

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.2 - phase: 0
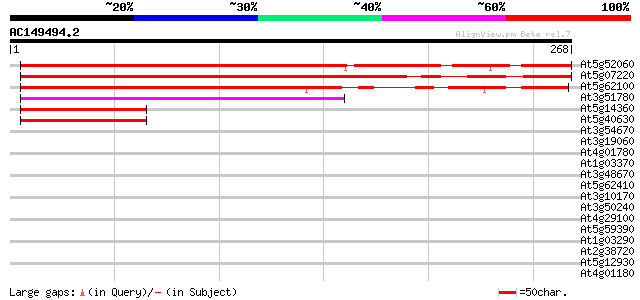
(268 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g52060 putative protein 263 7e-71
At5g07220 unknown protein 250 6e-67
At5g62100 unknown protein 228 3e-60
At3g51780 unknown protein 108 3e-24
At5g14360 putative protein 55 3e-08
At5g40630 unknown protein 49 2e-06
At3g54670 structural maintenance of chromosomes (SMC) - like pro... 37 0.011
At3g19060 hypothetical protein 37 0.014
At4g01780 hypothetical protein 35 0.055
At1g03370 unknown protein 34 0.071
At3g48670 unknown protein 33 0.12
At5g62410 chromosomal protein - like 33 0.16
At3g10170 hypothetical protein 33 0.16
At3g50240 kinesin-related protein (kicp-02) 33 0.21
At4g29100 putative bHLH transcription factor (bHLH068) 32 0.27
At5g59390 transcriptional regulator - like protein 32 0.35
At1g03290 unknown protein 32 0.35
At2g38720 hypothetical protein 32 0.46
At5g12930 unknown protein 30 1.0
At4g01180 hypothetical protein 30 1.0
>At5g52060 putative protein
Length = 326
Score = 263 bits (672), Expect = 7e-71
Identities = 141/267 (52%), Positives = 187/267 (69%), Gaps = 17/267 (6%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKMLTGPTG+HHQDQK+ YK+KERDSKAFLD+ GVKDKSK+V++EDP++QEKR+LEM
Sbjct: 73 GELKKMLTGPTGIHHQDQKLMYKDKERDSKAFLDVSGVKDKSKMVLIEDPLSQEKRFLEM 132
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RK K E+A+K+IS+ISLEVDRL G+VSA E + KGGK+ E D++++IE LMN+L+KLD
Sbjct: 133 RKIAKTEKASKAISDISLEVDRLGGRVSAFEMVTKKGGKIAEKDLVTVIELLMNELIKLD 192
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS--NGGHVPKKKPQQKVKLPPIDEQLE 183
IVA+GDVKLQRKMQVKRVQ YVETLD LK+KNS NG +K+ +L PI E
Sbjct: 193 AIVAEGDVKLQRKMQVKRVQNYVETLDALKVKNSMANG---QQKQSSTAQRLAPIQEHNN 249
Query: 184 GMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVV--TTKWETFDSLPPL 241
+Q ++ Q Q+++ +P + V+ + KWETFD P
Sbjct: 250 EERQEQKPIQSLMDMPIQYKEKK-----QEIEEEPRNSGEGPFVLDSSAKWETFDHHP-- 302
Query: 242 IPVTSASSSSSSTNNSVHPKFKWEHFE 268
+ SS+++ NN++ P+F WE F+
Sbjct: 303 ---VTPLSSTTAKNNAIPPRFNWEFFD 326
>At5g07220 unknown protein
Length = 303
Score = 250 bits (638), Expect = 6e-67
Identities = 139/263 (52%), Positives = 178/263 (66%), Gaps = 26/263 (9%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKML+ GLHH+D K+ YK+KERDSK FLD+ GVKD+SKLVV EDPI+QEKR L
Sbjct: 67 GELKKMLSDQVGLHHEDMKVLYKDKERDSKMFLDLCGVKDRSKLVVKEDPISQEKRLLAK 126
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RKN +E+A+KSIS+IS EVDRLAGQVSA ET+I+KGGKV E +++LIE LMNQLL+LD
Sbjct: 127 RKNAAIEKASKSISDISFEVDRLAGQVSAFETVINKGGKVEEKSLVNLIEMLMNQLLRLD 186
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGM 185
I+ADGDVKL RKMQV+RVQKYVE LD+LK+KNS K + K + EQ + +
Sbjct: 187 AIIADGDVKLMRKMQVQRVQKYVEALDLLKVKNSAKKVEVNKSVRHKPQTQTRFEQRDLL 246
Query: 186 SIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVT 245
S E++ RNSN S++S + VV +KWE FDS
Sbjct: 247 SFVEE------EEEEPRNSNA------------SSSSGTPAVVASKWEMFDS-------- 280
Query: 246 SASSSSSSTNNSVHPKFKWEHFE 268
++++ ++ T V P+FKWE F+
Sbjct: 281 ASTAKAAETVKPVPPRFKWEFFD 303
>At5g62100 unknown protein
Length = 296
Score = 228 bits (580), Expect = 3e-60
Identities = 129/274 (47%), Positives = 179/274 (65%), Gaps = 51/274 (18%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKK+L+G TG+HHQD +I YK+KERDSK FLD+ GVKD+SKL+++EDPI+QEKR LE+
Sbjct: 61 GELKKILSGATGVHHQDMQIIYKDKERDSKMFLDLSGVKDRSKLILIEDPISQEKRLLEL 120
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RK E+++K+IS+IS +V+RLAGQ+SA +T+I KGGKV E ++ +L+E LMNQL+KLD
Sbjct: 121 RKIATKEKSSKAISDISFQVERLAGQLSAFDTVIGKGGKVEEKNLENLMEMLMNQLVKLD 180
Query: 126 GIVADGDVKLQRKMQ-----------VKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVK 174
I DGDVKL++KMQ +R+ KYVE LD+LKIKNS ++PQ K K
Sbjct: 181 AISGDGDVKLKKKMQNLMIRFTNCWKEERLHKYVEALDLLKIKNS-------RQPQTKPK 233
Query: 175 LPPIDEQLEGMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSE-VVVTTKWE 233
P +++ +++ KP+ +S+S V++TT+WE
Sbjct: 234 -------------------PQYKEREMLT------FYEEASRKPTASSSSPPVIITTRWE 268
Query: 234 TFDSLPPLIPVTSASSSSSSTNNSVHPKFKWEHF 267
TFDS +SAS+++ VHPKFKWE F
Sbjct: 269 TFDS-------SSASTATLQPVRPVHPKFKWELF 295
>At3g51780 unknown protein
Length = 269
Score = 108 bits (270), Expect = 3e-24
Identities = 62/155 (40%), Positives = 91/155 (58%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G++KK L TGL + KI ++ ERD L GVKD SKLVV+ + +
Sbjct: 70 GDVKKALVQKTGLEASELKILFRGVERDDAEQLQAAGVKDASKLVVVVEDTNKRVEQQPP 129
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
+ME+A +++ ++ EVD+L+ +V ALE ++ G +V + E LM QLLKLD
Sbjct: 130 VVTKEMEKAIAAVNAVTGEVDKLSDRVVALEVAVNGGTQVAVREFDMAAELLMRQLLKLD 189
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSN 160
GI A+GD K+QRK +V+R+Q E +D LK + SN
Sbjct: 190 GIEAEGDAKVQRKAEVRRIQNLQEAVDKLKARCSN 224
>At5g14360 putative protein
Length = 163
Score = 55.5 bits (132), Expect = 3e-08
Identities = 25/60 (41%), Positives = 43/60 (71%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELK +L+ TGL + Q++ +K KER+ +L +VGV DK K++++EDP ++K+ L++
Sbjct: 92 GELKMVLSLLTGLEPKQQRLVFKGKEREDHEYLHMVGVGDKDKVLLLEDPGFKDKKLLDL 151
>At5g40630 unknown protein
Length = 165
Score = 49.3 bits (116), Expect = 2e-06
Identities = 18/60 (30%), Positives = 41/60 (68%)
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELK M+ +G+ ++Q++ ++ KER+ + +L ++GV D K+ +++DP +E +++ +
Sbjct: 102 GELKMMIAIVSGIEAKEQRLLFRGKEREDREYLHMIGVGDGDKVFLLQDPAFKELKHIHL 161
>At3g54670 structural maintenance of chromosomes (SMC) - like
protein
Length = 1265
Score = 37.0 bits (84), Expect = 0.011
Identities = 46/160 (28%), Positives = 78/160 (48%), Gaps = 17/160 (10%)
Query: 29 NKERD-SKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEVDR 87
N E D KA D+ K K V+ E EK E K K+E+A K + EI+ +
Sbjct: 240 NIENDIEKANEDVDSEKSNRKDVMRE----LEKFEREAGKR-KVEQA-KYLKEIAQREKK 293
Query: 88 LAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQRK----MQVKR 143
+A + S L I+S K V+ ++L E++ K++ D D + + K ++++
Sbjct: 294 IAEKSSKLGKIVSIPWKSVQPELLRFKEEIARIKAKIETNRKDVDKRKKEKGKHSKEIEQ 353
Query: 144 VQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLE 183
+QK ++ L+ K++ N KK+ KLP +D QL+
Sbjct: 354 MQKSIKELNK-KMELFN-----KKRQDSSGKLPMLDSQLQ 387
>At3g19060 hypothetical protein
Length = 1647
Score = 36.6 bits (83), Expect = 0.014
Identities = 34/143 (23%), Positives = 64/143 (43%), Gaps = 29/143 (20%)
Query: 70 KMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVA 129
+ME A S+ E+ +E+ L G+ L K E LS + L Q+ L+ +++
Sbjct: 591 EMESALASLKEVQVEMANLKGEKEEL--------KASEKRSLSNLNDLAAQICNLNTVMS 642
Query: 130 DGDVKLQRKMQ----------VKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPID 179
+ + + + KM+ ++ Q+YVE L +L KK + + + D
Sbjct: 643 NMEEQYEHKMETLEHEIAKMKIEADQEYVENLCIL-----------KKFEEAQGTIREAD 691
Query: 180 EQLEGMSIGNHKLQPSLEQQSQR 202
+ + I N K++ LE+Q +R
Sbjct: 692 ITVNELVIANEKMRFDLEKQKKR 714
>At4g01780 hypothetical protein
Length = 456
Score = 34.7 bits (78), Expect = 0.055
Identities = 51/216 (23%), Positives = 94/216 (42%), Gaps = 36/216 (16%)
Query: 48 KLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGK--V 105
++ + ED ++K L K I++ER + I LEV++L GQ++ ++ + S G V
Sbjct: 215 RMKLSEDLEQRQKEELH-EKIIRLERQIDQVQAIELEVEQLKGQLNVMKHMASDGDAQVV 273
Query: 106 VETDVL--SLIEK--------LMNQLLKLDGIVADGDVKLQRK----------------- 138
E D++ L+EK NQ L L + +++ RK
Sbjct: 274 KEVDIIFKDLVEKEAELADLNKFNQTLILRERRTNDELQEARKELVNCMGELVRKPFVDA 333
Query: 139 MQVKRVQKYVE--TLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQPSL 196
MQ K Q+ VE +++L++ + + P P ++VKL D ++E + + KL+
Sbjct: 334 MQQKYCQEDVEDRAVEVLQLW-EHYINDPDWHPYKRVKLENQDREVEVIDDRDEKLR--- 389
Query: 197 EQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKW 232
E ++ + V + L N + + T W
Sbjct: 390 ELKADLGDGPYNAVTKALLEINEYNPSGRYITTELW 425
>At1g03370 unknown protein
Length = 1859
Score = 34.3 bits (77), Expect = 0.071
Identities = 32/150 (21%), Positives = 70/150 (46%), Gaps = 14/150 (9%)
Query: 26 FYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEV 85
F + KER ++ DI + + K+++ E+ Q ++ ++K + + ++ E +L+
Sbjct: 614 FREAKERGERSLKDIHSWEGQ-KIMLQEELKGQREKVTVLQKEVTKAKNRQNQIEAALKQ 672
Query: 86 DRLA-GQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVKRV 144
+R A G++SA ++I K ET L + K+ + +K G + K + +
Sbjct: 673 ERTAKGKLSAQASLIRK-----ETKELEALGKVEEERIK-------GKAETDVKYYIDNI 720
Query: 145 QKYVETLDMLKIKNSNGGHVPKKKPQQKVK 174
++ + LK+K+ + KK + K
Sbjct: 721 KRLEREISELKLKSDYSRIIALKKGSSESK 750
>At3g48670 unknown protein
Length = 647
Score = 33.5 bits (75), Expect = 0.12
Identities = 29/124 (23%), Positives = 62/124 (49%), Gaps = 12/124 (9%)
Query: 45 DKSKLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGK 104
D+ + ED Q++ E K I++ER I LEV++L GQ++ ++ + S G
Sbjct: 392 DEEVKKLAEDQRRQKEELHE--KIIRLERQRDQKQAIELEVEQLKGQLNVMKHMASDG-- 447
Query: 105 VVETDVLSLIEKLMNQLLKLDGIVADGD------VKLQRKMQVKRVQKYVETLDMLKIKN 158
+ +V+ ++ + L + + +AD D + +R+ + + + E ++++K N
Sbjct: 448 --DAEVVKEVDIIFKDLGEKEAQLADLDKFNQTLILRERRTNDELQEAHKELVNIMKEWN 505
Query: 159 SNGG 162
+N G
Sbjct: 506 TNIG 509
>At5g62410 chromosomal protein - like
Length = 1175
Score = 33.1 bits (74), Expect = 0.16
Identities = 30/106 (28%), Positives = 49/106 (45%), Gaps = 10/106 (9%)
Query: 45 DKSKLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGK 104
+K KLV+ E+ + QE+ LE + IS ++ EVD +V AL+ I +
Sbjct: 816 EKEKLVMEEEAMKQEQSSLESH----LTSLETQISTLTSEVDEQRAKVDALQKIHDE--- 868
Query: 105 VVETDVLSLIEKLMNQL-LKLDGIVADGDVKLQRKMQVKRVQKYVE 149
L LI M + ++ G V D + LQ+ +K +K +E
Sbjct: 869 --SLAELKLIHAKMKECDTQISGFVTDQEKCLQKLSDMKLERKKLE 912
>At3g10170 hypothetical protein
Length = 634
Score = 33.1 bits (74), Expect = 0.16
Identities = 37/217 (17%), Positives = 103/217 (47%), Gaps = 20/217 (9%)
Query: 18 LHHQDQKIFYKNKERD-SKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM----------- 65
L +++K + +KE+ ++A + + + ++ + +++EK+ LE
Sbjct: 403 LEMEEEKAIWSSKEKALTEAVEEKIRLYKNIQIESLSKEMSEEKKELESCRLECVTLADR 462
Query: 66 ----RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQL 121
+N K ++ +S E SLE+DRL ++ + + + + +V+++D+ L ++ +
Sbjct: 463 LRCSEENAKQDK--ESSLEKSLEIDRLGDELRSADAVSKQSQEVLKSDIDILKSEVQHAC 520
Query: 122 LKLDGIVADGD-VKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDE 180
D + D V +R+ + R+++ + L + + K+K + K++L +
Sbjct: 521 KMSDTFQREMDYVTSERQGLLARIEELSKELASSN-RWQDAAAENKEKAKLKMRLRGMQA 579
Query: 181 QLEGMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHK 217
+L+ +S+ + E +++ ++++ ++L K
Sbjct: 580 RLDAISLRYKQSVQESELMNRKFKEASAKLKEKLASK 616
>At3g50240 kinesin-related protein (kicp-02)
Length = 1051
Score = 32.7 bits (73), Expect = 0.21
Identities = 54/247 (21%), Positives = 100/247 (39%), Gaps = 43/247 (17%)
Query: 58 QEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVV-------ETDV 110
QE + +++ K E AAK + E+ + Q L+ + + + E ++
Sbjct: 604 QENQVEVLKQKQKSEDAAK---RLKTEIQCIKAQKVQLQQKMKQEAEQFRQWKASQEKEL 660
Query: 111 LSLIE---KLMNQLLKLDGIVADGDVKLQRK-----MQVKRVQKYVETL-----DMLKIK 157
L L + K ++ LKL+ + + LQRK M KR+++ +E D+ I
Sbjct: 661 LQLKKEGRKTEHERLKLEALNRRQKMVLQRKTEEAAMATKRLKELLEARKSSPHDISVIA 720
Query: 158 NSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQPSLEQQSQ--------RNSNGNSQ 209
N G P ++ +K +D +LE M+ H+++ E+Q Q S
Sbjct: 721 N---GQPPSRQTNEKSLRKWLDNELEVMA-KVHQVRFQYEKQIQVRAALAVELTSLRQEM 776
Query: 210 VFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSAS--------SSSSSTNNSVHPK 261
F H+ + T+ E SL ++ V+S + S + +S+H K
Sbjct: 777 EFPSNSHQEKNGQFRFLSPNTRLERIASLESMLDVSSNALTAMGSQLSEAEEREHSLHAK 836
Query: 262 FKWEHFE 268
+W H +
Sbjct: 837 PRWNHIQ 843
>At4g29100 putative bHLH transcription factor (bHLH068)
Length = 407
Score = 32.3 bits (72), Expect = 0.27
Identities = 23/85 (27%), Positives = 43/85 (50%), Gaps = 8/85 (9%)
Query: 67 KNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLS----LIEKLMNQLL 122
K +++ + S S + + ++L G+++AL ++S GK VLS I L +Q+
Sbjct: 248 KKPRLQPSPSSQSTLKVRKEKLGGRIAALHQLVSPFGKTDTASVLSEAIGYIRFLQSQIE 307
Query: 123 KLD----GIVADGDVKLQRKMQVKR 143
L G A G+++ Q+ +Q R
Sbjct: 308 ALSHPYFGTTASGNMRHQQHLQGDR 332
>At5g59390 transcriptional regulator - like protein
Length = 561
Score = 32.0 bits (71), Expect = 0.35
Identities = 25/115 (21%), Positives = 55/115 (47%), Gaps = 4/115 (3%)
Query: 58 QEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKL 117
+EK L R ++ME E+ LE+++L G + ++ ++ G + D++ I K
Sbjct: 310 KEKEKLHKRI-MEMEAKLNETQELELEIEKLKGTTNVMKHMVGSDG---DKDIVEKIAKT 365
Query: 118 MNQLLKLDGIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQK 172
QL + + + + L RK + + +M+++ N+N + ++K + K
Sbjct: 366 QIQLDAQETALHEKMMTLARKERATNDEYQDVLKEMIQVWNANEELMKQEKIRVK 420
>At1g03290 unknown protein
Length = 571
Score = 32.0 bits (71), Expect = 0.35
Identities = 43/197 (21%), Positives = 79/197 (39%), Gaps = 29/197 (14%)
Query: 70 KMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVA 129
K+E K + D AG+V ++I++ K +E +L+L E+ L LD +
Sbjct: 340 KVEELKKMLEHAKEANDMHAGEVYGEKSILATEVKELENRLLNLSEERNKSLAILDEMRG 399
Query: 130 DGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMS--I 187
+++L +++K+ + +KK ++ L + EQ M +
Sbjct: 400 SLEIRLAAALELKKTAE------------------KEKKDKEDSALKALAEQEANMEKVV 441
Query: 188 GNHKLQPSLEQQSQRNSN------GNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPL 241
KL L+Q+++ NS Q+ LQ + S ++ K+E L
Sbjct: 442 QESKL---LQQEAEENSKLRDFLMDRGQIVDTLQGEISVICQDVKLLKEKFENRVPLTKS 498
Query: 242 IPVTSASSSSSSTNNSV 258
I + SS SS + V
Sbjct: 499 ISSSFTSSCGSSMKSLV 515
>At2g38720 hypothetical protein
Length = 587
Score = 31.6 bits (70), Expect = 0.46
Identities = 25/110 (22%), Positives = 54/110 (48%), Gaps = 11/110 (10%)
Query: 46 KSKLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKV 105
K K +++ I+ K LE +K +R K +SE ++ + ++ + +S G +V
Sbjct: 85 KKKEGSLKEQISSVKPVLEDLL-MKKDRRRKELSETLNQIAEITSNIAGNDYTVSSGSEV 143
Query: 106 VETDVLSLIEKLMNQLLKLDGIVAD-GDVKLQRKMQVKRVQKYVETLDML 154
E+D+ KLD + AD D++ ++ +++++V Y+ + L
Sbjct: 144 DESDLTQ---------RKLDELRADLQDLRNEKAVRLQKVNSYISAVHEL 184
>At5g12930 unknown protein
Length = 439
Score = 30.4 bits (67), Expect = 1.0
Identities = 47/209 (22%), Positives = 87/209 (41%), Gaps = 9/209 (4%)
Query: 54 DPIAQEKRYLEMRKNIKMERAAKSISEISLEVDRLAG--QVSALETIISKGGKVVET--- 108
D ++ ++ L +RK + + A L++ AG + + LE IIS+ +++
Sbjct: 229 DSLSLDQDMLSLRKECQEKDATIKDLTSFLQLTNKAGSKRETELEEIISRKKTIIKKLKR 288
Query: 109 DVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKK 168
DVL L +K+ QL +L V + + R+ + +D+L +S+
Sbjct: 289 DVLVLEDKV-TQLTRLRRSSYSPAVSNTHEFPM-RMDNLLYDMDVLTASSSSDSEATVNT 346
Query: 169 PQQKVKLPPIDE-QLEGMSIG-NHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEV 226
PQ+ V P+D + E ++G +K P+ S S V + + +S
Sbjct: 347 PQRAVLEAPVDSVKEEPATLGQTNKSAPAKSSTSLVKSVKPPSVVSPSTTRKPVSVSSSS 406
Query: 227 VVTTKWETFDSLPPLIPVTSASSSSSSTN 255
V T DS P P+ + SS ++
Sbjct: 407 RVRRGSSTGDSKKPRRPIQTIPRDSSGSH 435
>At4g01180 hypothetical protein
Length = 554
Score = 30.4 bits (67), Expect = 1.0
Identities = 24/116 (20%), Positives = 55/116 (46%), Gaps = 4/116 (3%)
Query: 58 QEKRYLEMRKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKL 117
+EK L R ++ME E+ LE+++L G + ++ ++ G + D++ I K
Sbjct: 300 KEKEKLHKRI-MEMEAKLNETQELELEIEKLKGTTNVMKHMVGCDG---DKDIVEKIAKT 355
Query: 118 MNQLLKLDGIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKV 173
+L + + + + L RK + + +M+K+ +N + ++K + K+
Sbjct: 356 QIELDARETALHEKMMTLARKERATNDEYQDARKEMIKVWKANEELMKQEKIRVKI 411
Score = 28.5 bits (62), Expect = 3.9
Identities = 21/83 (25%), Positives = 41/83 (49%), Gaps = 4/83 (4%)
Query: 135 LQRKMQVKRVQKYVETLDMLKIKN--SNGGHVPKKKPQQKVKLPP-IDEQLEGMSIGNHK 191
+++K +K + + VE D K+ + N +K Q+K +L +DE LE + N
Sbjct: 150 VKKKRDLKSISQIVEE-DQRKLYHLFENMCQTIEKNKQRKQQLEQKVDETLESLEFHNLM 208
Query: 192 LQPSLEQQSQRNSNGNSQVFQQL 214
L S +++ Q+ + +QQ+
Sbjct: 209 LNNSYQEEIQKMEKNMQEFYQQV 231
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,772,589
Number of Sequences: 26719
Number of extensions: 235873
Number of successful extensions: 1033
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 999
Number of HSP's gapped (non-prelim): 64
length of query: 268
length of database: 11,318,596
effective HSP length: 98
effective length of query: 170
effective length of database: 8,700,134
effective search space: 1479022780
effective search space used: 1479022780
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149494.2


