ + T+ + T Q P+
Sbjct: 24 VSGLCILSYVLGSWQTNTVPTSSSEAYSRMGCDETSTTTRAQTTQTQTNPSSDDTSSSLS 83
Query: 70 ------LDFQSHHNSSDTIIALSSETFN-FPRCGVNFTEYTPCEDPTRSLRYKRSRMIYR 122
LDF+SHH + ++++T F C ++ +EYTPCED R R+ R+ M YR
Sbjct: 84 SSEPVELDFESHHKLE---LKITNQTVKYFEPCDMSLSEYTPCEDRERGRRFDRNMMKYR 140
Query: 123 ERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGD 182
ERHCP K +E L C +PPP YK PF WP SRD AWY N+PH+EL++EKA+QNWI+ +G+
Sbjct: 141 ERHCPSK-DELLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGE 199
Query: 183 RFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSL 242
RF FPGGGTMFP GA AYIDDI +LI L DG+IRTA+DTGCGVAS+GAYL R+I+ +S
Sbjct: 200 RFRFPGGGTMFPRGADAYIDDIARLIPLTDGAIRTAIDTGCGVASFGAYLLKRDIVAMSF 259
Query: 243 APRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEV 302
APRDTHEAQVQFALERGVPA+IG++ S+RLP+P+RAFD++HCSRCLIPW + DG++L EV
Sbjct: 260 APRDTHEAQVQFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNDGLYLTEV 319
Query: 303 DRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAI 362
DRVLRPGGYWILSGPPINW K+ +GW+R+++DL QEQ IE A+SLCW K+ EK D++I
Sbjct: 320 DRVLRPGGYWILSGPPINWKKYWKGWERSQEDLKQEQDSIEDAARSLCWKKVTEKGDLSI 379
Query: 363 WQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKN 422
WQKPINH++C +++ P C + PD AWY DL++C+ P+P+ ++ +E AGG L++
Sbjct: 380 WQKPINHVECNKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTPLPEANSSDEFAGGALED 439
Query: 423 WPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNAN 482
WP R +VPPRI GTI + +E + +DNE+WK+RI +YK++ +L R+RN++DMNA
Sbjct: 440 WPNRAFAVPPRIIGGTIPDINAEKFREDNEVWKERISYYKQIMPELSRGRFRNIMDMNAY 499
Query: 483 LGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHAD 542
LGGFA+A++K P WVMNVVPV A+ TLG I+ERG IGTY DWCE STYPRTYDLIHA
Sbjct: 500 LGGFAAAMMKYPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEGFSTYPRTYDLIHAG 559
Query: 543 SLFSLYNGR 551
LFS+Y R
Sbjct: 560 GLFSIYENR 568
>At1g26850 unknown protein
Length = 616
Score = 615 bits (1585), Expect = e-176
Identities = 292/532 (54%), Positives = 378/532 (70%), Gaps = 13/532 (2%)
Query: 25 FLIAFLCAAFYLLGAYQQRASF---TSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDT 81
F++ LC FY+LGA+Q R+ F S+ + + C I P+L+F++HH +
Sbjct: 19 FIVFSLCCFFYILGAWQ-RSGFGKGDSIALEMTNSGADCNIV----PSLNFETHHAGESS 73
Query: 82 IIALS--SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVP 139
++ S ++ F C +T+YTPC+D R++ + R MIYRERHC + E+ L C +P
Sbjct: 74 LVGASEAAKVKAFEPCDGRYTDYTPCQDQRRAMTFPRDSMIYRERHCAPENEK-LHCLIP 132
Query: 140 PPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGA 199
P GY TPF+WP SRD YAN P++ LTVEKA+QNWI+Y+GD F FPGGGT FP GA
Sbjct: 133 APKGYVTPFSWPKSRDYVPYANAPYKALTVEKAIQNWIQYEGDVFRFPGGGTQFPQGADK 192
Query: 200 YIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERG 259
YID + +I +++G++RTALDTGCGVASWGAYL SRN+ +S APRD+HEAQVQFALERG
Sbjct: 193 YIDQLASVIPMENGTVRTALDTGCGVASWGAYLWSRNVRAMSFAPRDSHEAQVQFALERG 252
Query: 260 VPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPI 319
VPA+IGVL + +LP+P+RAFD++HCSRCLIPW DG++L EVDRVLRPGGYWILSGPPI
Sbjct: 253 VPAVIGVLGTIKLPYPTRAFDMAHCSRCLIPWGANDGMYLMEVDRVLRPGGYWILSGPPI 312
Query: 320 NWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLA 379
NW +++ WQR K+DL +EQ KIE+ AK LCW K E +IAIWQK +N CRS R+
Sbjct: 313 NWKVNYKAWQRPKEDLQEEQRKIEEAAKLLCWEKKYEHGEIAIWQKRVNDEACRS-RQDD 371
Query: 380 TDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTI 439
FC ++ D WY ++ C+ P P+ S+ +E AGG L+ +P RL +VPPRI G+I
Sbjct: 372 PRANFC-KTDDTDDVWYKKMEACITPYPETSSSDEVAGGELQAFPDRLNAVPPRISSGSI 430
Query: 440 EGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMN 499
GVT + Y DN WKK + YK++N+ L T RYRN++DMNA GGFA+AL +WVMN
Sbjct: 431 SGVTVDAYEDDNRQWKKHVKAYKRINSLLDTGRYRNIMDMNAGFGGFAAALESQKLWVMN 490
Query: 500 VVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
VVP A+ + LG +YERGLIG YHDWCEA STYPRTYDLIHA+ LFSLY +
Sbjct: 491 VVPTIAEKNRLGVVYERGLIGIYHDWCEAFSTYPRTYDLIHANHLFSLYKNK 542
Score = 36.6 bits (83), Expect = 0.038
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 5/108 (4%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G + A L+S+ + +++ P + ++ ERG+ +
Sbjct: 459 LDTGRYRNIMDMNAGFGGFAAALESQKLWVMNVVPTIAEKNRLGVVYERGLIGIYHDWCE 518
Query: 270 KRLPFPSRAFDISHCSRCLIPW---AEYDGIFLNEVDRVLRPGGYWIL 314
+P R +D+ H + + D I L E+DR+LRP G I+
Sbjct: 519 AFSTYP-RTYDLIHANHLFSLYKNKCNADDILL-EMDRILRPEGAVII 564
>At4g18030 unknown protein
Length = 621
Score = 600 bits (1546), Expect = e-172
Identities = 286/523 (54%), Positives = 363/523 (68%), Gaps = 9/523 (1%)
Query: 26 LIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIAL 85
++ LC FYLLGA+Q+ + IT + V LDF+ HHN+
Sbjct: 21 VVVGLCCFFYLLGAWQKSGFGKGDSIAMEITKQAQCTDIVTD--LDFEPHHNTVKIPHKA 78
Query: 86 SSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYK 145
+ +F C V +YTPC++ R++++ R MIYRERHCP E+ L+C VP P GY
Sbjct: 79 DPKPVSFKPCDVKLKDYTPCQEQDRAMKFPRENMIYRERHCPPDNEK-LRCLVPAPKGYM 137
Query: 146 TPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIG 205
TPF WP SRD YAN P + LTVEKA QNW+++ G+ F FPGGGTMFP GA AYI+++
Sbjct: 138 TPFPWPKSRDYVHYANAPFKSLTVEKAGQNWVQFQGNVFKFPGGGTMFPQGADAYIEELA 197
Query: 206 KLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
+I +KDGS+RTALDTGCGVASWGAY+ RN++T+S APRD HEAQVQFALERGVPA+I
Sbjct: 198 SVIPIKDGSVRTALDTGCGVASWGAYMLKRNVLTMSFAPRDNHEAQVQFALERGVPAIIA 257
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHH 325
VL S LP+P+RAFD++ CSRCLIPW +G +L EVDRVLRPGGYW+LSGPPINW H
Sbjct: 258 VLGSILLPYPARAFDMAQCSRCLIPWTANEGTYLMEVDRVLRPGGYWVLSGPPINWKTWH 317
Query: 326 RGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFC 385
+ W RTK +LN EQ +IE +A+SLCW K EK DIAI++K IN C + + T +
Sbjct: 318 KTWNRTKAELNAEQKRIEGIAESLCWEKKYEKGDIAIFRKKINDRSCDRSTPVDTCK--- 374
Query: 386 GPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSE 445
+++ D WY +++TC+ P P+VSN+EE AGG LK +P+RL +VPP I G I GV E
Sbjct: 375 --RKDTDDVWYKEIETCVTPFPKVSNEEEVAGGKLKKFPERLFAVPPSISKGLINGVDEE 432
Query: 446 GYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQA 505
Y +D LWKKR+ YK++N +G+ RYRN++DMNA LGGFA+AL WVMNV+P
Sbjct: 433 SYQEDINLWKKRVTGYKRINRLIGSTRYRNVMDMNAGLGGFAAALESPKSWVMNVIPTIN 492
Query: 506 KVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLY 548
K +TL +YERGLIG YHDWCE STYPRTYD IHA +FSLY
Sbjct: 493 K-NTLSVVYERGLIGIYHDWCEGFSTYPRTYDFIHASGVFSLY 534
>At2g43200 hypothetical protein
Length = 611
Score = 532 bits (1370), Expect = e-151
Identities = 246/463 (53%), Positives = 319/463 (68%), Gaps = 5/463 (1%)
Query: 86 SSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYK 145
SS + FP C NFT Y PC DP+ + +Y R RERHCP +E +C VP P GYK
Sbjct: 85 SSLSSYFPLCPKNFTNYLPCHDPSTARQYSIERHYRRERHCPDIAQEKFRCLVPKPTGYK 144
Query: 146 TPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIG 205
TPF WP SR AW+ NVP + L K QNW+R +GDRF FPGGGT FP G Y+D I
Sbjct: 145 TPFPWPESRKYAWFRNVPFKRLAELKKTQNWVRLEGDRFVFPGGGTSFPGGVKDYVDVIL 204
Query: 206 KLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
++ L GSIRT LD GCGVAS+GA+L + I+T+S+APRD HEAQVQFALERG+PA++G
Sbjct: 205 SVLPLASGSIRTVLDIGCGVASFGAFLLNYKILTMSIAPRDIHEAQVQFALERGLPAMLG 264
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHH 325
VL++ +LP+PSR+FD+ HCSRCL+ W YDG++L EVDRVLRP GYW+LSGPP+
Sbjct: 265 VLSTYKLPYPSRSFDMVHCSRCLVNWTSYDGLYLMEVDRVLRPEGYWVLSGPPVASRVKF 324
Query: 326 RGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFC 385
+ +R K+L + K+ V + LCW K+ E + IW+KP NHL CR K C
Sbjct: 325 KNQKRDSKELQNQMEKLNDVFRRLCWEKIAESYPVVIWRKPSNHLQCRKRLKALKFPGLC 384
Query: 386 GPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSE 445
+PD AWY +++ C+ P+P V++ +T VLKNWP+RL V PR+ G+I+G T
Sbjct: 385 S-SSDPDAAWYKEMEPCITPLPDVNDTNKT---VLKNWPERLNHV-PRMKTGSIQGTTIA 439
Query: 446 GYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQA 505
G+ D LW++R+ +Y L +YRN++DMNA LGGFA+AL+K P+WVMNVVP
Sbjct: 440 GFKADTNLWQRRVLYYDTKFKFLSNGKYRNVIDMNAGLGGFAAALIKYPMWVMNVVPFDL 499
Query: 506 KVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLY 548
K +TLG +Y+RGLIGTY +WCEA+STYPRTYDLIHA+ +FSLY
Sbjct: 500 KPNTLGVVYDRGLIGTYMNWCEALSTYPRTYDLIHANGVFSLY 542
>At1g31850 unknown protein
Length = 603
Score = 515 bits (1326), Expect = e-146
Identities = 245/529 (46%), Positives = 348/529 (65%), Gaps = 26/529 (4%)
Query: 27 IAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNSSDTIIALS 86
IAF +FYL G + + K + + + + +PT +
Sbjct: 25 IAFCGFSFYLGGIFCSERD--KIVAKDVTRTTTKAVASPKEPTAT------------PIQ 70
Query: 87 SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYKT 146
++ +FP CG F +YTPC DP R +Y R+ + ERHCP E++ +C +PPP GYK
Sbjct: 71 IKSVSFPECGSEFQDYTPCTDPKRWKKYGVHRLSFLERHCPPVYEKN-ECLIPPPDGYKP 129
Query: 147 PFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGK 206
P WP SR+ WY NVP+ + +K+ Q+W++ +GD+F FPGGGTMFP G Y+D +
Sbjct: 130 PIRWPKSREQCWYRNVPYDWINKQKSNQHWLKKEGDKFHFPGGGTMFPRGVSHYVDLMQD 189
Query: 207 LI-NLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
LI +KDG++RTA+DTGCGVASWG L R I++LSLAPRD HEAQVQFALERG+PA++G
Sbjct: 190 LIPEMKDGTVRTAIDTGCGVASWGGDLLDRGILSLSLAPRDNHEAQVQFALERGIPAILG 249
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHH 325
+++++RLPFPS AFD++HCSRCLIPW E+ GI+L E+ R++RPGG+W+LSGPP+N+N+
Sbjct: 250 IISTQRLPFPSNAFDMAHCSRCLIPWTEFGGIYLLEIHRIVRPGGFWVLSGPPVNYNRRW 309
Query: 326 RGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDC--RSARKLATDRP 383
RGW T +D + K++ + S+C+ K +KDDIA+WQK ++ C + A+ + P
Sbjct: 310 RGWNTTMEDQKSDYNKLQSLLTSMCFKKYAQKDDIAVWQK-LSDKSCYDKIAKNMEAYPP 368
Query: 384 FCGPQENPDKAWYTDLKTCLM-PVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGV 442
C PD AWYT L+ C++ P P+V +++ G + WP+RL P RI G + G
Sbjct: 369 KCDDSIEPDSAWYTPLRPCVVAPTPKV---KKSGLGSIPKWPERLHVAPERI--GDVHGG 423
Query: 443 TSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVP 502
++ D+ WK R+ HYKKV LGT + RN++DMN GGF++AL+++P+WVMNVV
Sbjct: 424 SANSLKHDDGKWKNRVKHYKKVLPALGTDKIRNVMDMNTVYGGFSAALIEDPIWVMNVVS 483
Query: 503 VQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
++L +++RGLIGTYHDWCEA STYPRTYDL+H DSLF+L + R
Sbjct: 484 -SYSANSLPVVFDRGLIGTYHDWCEAFSTYPRTYDLLHLDSLFTLESHR 531
>At4g19120 ERD3 protein (ERD3)
Length = 600
Score = 491 bits (1263), Expect = e-139
Identities = 231/479 (48%), Positives = 321/479 (66%), Gaps = 14/479 (2%)
Query: 73 QSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEE 132
++ +S D +L ++ +F C ++ +YTPC DP + +Y R+ + ERHCP +
Sbjct: 51 KAESSSLDVDDSLQVKSVSFSECSSDYQDYTPCTDPRKWKKYGTHRLTFMERHCPPVFDR 110
Query: 133 DLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTM 192
+C VPPP GYK P WP S+D WY NVP+ + +K+ QNW+R +G++F FPGGGTM
Sbjct: 111 K-QCLVPPPDGYKPPIRWPKSKDECWYRNVPYDWINKQKSNQNWLRKEGEKFIFPGGGTM 169
Query: 193 FPNGAGAYIDDIGKLI-NLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQ 251
FP+G AY+D + LI +KDG+IRTA+DTGCGVASWG L R I+T+SLAPRD HEAQ
Sbjct: 170 FPHGVSAYVDLMQDLIPEMKDGTIRTAIDTGCGVASWGGDLLDRGILTVSLAPRDNHEAQ 229
Query: 252 VQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGY 311
VQFALERG+PA++G+++++RLPFPS +FD++HCSRCLIPW E+ G++L EV R+LRPGG+
Sbjct: 230 VQFALERGIPAILGIISTQRLPFPSNSFDMAHCSRCLIPWTEFGGVYLLEVHRILRPGGF 289
Query: 312 WILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLD 371
W+LSGPP+N+ +GW T ++ K++++ S+C+ +KDDIA+WQK ++L
Sbjct: 290 WVLSGPPVNYENRWKGWDTTIEEQRSNYEKLQELLSSMCFKMYAKKDDIAVWQKSPDNL- 348
Query: 372 CRSARKLATD----RPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRL 427
KL+ D P C PD AWYT L+ C++ VP K+ K WP+RL
Sbjct: 349 --CYNKLSNDPDAYPPKCDDSLEPDSAWYTPLRPCVV-VPSPKLKKTDLESTPK-WPERL 404
Query: 428 ESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFA 487
+ P RI + G + D+ WK R HYKK+ +G+ + RN++DMN GG A
Sbjct: 405 HTTPERI--SDVPGGNGNVFKHDDSKWKTRAKHYKKLLPAIGSDKIRNVMDMNTAYGGLA 462
Query: 488 SALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
+ALV +P+WVMNVV A +TL +++RGLIGTYHDWCEA STYPRTYDL+H D LF+
Sbjct: 463 AALVNDPLWVMNVVSSYA-ANTLPVVFDRGLIGTYHDWCEAFSTYPRTYDLLHVDGLFT 520
>At4g00740 unknown protein
Length = 600
Score = 414 bits (1064), Expect = e-116
Identities = 203/454 (44%), Positives = 276/454 (60%), Gaps = 21/454 (4%)
Query: 95 CGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASR 154
C + PCEDP R+ + R YRERHCP+ E L C +PPP GYK P WP S
Sbjct: 84 CPAEAVAHMPCEDPRRNSQLSREMNFYRERHCPLPEETPL-CLIPPPSGYKIPVPWPESL 142
Query: 155 DVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKDGS 214
W+AN+P+ ++ K Q W++ +G+ F FPGGGTMFP GAG YI+ + + I L G+
Sbjct: 143 HKIWHANMPYNKIADRKGHQGWMKREGEYFTFPGGGTMFPGGAGQYIEKLAQYIPLNGGT 202
Query: 215 IRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPF 274
+RTALD GCGVAS+G L S+ I+ LS APRD+H++Q+QFALERGVPA + +L ++RLPF
Sbjct: 203 LRTALDMGCGVASFGGTLLSQGILALSFAPRDSHKSQIQFALERGVPAFVAMLGTRRLPF 262
Query: 275 PSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKD 334
P+ +FD+ HCSRCLIP+ Y+ + EVDR+LRPGGY ++SGPP+ W K + W
Sbjct: 263 PAYSFDLMHCSRCLIPFTAYNATYFIEVDRLLRPGGYLVISGPPVQWPKQDKEW------ 316
Query: 335 LNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKA 394
++ VA++LC+ + + IW+KP+ S + + C P A
Sbjct: 317 -----ADLQAVARALCYELIAVDGNTVIWKKPVGDSCLPSQNEFGLE--LCDESVPPSDA 369
Query: 395 WYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELW 454
WY LK C+ V K E A G + WP+RL VP R + + + D W
Sbjct: 370 WYFKLKRCVTRPSSV--KGEHALGTISKWPERLTKVPSR---AIVMKNGLDVFEADARRW 424
Query: 455 KKRIPHYK-KVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAI 513
+R+ +Y+ +N +L + RN++DMNA GGFA+ L +PVWVMNV+P + K TL I
Sbjct: 425 ARRVAYYRDSLNLKLKSPTVRNVMDMNAFFGGFAATLASDPVWVMNVIPAR-KPLTLDVI 483
Query: 514 YERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSL 547
Y+RGLIG YHDWCE STYPRTYD IH + SL
Sbjct: 484 YDRGLIGVYHDWCEPFSTYPRTYDFIHVSGIESL 517
>At1g04430 unknown protein
Length = 623
Score = 399 bits (1025), Expect = e-111
Identities = 217/530 (40%), Positives = 307/530 (56%), Gaps = 33/530 (6%)
Query: 29 FLCAAFYLLGAYQQRASFTSLTKKAI-ITSPSCTIQQVNKPTLDFQSHHNSSDTIIALSS 87
F+C F G+ Q AS + + S + N T S N+ D+++ S
Sbjct: 24 FVCFLFMYYGSSSQGASALEYGRSLRKLGSSYLSGDDDNGDTKQDDSVANAEDSLVVAKS 83
Query: 88 ETFNFPRCGVNFTEYTPCEDPT----RSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHG 143
FP C +E PC D L+ S M + ERHCP E C +PPP G
Sbjct: 84 ----FPVCDDRHSEIIPCLDRNFIYQMRLKLDLSLMEHYERHCPPP-ERRFNCLIPPPSG 138
Query: 144 YKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDD 203
YK P WP SRD W AN+PH L EK+ QNW+ G++ FPGGGT F GA YI
Sbjct: 139 YKVPIKWPKSRDEVWKANIPHTHLAKEKSDQNWMVEKGEKISFPGGGTHFHYGADKYIAS 198
Query: 204 IGKLINLK------DGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALE 257
I ++N +G +RT LD GCGVAS+GAYL + +I+T+SLAP D H+ Q+QFALE
Sbjct: 199 IANMLNFSNDVLNDEGRLRTVLDVGCGVASFGAYLLASDIMTMSLAPNDVHQNQIQFALE 258
Query: 258 RGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGP 317
RG+PA +GVL +KRLP+PSR+F+ +HCSRC I W + DG+ L E+DRVLRPGGY+ S P
Sbjct: 259 RGIPAYLGVLGTKRLPYPSRSFEFAHCSRCRIDWLQRDGLLLLELDRVLRPGGYFAYSSP 318
Query: 318 PINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARK 377
+ + +++L + ++ + + +CW ++++ +WQKP+++ DC R+
Sbjct: 319 --------EAYAQDEENL-KIWKEMSALVERMCWRIAVKRNQTVVWQKPLSN-DCYLERE 368
Query: 378 LATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMG 437
T P C +PD ++ C+ P + +K T G L WP RL S PPR+
Sbjct: 369 PGTQPPLCRSDADPDAVAGVSMEACITPYSKHDHK--TKGSGLAPWPARLTSSPPRL--- 423
Query: 438 TIEGVTSEGYSKDNELWKKRIPHY-KKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVW 496
G +++ + KD ELWK+++ Y +++++ + RN++DM A++G FA+AL VW
Sbjct: 424 ADFGYSTDMFEKDTELWKQQVDSYWNLMSSKVKSNTVRNIMDMKAHMGSFAAALKDKDVW 483
Query: 497 VMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
VMNVV +TL IY+RGLIGT H+WCEA STYPRTYDL+HA S+FS
Sbjct: 484 VMNVVSPDGP-NTLKLIYDRGLIGTNHNWCEAFSTYPRTYDLLHAWSIFS 532
Score = 28.9 bits (63), Expect = 7.8
Identities = 27/116 (23%), Positives = 53/116 (45%), Gaps = 21/116 (18%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
+K ++R +D + S+ A L+ +++ +++ D ++ +RG LIG +
Sbjct: 455 VKSNTVRNIMDMKAHMGSFAAALKDKDVWVMNVVSPDGPNT-LKLIYDRG---LIGTNHN 510
Query: 270 KRLPFPS--RAFDISHCSRCLIPWAEYDGI---------FLNEVDRVLRPGGYWIL 314
F + R +D+ H W+ + I L E+DR+LRP G+ I+
Sbjct: 511 WCEAFSTYPRTYDLLHA------WSIFSDIKSKGCSAEDLLIEMDRILRPTGFVII 560
>At5g14430 unknown protein
Length = 612
Score = 394 bits (1011), Expect = e-110
Identities = 213/481 (44%), Positives = 285/481 (58%), Gaps = 36/481 (7%)
Query: 83 IALSSETFNFPR----CGVNFTEYTPCEDPTRSLRYKR------SRMIYRERHCPVKGEE 132
I L+ F P+ C +E PC D R+L Y+ S M + E HCP E
Sbjct: 67 IVLAVSRFEVPKSVPICDSRHSELIPCLD--RNLHYQLKLKLNLSLMEHYEHHCP-PSER 123
Query: 133 DLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTM 192
C VPPP GYK P WP SRD W AN+PH L EK+ QNW+ +GD+ FPGGGT
Sbjct: 124 RFNCLVPPPVGYKIPLRWPVSRDEVWKANIPHTHLAQEKSDQNWMVVNGDKINFPGGGTH 183
Query: 193 FPNGAGAYIDDIGKLINL------KDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRD 246
F NGA YI + +++ GSIR LD GCGVAS+GAYL S +II +SLAP D
Sbjct: 184 FHNGADKYIVSLAQMLKFPGDKLNNGGSIRNVLDVGCGVASFGAYLLSHDIIAMSLAPND 243
Query: 247 THEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVL 306
H+ Q+QFALERG+P+ +GVL +KRLP+PSR+F+++HCSRC I W + DGI L E+DR+L
Sbjct: 244 VHQNQIQFALERGIPSTLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRLL 303
Query: 307 RPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKP 366
RPGGY++ S P + + +K N + + K +CW + ++D IW KP
Sbjct: 304 RPGGYFVYSSP-----EAYAHDPENRKIGN----AMHDLFKRMCWKVVAKRDQSVIWGKP 354
Query: 367 INHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQR 426
I++ C R P C ++PD W +K C+ P +KE +G L WP+R
Sbjct: 355 ISN-SCYLKRDPGVLPPLCPSGDDPDATWNVSMKACISPYSVRMHKERWSG--LVPWPRR 411
Query: 427 LESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKR-YRNLVDMNANLGG 485
L + PPR+ GVT E + +D E W+ R+ Y K+ + K RN++DM++NLGG
Sbjct: 412 LTAPPPRLEE---IGVTPEQFREDTETWRLRVIEYWKLLKPMVQKNSIRNVMDMSSNLGG 468
Query: 486 FASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLF 545
FA+AL VWVMNV+PVQ+ + IY+RGLIG HDWCEA TYPRT+DLIHA + F
Sbjct: 469 FAAALNDKDVWVMNVMPVQSS-PRMKIIYDRGLIGATHDWCEAFDTYPRTFDLIHAWNTF 527
Query: 546 S 546
+
Sbjct: 528 T 528
Score = 30.0 bits (66), Expect = 3.5
Identities = 27/110 (24%), Positives = 49/110 (44%), Gaps = 9/110 (8%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
++ SIR +D + + A L +++ +++ P + +++ +RG LIG
Sbjct: 451 VQKNSIRNVMDMSSNLGGFAAALNDKDVWVMNVMPVQS-SPRMKIIYDRG---LIGATHD 506
Query: 270 KRLPFPS--RAFDISHCSRCLIPWAEYDGIF---LNEVDRVLRPGGYWIL 314
F + R FD+ H F L E+DR+LRP G+ I+
Sbjct: 507 WCEAFDTYPRTFDLIHAWNTFTETQARGCSFEDLLIEMDRILRPEGFVII 556
>At4g14360
Length = 608
Score = 390 bits (1001), Expect = e-108
Identities = 204/469 (43%), Positives = 281/469 (59%), Gaps = 32/469 (6%)
Query: 91 NFPRCGVNFTEYTPCEDPTRSLRYKR------SRMIYRERHCPVKGEEDLKCRVPPPHGY 144
+FP C +E PC D R+L Y+ S M + ERHCP E C +PPP+GY
Sbjct: 75 SFPVCDDRHSELIPCLD--RNLIYQMRLKLDLSLMEHYERHCPPP-ERRFNCLIPPPNGY 131
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
K P WP SRD W N+PH L EK+ QNW+ GD+ FPGGGT F GA YI +
Sbjct: 132 KVPIKWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGDKINFPGGGTHFHYGADKYIASM 191
Query: 205 GKLINLKD------GSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
++N + G +RT D GCGVAS+G YL S +I+T+SLAP D H+ Q+QFALER
Sbjct: 192 ANMLNYPNNVLNNGGRLRTVFDVGCGVASFGGYLLSSDILTMSLAPNDVHQNQIQFALER 251
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPP 318
G+PA +GVL +KRLP+PSR+F++SHCSRC I W + DGI L E+DRVLRPGGY+ S P
Sbjct: 252 GIPASLGVLGTKRLPYPSRSFELSHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSP- 310
Query: 319 INWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKL 378
+ + ++DL + ++ + + +CW +++ IWQKP+ + DC R+
Sbjct: 311 -------EAYAQDEEDL-RIWREMSALVERMCWKIAAKRNQTVIWQKPLTN-DCYLEREP 361
Query: 379 ATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGT 438
T P C +PD W +++ C+ +K T G L WP RL S PPR+
Sbjct: 362 GTQPPLCRSDNDPDAVWGVNMEACITSYSDHDHK--TKGSGLAPWPARLTSPPPRL---A 416
Query: 439 IEGVTSEGYSKDNELWKKRIPHY-KKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWV 497
G ++ + KD ELW++R+ Y ++ ++ + RN++DM A++G FA+AL + VWV
Sbjct: 417 DFGYSTGMFEKDTELWRQRVDTYWDLLSPRIESDTVRNIMDMKASMGSFAAALKEKDVWV 476
Query: 498 MNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
MNVVP +TL IY+RGL+G H WCEA STYPRTYDL+HA + S
Sbjct: 477 MNVVPEDGP-NTLKLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIIS 524
Score = 34.3 bits (77), Expect = 0.19
Identities = 31/155 (20%), Positives = 66/155 (42%), Gaps = 27/155 (17%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
++ ++R +D + S+ A L+ +++ +++ P D ++ +RG+ +
Sbjct: 447 IESDTVRNIMDMKASMGSFAAALKEKDVWVMNVVPEDGPNT-LKLIYDRGLMGAVHSWCE 505
Query: 270 KRLPFPSRAFDISHCSRCLIP-----WAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKH 324
+P R +D+ H + +E D L E+DR+LRP G+ I+
Sbjct: 506 AFSTYP-RTYDLLHAWDIISDIKKKGCSEVD--LLLEMDRILRPSGFIII---------- 552
Query: 325 HRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDD 359
+D + ++K K+L W ++ K D
Sbjct: 553 --------RDKQRVVDFVKKYLKALHWEEVGTKTD 579
>At2g39750 unknown protein
Length = 694
Score = 387 bits (995), Expect = e-108
Identities = 209/469 (44%), Positives = 287/469 (60%), Gaps = 28/469 (5%)
Query: 92 FPRCGVNFTEYTPCEDPTRSLRYKRS--RMIYRERHCPVKGEEDLKCRVPPPHGYKTPFT 149
F C + EY PC D T ++ +S R ERHCP KG+ L C VPPP GY+ P
Sbjct: 177 FGMCPESMREYIPCLDNTDVIKKLKSTERGERFERHCPEKGK-GLNCLVPPPKGYRQPIP 235
Query: 150 WPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLIN 209
WP SRD W++NVPH L +K QNWI D ++F FPGGGT F +GA Y+D + K+++
Sbjct: 236 WPKSRDEVWFSNVPHTRLVEDKGGQNWISRDKNKFKFPGGGTQFIHGADQYLDQMSKMVS 295
Query: 210 -LKDGS-IRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVL 267
+ G IR A+D GCGVAS+GAYL SR+++T+S+AP+D HE Q+QFALERGVPA+
Sbjct: 296 DITFGKHIRVAMDVGCGVASFGAYLLSRDVMTMSVAPKDVHENQIQFALERGVPAMAAAF 355
Query: 268 ASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRG 327
A++RL +PS+AFD+ HCSRC I W DGI L E++R+LR GGY+ + P+ KH
Sbjct: 356 ATRRLLYPSQAFDLIHCSRCRINWTRDDGILLLEINRMLRAGGYFAWAAQPV--YKH--- 410
Query: 328 WQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDD-IAIWQKPINHLDCRSARKLATDRPFCG 386
+ L ++ T++ + SLCW KL++K+ +AIWQKP N+ DC +R+ T P C
Sbjct: 411 ----EPALEEQWTEMLNLTISLCW-KLVKKEGYVAIWQKPFNN-DCYLSREAGTKPPLCD 464
Query: 387 PQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEG--VTS 444
++PD WYT+LK C+ +P E+ GG + WP RL + P R+ +
Sbjct: 465 ESDDPDNVWYTNLKPCISRIP-----EKGYGGNVPLWPARLHTPPDRLQTIKFDSYIARK 519
Query: 445 EGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASAL--VKNPVWVMNVVP 502
E + +++ W + I Y + K RN++DM A GGFA+AL K WV++VVP
Sbjct: 520 ELFKAESKYWNEIIGGYVRALKWKKMK-LRNVLDMRAGFGGFAAALNDHKLDCWVLSVVP 578
Query: 503 VQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
V +TL IY+RGL+G HDWCE TYPRTYD +HA LFS+ R
Sbjct: 579 VSGP-NTLPVIYDRGLLGVMHDWCEPFDTYPRTYDFLHASGLFSIERKR 626
Score = 30.0 bits (66), Expect = 3.5
Identities = 43/167 (25%), Positives = 66/167 (38%), Gaps = 44/167 (26%)
Query: 198 GAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNII--TLSLAPRDTHEAQVQFA 255
G Y+ + + K +R LD G + A L + LS+ P +
Sbjct: 534 GGYV----RALKWKKMKLRNVLDMRAGFGGFAAALNDHKLDCWVLSVVPVSGPNT-LPVI 588
Query: 256 LERGVPALIGVLASKRLPFPS--RAFDISHCS--------RCLIPWAEYDGIFLNEVDRV 305
+RG L+GV+ PF + R +D H S RC E I L E+DR+
Sbjct: 589 YDRG---LLGVMHDWCEPFDTYPRTYDFLHASGLFSIERKRC-----EMSTILL-EMDRI 639
Query: 306 LRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWN 352
LRPGG R + R D+ E I+++ K++ W+
Sbjct: 640 LRPGG---------------RAYIRDSIDVMDE---IQEITKAMGWH 668
>At5g06050 ankyrin-like protein
Length = 682
Score = 384 bits (987), Expect = e-107
Identities = 214/499 (42%), Positives = 294/499 (58%), Gaps = 38/499 (7%)
Query: 69 TLDFQSHHNSSD------TIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRS--RMI 120
TL Q+ SSD T +S F C N TEY PC D +++ S R
Sbjct: 122 TLGNQTEFESSDDDDIKSTTARVSVRKFEI--CSENMTEYIPCLDNVEAIKRLNSTARGE 179
Query: 121 YRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYD 180
ER+CP G L C VP P GY++P WP SRD W+ NVPH +L +K QNWI +
Sbjct: 180 RFERNCPNDGM-GLNCTVPIPQGYRSPIPWPRSRDEVWFNNVPHTKLVEDKGGQNWIYKE 238
Query: 181 GDRFFFPGGGTMFPNGAGAYIDDIGKLI-NLKDGS-IRTALDTGCGVASWGAYLQSRNII 238
D+F FPGGGT F +GA Y+D I ++I ++ G+ R LD GCGVAS+GAYL SRN++
Sbjct: 239 NDKFKFPGGGTQFIHGADQYLDQISQMIPDISFGNHTRVVLDIGCGVASFGAYLMSRNVL 298
Query: 239 TLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIF 298
T+S+AP+D HE Q+QFALERGVPA++ ++RL +PS+AFD+ HCSRC I W DGI
Sbjct: 299 TMSIAPKDVHENQIQFALERGVPAMVAAFTTRRLLYPSQAFDLVHCSRCRINWTRDDGIL 358
Query: 299 LNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKD 358
L EV+R+LR GGY++ + P+ KH +K L ++ ++ + LCW + ++
Sbjct: 359 LLEVNRMLRAGGYFVWAAQPV--YKH-------EKALEEQWEEMLNLTTRLCWVLVKKEG 409
Query: 359 DIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGG 418
IAIWQKP+N+ C +R P C +++PD WY DLK C+ + +E G
Sbjct: 410 YIAIWQKPVNN-TCYLSRGAGVSPPLCNSEDDPDNVWYVDLKACITRI-----EENGYGA 463
Query: 419 VLKNWPQRLESVPPRIHMGTIEG--VTSEGYSKDNELWKKRIPHYKKVNN--QLGTKRYR 474
L WP RL + P R+ I+ E + +++ WK+ I +Y + Q+G R
Sbjct: 464 NLAPWPARLLTPPDRLQTIQIDSYIARKELFVAESKYWKEIISNYVNALHWKQIG---LR 520
Query: 475 NLVDMNANLGGFASAL--VKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTY 532
N++DM A GGFA+AL +K WV+NV+PV +TL IY+RGL+G HDWCE TY
Sbjct: 521 NVLDMRAGFGGFAAALAELKVDCWVLNVIPVSGP-NTLPVIYDRGLLGVMHDWCEPFDTY 579
Query: 533 PRTYDLIHADSLFSLYNGR 551
PRTYDL+HA LFS+ R
Sbjct: 580 PRTYDLLHAAGLFSIERKR 598
>At3g23300 ankyrin repeat protein
Length = 611
Score = 381 bits (978), Expect = e-106
Identities = 200/469 (42%), Positives = 279/469 (58%), Gaps = 32/469 (6%)
Query: 91 NFPRCGVNFTEYTPCEDPTRSLRYKR------SRMIYRERHCPVKGEEDLKCRVPPPHGY 144
+FP C +E PC D R+L Y+ S M + ERHCP E C +PPP GY
Sbjct: 78 SFPVCDDRHSELIPCLD--RNLIYQMRLKLDLSLMEHYERHCPPP-ERRFNCLIPPPPGY 134
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
K P WP SRD W N+PH L EK+ QNW+ G++ FPGGGT F GA YI +
Sbjct: 135 KIPIKWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGEKINFPGGGTHFHYGADKYIASM 194
Query: 205 GKLINLKD------GSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
++N + G +RT LD GCGVAS+G YL + I+T+SLAP D H+ Q+QFALER
Sbjct: 195 ANMLNFPNNVLNNGGRLRTFLDVGCGVASFGGYLLASEIMTMSLAPNDVHQNQIQFALER 254
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPP 318
G+PA +GVL +KRLP+PSR+F+++HCSRC I W + DGI L E+DRVLRPGGY+ S P
Sbjct: 255 GIPAYLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSP- 313
Query: 319 INWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKL 378
+ + ++DL + ++ + +CW +++ IWQKP+ + DC R+
Sbjct: 314 -------EAYAQDEEDL-RIWREMSALVGRMCWTIAAKRNQTVIWQKPLTN-DCYLGREP 364
Query: 379 ATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGT 438
T P C +PD + +++ C+ +K T G L WP RL S PPR+
Sbjct: 365 GTQPPLCNSDSDPDAVYGVNMEACITQYSDHDHK--TKGSGLAPWPARLTSPPPRL---A 419
Query: 439 IEGVTSEGYSKDNELWKKRIPHY-KKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWV 497
G +++ + KD E W++R+ Y ++ ++ + RN++DM A++G FA+AL + VWV
Sbjct: 420 DFGYSTDIFEKDTETWRQRVDTYWDLLSPKIQSDTVRNIMDMKASMGSFAAALKEKDVWV 479
Query: 498 MNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
MNVVP +TL IY+RGL+G H WCEA STYPRTYDL+HA + S
Sbjct: 480 MNVVPEDGP-NTLKLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIIS 527
Score = 32.0 bits (71), Expect = 0.93
Identities = 23/114 (20%), Positives = 50/114 (43%), Gaps = 17/114 (14%)
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
++ ++R +D + S+ A L+ +++ +++ P D ++ +RG+ +
Sbjct: 450 IQSDTVRNIMDMKASMGSFAAALKEKDVWVMNVVPEDGPNT-LKLIYDRGLMGAVHSWCE 508
Query: 270 KRLPFPSRAFDISHCSRCLIPWAEYDGI---------FLNEVDRVLRPGGYWIL 314
+P R +D+ H W I L E+DR+LRP G+ ++
Sbjct: 509 AFSTYP-RTYDLLHA------WDIISDIKKRGCSAEDLLLEMDRILRPSGFILI 555
>At1g77260 unknown protein
Length = 655
Score = 374 bits (959), Expect = e-103
Identities = 199/465 (42%), Positives = 276/465 (58%), Gaps = 28/465 (6%)
Query: 95 CGVNFTEYTPCEDPTRSLRYKRS--RMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPA 152
C +Y PC D ++ + R ERHCP ++ L C +PPP GYK P WP
Sbjct: 146 CDKTKIDYIPCLDNEEEIKRLNNTDRGENYERHCP---KQSLDCLIPPPDGYKKPIQWPQ 202
Query: 153 SRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLI-NLK 211
SRD W+ NVPH L +K QNWIR + D+F FPGGGT F +GA Y+D I ++I ++
Sbjct: 203 SRDKIWFNNVPHTRLVEDKGGQNWIRREKDKFVFPGGGTQFIHGADQYLDQISQMIPDIT 262
Query: 212 DGS-IRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASK 270
GS R ALD GCGVAS+GA+L RN TLS+AP+D HE Q+QFALERGVPA++ V A++
Sbjct: 263 FGSRTRVALDIGCGVASFGAFLMQRNTTTLSVAPKDVHENQIQFALERGVPAMVAVFATR 322
Query: 271 RLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQR 330
RL +PS++F++ HCSRC I W DGI L EV+R+LR GGY++ + P+ KH
Sbjct: 323 RLLYPSQSFEMIHCSRCRINWTRDDGILLLEVNRMLRAGGYFVWAAQPV--YKH------ 374
Query: 331 TKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQEN 390
+ +L ++ ++ + +CW + ++ IA+W+KP+N+ C +R+ T P C P ++
Sbjct: 375 -EDNLQEQWKEMLDLTNRICWELIKKEGYIAVWRKPLNN-SCYVSREAGTKPPLCRPDDD 432
Query: 391 PDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTS--EGYS 448
PD WY D+K C+ +P + G + WP RL P R+ ++ S E
Sbjct: 433 PDDVWYVDMKPCITRLP-----DNGYGANVSTWPARLHDPPERLQSIQMDAYISRKEIMK 487
Query: 449 KDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASAL--VKNPVWVMNVVPVQAK 506
++ W + + Y +V K RN++DM A GGFA+AL + WVMN+VPV
Sbjct: 488 AESRFWLEVVESYVRVFRWKEFK-LRNVLDMRAGFGGFAAALNDLGLDCWVMNIVPVSG- 545
Query: 507 VDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
+TL IY+RGL G HDWCE TYPRTYDLIHA LFS+ R
Sbjct: 546 FNTLPVIYDRGLQGAMHDWCEPFDTYPRTYDLIHAAFLFSVEKKR 590
>At3g51070 putative protein
Length = 895
Score = 364 bits (935), Expect = e-101
Identities = 208/462 (45%), Positives = 277/462 (59%), Gaps = 34/462 (7%)
Query: 100 TEYTPCEDPTRSLRYKRSRMIY--RERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVA 157
T+Y PC D ++ RSR + RERHCP E+ C VP P GYK WP SRD
Sbjct: 380 TDYIPCLDNEEAIMKLRSRRHFEHRERHCP---EDPPTCLVPLPEGYKEAIKWPESRDKI 436
Query: 158 WYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGK-LINLKDGS-I 215
WY NVPH +L K QNW++ G+ FPGGGT F +GA YID + + L N+ G
Sbjct: 437 WYHNVPHTKLAEVKGHQNWVKVTGEFLTFPGGGTQFIHGALHYIDFLQQSLKNIAWGKRT 496
Query: 216 RTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFP 275
R LD GCGVAS+G +L R++I +SLAP+D HEAQVQFALER +PA+ V+ SKRLPFP
Sbjct: 497 RVILDVGCGVASFGGFLFERDVIAMSLAPKDEHEAQVQFALERKIPAISAVMGSKRLPFP 556
Query: 276 SRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDL 335
SR FD+ HC+RC +PW G+ L E++R+LRPGGY++ S P+ +Q+ ++D+
Sbjct: 557 SRVFDLIHCARCRVPWHNEGGMLLLELNRMLRPGGYFVWSATPV--------YQKLEEDV 608
Query: 336 NQEQTKIEKVAKSLCWNKL-IEKDDI-----AIWQKPINHLDCRSARKLATDRPFCGPQE 389
Q ++ + KSLCW + I KD + AI+QKP + +C RK P C +
Sbjct: 609 -QIWKEMSALTKSLCWELVTINKDKLNGIGAAIYQKPATN-ECYEKRK-HNKPPLCKNND 665
Query: 390 NPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIH---MGTIEGVTSEG 446
+ + AWY L+ C+ VP +N E NWP+RL++ P ++ MG
Sbjct: 666 DANAAWYVPLQACMHKVP--TNVVERGSKWPVNWPRRLQTPPYWLNSSQMGIYGKPAPRD 723
Query: 447 YSKDNELWKKRIPHYKKVNNQLGT--KRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQ 504
++ D E WK + K N++G RN++DM A GGFA+AL VWVMNVV +
Sbjct: 724 FTTDYEHWKHVVS--KVYMNEIGISWSNVRNVMDMRAVYGGFAAALKDLQVWVMNVVNIN 781
Query: 505 AKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
+ DTL IYERGL G YHDWCE+ STYPR+YDL+HAD LFS
Sbjct: 782 SP-DTLPIIYERGLFGIYHDWCESFSTYPRSYDLLHADHLFS 822
>At1g29470 unknown protein
Length = 768
Score = 358 bits (918), Expect = 6e-99
Identities = 201/463 (43%), Positives = 283/463 (60%), Gaps = 39/463 (8%)
Query: 101 EYTPCEDPTRSLRYKRSRMIY--RERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAW 158
+Y PC D +++R S Y RERHCP EE +C V P GYK WP SR+ W
Sbjct: 248 DYIPCLDNWQAIRKLHSTKHYEHRERHCP---EESPRCLVSLPEGYKRSIKWPKSREKIW 304
Query: 159 YANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLI-NLKDGS-IR 216
Y N+PH +L K QNW++ G+ FPGGGT F NGA YID + + ++ G+ R
Sbjct: 305 YTNIPHTKLAEVKGHQNWVKMSGEYLTFPGGGTQFKNGALHYIDFLQESYPDIAWGNRTR 364
Query: 217 TALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPS 276
LD GCGVAS+G YL R+++ LS AP+D HEAQVQFALERG+PA+ V+ +KRLPFP
Sbjct: 365 VILDVGCGVASFGGYLFDRDVLALSFAPKDEHEAQVQFALERGIPAMSNVMGTKRLPFPG 424
Query: 277 RAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLN 336
FD+ HC+RC +PW G L E++R LRPGG+++ S P+ +++T++D+
Sbjct: 425 SVFDLIHCARCRVPWHIEGGKLLLELNRALRPGGFFVWSATPV--------YRKTEEDVG 476
Query: 337 QEQTKIEKVAKSLCWNKL-IEKDDI-----AIWQKPINHLDCRSARKLATDRPFCGPQEN 390
+ + K+ K++CW + I+KD++ AI+QKP+++ C + R + P C ++
Sbjct: 477 IWKA-MSKLTKAMCWELMTIKKDELNEVGAAIYQKPMSN-KCYNERS-QNEPPLCKDSDD 533
Query: 391 PDKAWYTDLKTCLMPVPQVSNKEETAGGVL-KNWPQRLESVPPRIHMGTIEGV----TSE 445
+ AW L+ C+ V + S+K G V ++WP+R+E+VP + + EGV E
Sbjct: 534 QNAAWNVPLEACIHKVTEDSSKR---GAVWPESWPERVETVPQ--WLDSQEGVYGKPAQE 588
Query: 446 GYSKDNELWKKRIPHYKKVNNQLGT--KRYRNLVDMNANLGGFASALVKNPVWVMNVVPV 503
++ D+E WK + K N +G RN++DM A GGFA+AL +WVMNVVP+
Sbjct: 589 DFTADHERWKTIVS--KSYLNGMGIDWSYVRNVMDMRAVYGGFAAALKDLKLWVMNVVPI 646
Query: 504 QAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
+ DTL IYERGL G YHDWCE+ STYPRTYDL+HAD LFS
Sbjct: 647 DSP-DTLPIIYERGLFGIYHDWCESFSTYPRTYDLLHADHLFS 688
Score = 31.6 bits (70), Expect = 1.2
Identities = 30/140 (21%), Positives = 57/140 (40%), Gaps = 22/140 (15%)
Query: 215 IRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPF 274
+R +D + A L+ + +++ P D+ + + ERG+ + +
Sbjct: 616 VRNVMDMRAVYGGFAAALKDLKLWVMNVVPIDSPDT-LPIIYERGLFGIYHDWCESFSTY 674
Query: 275 PSRAFDISHCSRCLIPWAEYDGIF--LNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTK 332
P R +D+ H + + + EVDR+LRP G +I+
Sbjct: 675 P-RTYDLLHADHLFSSLKKRCNLVGVMAEVDRILRPQGTFIV------------------ 715
Query: 333 KDLNQEQTKIEKVAKSLCWN 352
+D + +IEK+ KS+ WN
Sbjct: 716 RDDMETIGEIEKMVKSMKWN 735
>At2g34300 unknown protein
Length = 770
Score = 355 bits (910), Expect = 5e-98
Identities = 206/500 (41%), Positives = 288/500 (57%), Gaps = 40/500 (8%)
Query: 70 LDFQSHHNSSDTIIALSSETFNFPRCGVNF-TEYTPCEDPTRSLRYKRSRMIY--RERHC 126
++ Q+ + + I+ ++ + C V +Y PC D ++++ + M Y RERHC
Sbjct: 218 VESQNEKKAQQSSISKDQSSYGWKTCNVTAGPDYIPCLDNWQAIKKLHTTMHYEHRERHC 277
Query: 127 PVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFF 186
P EE C V P GYK WP SR+ WY NVPH +L K QNW++ G+ F
Sbjct: 278 P---EESPHCLVSLPDGYKRSIKWPKSREKIWYNNVPHTKLAEIKGHQNWVKMSGEHLTF 334
Query: 187 PGGGTMFPNGAGAYIDDIGKLINLKDGSIRTA--LDTGCGVASWGAYLQSRNIITLSLAP 244
PGGGT F NGA YID I + RT LD GCGVAS+G YL R+++ LS AP
Sbjct: 335 PGGGTQFKNGALHYIDFIQQSHPAIAWGNRTRVILDVGCGVASFGGYLFERDVLALSFAP 394
Query: 245 RDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDR 304
+D HEAQVQFALERG+PA++ V+ +KRLPFP FD+ HC+RC +PW G L E++R
Sbjct: 395 KDEHEAQVQFALERGIPAMLNVMGTKRLPFPGSVFDLIHCARCRVPWHIEGGKLLLELNR 454
Query: 305 VLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKL-IEKDDI--- 360
LRPGG+++ S P+ +++ ++D + + ++ K++CW + I+KD +
Sbjct: 455 ALRPGGFFVWSATPV--------YRKNEEDSGIWKA-MSELTKAMCWKLVTIKKDKLNEV 505
Query: 361 --AIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGG 418
AI+QKP ++ C + R + P C ++ + AW L+ C+ V + S+K G
Sbjct: 506 GAAIYQKPTSN-KCYNKRP-QNEPPLCKDSDDQNAAWNVPLEACMHKVTEDSSKR---GA 560
Query: 419 VLKN-WPQRLESVPPRIHMGTIEGV----TSEGYSKDNELWKKRIPHYKKVNNQLGT--K 471
V N WP+R+E+ P + + EGV E ++ D E WK + K N +G
Sbjct: 561 VWPNMWPERVETAPE--WLDSQEGVYGKPAPEDFTADQEKWKTIVS--KAYLNDMGIDWS 616
Query: 472 RYRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMST 531
RN++DM A GGFA+AL +WVMNVVPV A DTL IYERGL G YHDWCE+ +T
Sbjct: 617 NVRNVMDMRAVYGGFAAALKDLKLWVMNVVPVDAP-DTLPIIYERGLFGIYHDWCESFNT 675
Query: 532 YPRTYDLIHADSLFSLYNGR 551
YPRTYDL+HAD LFS R
Sbjct: 676 YPRTYDLLHADHLFSTLRKR 695
Score = 32.7 bits (73), Expect = 0.54
Identities = 33/164 (20%), Positives = 68/164 (41%), Gaps = 27/164 (16%)
Query: 199 AYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
AY++D+G + ++R +D + A L+ + +++ P D + + ER
Sbjct: 606 AYLNDMG----IDWSNVRNVMDMRAVYGGFAAALKDLKLWVMNVVPVDAPDT-LPIIYER 660
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIF--LNEVDRVLRPGGYWILSG 316
G+ + +P R +D+ H + + + E+DR+LRP G +I+
Sbjct: 661 GLFGIYHDWCESFNTYP-RTYDLLHADHLFSTLRKRCNLVSVMAEIDRILRPQGTFII-- 717
Query: 317 PPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWN-KLIEKDD 359
+D + ++EK+ KS+ W K+ + D
Sbjct: 718 ----------------RDDMETLGEVEKMVKSMKWKVKMTQSKD 745
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,029,878
Number of Sequences: 26719
Number of extensions: 644973
Number of successful extensions: 1552
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1314
Number of HSP's gapped (non-prelim): 72
length of query: 556
length of database: 11,318,596
effective HSP length: 104
effective length of query: 452
effective length of database: 8,539,820
effective search space: 3859998640
effective search space used: 3859998640
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149494.6


