

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.9 - phase: 0
(1049 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
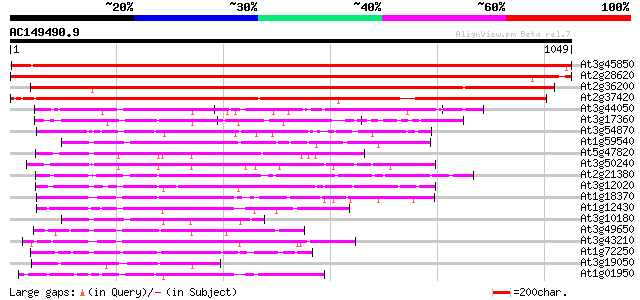
Score E
Sequences producing significant alignments: (bits) Value
At3g45850 kinesin-related protein - like 1558 0.0
At2g28620 putative kinesin-like spindle protein 1358 0.0
At2g36200 putative kinesin-related cytokinesis protein 838 0.0
At2g37420 putative kinesin heavy chain 834 0.0
At3g44050 kinesin -like protein 274 2e-73
At3g17360 kinesin-like protein 270 3e-72
At3g54870 kinesin-like protein 268 1e-71
At1g59540 zcf125 kinesin-like protein 253 4e-67
At5g47820 kinesin-like protein 251 1e-66
At3g50240 kinesin-related protein (kicp-02) 247 2e-65
At2g21380 putative kinesin heavy chain 234 3e-61
At3g12020 unknown protein 231 1e-60
At1g18370 AtNACK1 kinesin-like protein (AtNACK1) 230 3e-60
At1g12430 kinesin-like protein 229 8e-60
At3g10180 putative kinesin-like centromere protein 227 3e-59
At3g49650 kinesin-like protein 224 2e-58
At3g43210 kinesin -like protein 223 3e-58
At1g72250 kinesin, putative 223 6e-58
At3g19050 hypothetical protein 222 1e-57
At1g01950 unknown protein 218 2e-56
>At3g45850 kinesin-related protein - like
Length = 1058
Score = 1558 bits (4034), Expect = 0.0
Identities = 795/1057 (75%), Positives = 916/1057 (86%), Gaps = 12/1057 (1%)
Query: 3 VQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNED 62
+QQ+RGG+V LSP+QTPRS+DK AR+ RS++SNS + N DKEKGVNVQV++RCRP++ED
Sbjct: 4 IQQRRGGIVSLSPAQTPRSSDKSARESRSSESNSTNRN--DKEKGVNVQVILRCRPLSED 61
Query: 63 EMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYE 122
E R+HTPVVISCNE RREVAA QSIA K IDR F FDKVFGP SQQK+LYDQA+ PIV+E
Sbjct: 62 EARIHTPVVISCNENRREVAATQSIAGKHIDRHFAFDKVFGPASQQKDLYDQAICPIVFE 121
Query: 123 VLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEYS 182
VLEGYNCTIFAYGQTGTGKTYTMEGGA KKNGEFP+DAGVIPRAVKQIFDILEAQ AEYS
Sbjct: 122 VLEGYNCTIFAYGQTGTGKTYTMEGGARKKNGEFPSDAGVIPRAVKQIFDILEAQGAEYS 181
Query: 183 MKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEI 242
MKVTFLELYNEEI+DLLAPEET KFVDEKSKK IALMEDGKG V VRGLEEEIV TANEI
Sbjct: 182 MKVTFLELYNEEISDLLAPEETIKFVDEKSKKSIALMEDGKGSVFVRGLEEEIVSTANEI 241
Query: 243 YKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSE 302
YKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKE TPEGEEMIKCGKLNLVDLAGSE
Sbjct: 242 YKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKENTPEGEEMIKCGKLNLVDLAGSE 301
Query: 303 NISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKT 362
NISRSGAREGRAREAGEINKSLLTLGR INALVEHSGH+PYRDSKLTRLLR+SLGGKTKT
Sbjct: 302 NISRSGAREGRAREAGEINKSLLTLGRVINALVEHSGHIPYRDSKLTRLLRESLGGKTKT 361
Query: 363 CIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVY 422
C+IAT+SPSIHCLEETLSTLDYAHRAKNIKNKPE+NQKMMKSA++KDLYSEIDRLKQEVY
Sbjct: 362 CVIATISPSIHCLEETLSTLDYAHRAKNIKNKPEINQKMMKSAVMKDLYSEIDRLKQEVY 421
Query: 423 AAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAEL 482
AAREKNGIYIP+DRY+ EEAEKKAMAEKIER+EL ++SKDK +V+LQELYNSQQ+LTAEL
Sbjct: 422 AAREKNGIYIPKDRYIQEEAEKKAMAEKIERLELQSESKDKRVVDLQELYNSQQILTAEL 481
Query: 483 SAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAEL 542
S KLEKTEK LEETE +LFDLEE++RQANATIKEKEF+ISNLLKSEK LVERA +LR EL
Sbjct: 482 SEKLEKTEKKLEETEHSLFDLEEKYRQANATIKEKEFVISNLLKSEKSLVERAFQLRTEL 541
Query: 543 ENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDM 602
E+A+SDVSNLFSKIERKDKIE+ NR LIQKFQSQL QQLE LH+TV++SV QE QLK M
Sbjct: 542 ESASSDVSNLFSKIERKDKIEDGNRFLIQKFQSQLTQQLELLHKTVASSVTQQEVQLKHM 601
Query: 603 EKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAK 662
E+DM+SFVSTKSEATE+LR R+ +LK +YGSGI+ALDN+A +L N+Q T+ L SEV+K
Sbjct: 602 EEDMESFVSTKSEATEELRDRLSKLKRVYGSGIEALDNIAVKLDGNSQSTFSSLNSEVSK 661
Query: 663 HSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITM 722
HS LE++FKG A EAD LL DLQ+SL+KQE + FA QQR+AHSRAV+T RSVSK+T+
Sbjct: 662 HSHELENVFKGFASEADMLLQDLQSSLNKQEEKLITFAQQQRKAHSRAVDTARSVSKVTV 721
Query: 723 KFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNAR 782
+FF+T+D HA+ LT IVEE Q VN +KL E E KFEEC A EE+QLLEKVAE+LA+SNAR
Sbjct: 722 EFFKTLDTHATKLTGIVEEAQTVNHKKLSEFENKFEECAANEERQLLEKVAELLANSNAR 781
Query: 783 KKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVE 842
KK LVQMAV+DLRESA+ RT+ LQ E TMQDSTS +KAEW +HMEKTES++HEDTS+VE
Sbjct: 782 KKNLVQMAVHDLRESASTRTTTLQHEMSTMQDSTSSIKAEWSIHMEKTESSHHEDTSAVE 841
Query: 843 SGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARF 902
SGKK + EVL CL K E+ + QWR AQ+SL+SLE+ N SVD+IVRGGM+AN+ LR++F
Sbjct: 842 SGKKAMQEVLLNCLEKTEMSAHQWRKAQESLVSLERNNVASVDSIVRGGMDANENLRSQF 901
Query: 903 SSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVE 962
S++VS++L+ AN+ + +SID+SLQLD++AC +NSMI PCC DL ELK H ++I+E
Sbjct: 902 STAVSSSLDVFDAANSSLLTSIDHSLQLDNDACTKVNSMIIPCCEDLIELKSDHNHKIIE 961
Query: 963 ITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQKLA 1022
ITENAGKCLL+EY+VDEPSCSTP +R ++PS+ SIEELRTP+ EELL+AF D K K A
Sbjct: 962 ITENAGKCLLDEYVVDEPSCSTPKKRPIDIPSIESIEELRTPASEELLRAFRDEKLSKQA 1021
Query: 1023 NGDVKHIGSYEETQSVR----------DSRVPLTTIN 1049
NGD K ++ +R DSR PL+ +N
Sbjct: 1022 NGDAKQQQQQQQQHLIRASSLYEAAVSDSRYPLSAVN 1058
>At2g28620 putative kinesin-like spindle protein
Length = 1076
Score = 1358 bits (3514), Expect = 0.0
Identities = 708/1087 (65%), Positives = 859/1087 (78%), Gaps = 49/1087 (4%)
Query: 1 MEVQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSH--NKYDKEKGVNVQVLVRCRP 58
M+ + G SP QTPRST+K RD R DSNSNS+ +K +KEKGVN+QV+VRCRP
Sbjct: 1 MDSNNSKKGSSVKSPCQTPRSTEKSNRDFR-VDSNSNSNPVSKNEKEKGVNIQVIVRCRP 59
Query: 59 MNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSP 118
N +E RL TP V++CN+ ++EVA Q+IA KQID+TF+FDKVFGP SQQK+LY QAVSP
Sbjct: 60 FNSEETRLQTPAVLTCNDRKKEVAVAQNIAGKQIDKTFLFDKVFGPTSQQKDLYHQAVSP 119
Query: 119 IVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQS 178
IV+EVL+GYNCTIFAYGQTGTGKTYTMEGGA KKNGE P+DAGVIPRAVKQIFDILEAQS
Sbjct: 120 IVFEVLDGYNCTIFAYGQTGTGKTYTMEGGARKKNGEIPSDAGVIPRAVKQIFDILEAQS 179
Query: 179 A-EYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVC 237
A EYS+KV+FLELYNEE+TDLLAPEET KF D+KSKKP+ALMEDGKGGV VRGLEEEIV
Sbjct: 180 AAEYSLKVSFLELYNEELTDLLAPEET-KFADDKSKKPLALMEDGKGGVFVRGLEEEIVS 238
Query: 238 TANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVD 297
TA+EIYK+LEKGSAKRRTAETLLNKQSSRSHSIFS+TIHIKECTPEGEE++K GKLNLVD
Sbjct: 239 TADEIYKVLEKGSAKRRTAETLLNKQSSRSHSIFSVTIHIKECTPEGEEIVKSGKLNLVD 298
Query: 298 LAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLG 357
LAGSENISRSGAREGRAREAGEINKSLLTLGR INALVEHSGH+PYR+SKLTRLLRDSLG
Sbjct: 299 LAGSENISRSGAREGRAREAGEINKSLLTLGRVINALVEHSGHIPYRESKLTRLLRDSLG 358
Query: 358 GKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRL 417
GKTKTC+IATVSPS+HCLEETLSTLDYAHRAK+IKNKPEVNQKMMKSA++KDLYSEI+RL
Sbjct: 359 GKTKTCVIATVSPSVHCLEETLSTLDYAHRAKHIKNKPEVNQKMMKSAIMKDLYSEIERL 418
Query: 418 KQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQL 477
KQEVYAAREKNGIYIP++RY EEAEKKAMA+KIE+ME++ ++KDK +++LQELYNS+QL
Sbjct: 419 KQEVYAAREKNGIYIPKERYTQEEAEKKAMADKIEQMEVEGEAKDKQIIDLQELYNSEQL 478
Query: 478 LTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIE 537
+TA L KL+KTEK L ETEQ L DLEE+HRQA ATIKEKE+LISNLLKSEK LV+RA+E
Sbjct: 479 VTAGLREKLDKTEKKLYETEQALLDLEEKHRQAVATIKEKEYLISNLLKSEKTLVDRAVE 538
Query: 538 LRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQ 597
L+AEL NAASDVSNLF+KI RKDKIE+ NR LIQ FQSQL +QLE L+ +V+ SV QE+
Sbjct: 539 LQAELANAASDVSNLFAKIGRKDKIEDSNRSLIQDFQSQLLRQLELLNNSVAGSVSQQEK 598
Query: 598 QLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLK 657
QL+DME M SFVS K++ATE LR + +LK Y +GIK+LD++A L ++Q T DL
Sbjct: 599 QLQDMENVMVSFVSAKTKATETLRGSLAQLKEKYNTGIKSLDDIAGNLDKDSQSTLNDLN 658
Query: 658 SEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSV 717
SEV KHS ALED+FKG EA +LL LQ SLH QE ++AF QQR+ HSR++++ +SV
Sbjct: 659 SEVTKHSCALEDMFKGFTSEAYTLLEGLQGSLHNQEEKLSAFTQQQRDLHSRSMDSAKSV 718
Query: 718 SKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLA 777
S + + FF+T+D HA+ LT++ E+ Q VN+QKL KKFEE A EEKQ+LEKVAE+LA
Sbjct: 719 STVMLDFFKTLDTHANKLTKLAEDAQNVNEQKLSAFTKKFEESIANEEKQMLEKVAELLA 778
Query: 778 SSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHED 837
SSNARKK+LVQ+AV D+R+ ++ +T LQ+E MQDS S +K +W H+ + ES++ ++
Sbjct: 779 SSNARKKELVQIAVQDIRQGSSSQTGALQQEMSAMQDSASSIKVQWNSHIVQAESHHLDN 838
Query: 838 TSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQA 897
S+VE K+D+ ++ CL ++ G+QQW+ AQ+SL+ LEKRN + D+I+RG +E N+
Sbjct: 839 ISAVEVAKEDMQKMHLKCLENSKTGTQQWKTAQESLVDLEKRNVATADSIIRGAIENNEK 898
Query: 898 LRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHY 957
LR +FSS+VSTTL D +N +I SSID SLQLD +A ++NS I PC +L EL+ H
Sbjct: 899 LRTQFSSAVSTTLSDVDSSNREIISSIDNSLQLDKDASTDVNSTIVPCSENLKELRTHHD 958
Query: 958 NRIVEITENAGKCLLNEY----------------------------------MVDEPSCS 983
+ +VEI +N GKCL +EY VDE + S
Sbjct: 959 DNVVEIKQNTGKCLGHEYKVTRFDPFLYNHHIYMIELDKIVNRKLNSLKTSTQVDEATSS 1018
Query: 984 TPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQK-LANGDVKHIGSYEETQSVRDSR 1042
TP +R +N+P+V SIEEL+TPSFEELLKAF D KS K + NG+ KH V + R
Sbjct: 1019 TPRKREYNIPTVGSIEELKTPSFEELLKAFHDCKSPKQMQNGEAKH---------VSNGR 1069
Query: 1043 VPLTTIN 1049
PLT IN
Sbjct: 1070 PPLTAIN 1076
>At2g36200 putative kinesin-related cytokinesis protein
Length = 1056
Score = 838 bits (2165), Expect = 0.0
Identities = 453/986 (45%), Positives = 645/986 (64%), Gaps = 9/986 (0%)
Query: 40 NKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFD 99
+++DKEKGVNVQVL+RCRP ++DE+R + P V++CN+ +REVA Q+IA K IDR F FD
Sbjct: 3 SRHDKEKGVNVQVLLRCRPFSDDELRSNAPQVLTCNDLQREVAVSQNIAGKHIDRVFTFD 62
Query: 100 KVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKN----GE 155
KVFGP++QQK+LYDQAV PIV EVLEG+NCTIFAYGQTGTGKTYTMEG + G
Sbjct: 63 KVFGPSAQQKDLYDQAVVPIVNEVLEGFNCTIFAYGQTGTGKTYTMEGECRRSKSAPCGG 122
Query: 156 FPTDAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFV-DEKSKK 214
P +AGVIPRAVKQIFD LE Q AEYS+KVTFLELYNEEITDLLAPE+ ++ +EK KK
Sbjct: 123 LPAEAGVIPRAVKQIFDTLEGQQAEYSVKVTFLELYNEEITDLLAPEDLSRVAAEEKQKK 182
Query: 215 PIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSIT 274
P+ LMEDGKGGVLVRGLEEEIV +ANEI+ +LE+GS+KRRTAET LNKQSSRSHS+FSIT
Sbjct: 183 PLPLMEDGKGGVLVRGLEEEIVTSANEIFTLLERGSSKRRTAETFLNKQSSRSHSLFSIT 242
Query: 275 IHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINAL 334
IHIKE TPEGEE+IKCGKLNLVDLAGSENISRSGAR+GRAREAGEINKSLLTLGR I+AL
Sbjct: 243 IHIKEATPEGEELIKCGKLNLVDLAGSENISRSGARDGRAREAGEINKSLLTLGRVISAL 302
Query: 335 VEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNK 394
VEH GHVPYRDSKLTRLLRDSLGG+TKTCIIATVSP++HCLEETLSTLDYAHRAKNI+NK
Sbjct: 303 VEHLGHVPYRDSKLTRLLRDSLGGRTKTCIIATVSPAVHCLEETLSTLDYAHRAKNIRNK 362
Query: 395 PEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERM 454
PEVNQKMMKS +IKDLY EI+RLK EVYA+REKNG+Y+P++RY EE+E+K MAE+IE+M
Sbjct: 363 PEVNQKMMKSTLIKDLYGEIERLKAEVYASREKNGVYMPKERYYQEESERKVMAEQIEQM 422
Query: 455 ELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATI 514
++ K L ELQ+ Y Q ++L+ KL+ TEK+L +T + L E +++ +
Sbjct: 423 GGQIENYQKQLEELQDKYVGQVRECSDLTTKLDITEKNLSQTCKVLASTNEELKKSQYAM 482
Query: 515 KEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQ 574
KEK+F+IS KSE LV++A L++ LE A D S+L KI R+DK+ +NR ++ +Q
Sbjct: 483 KEKDFIISEQKKSENVLVQQACILQSNLEKATKDNSSLHQKIGREDKLSADNRKVVDNYQ 542
Query: 575 SQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSG 634
+L++Q+ L V++ + Q L+ + K QS + ++A +++ +V +++Y S
Sbjct: 543 VELSEQISNLFNRVASCLSQQNVHLQGVNKLSQSRLEAHNKAILEMKKKVKASRDLYSSH 602
Query: 635 IKALDNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEA 694
++A+ N+ K+N E++ + + ++++ SL ++LQ++L +
Sbjct: 603 LEAVQNVVRLHKANANACLEEVSALTTSSACSIDEFLASGDETTSSLFDELQSALSSHQG 662
Query: 695 NVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELE 754
+ FA + R+ +E T+ +S+ T FF+ + + + E + + +
Sbjct: 663 EMALFARELRQRFHTTMEQTQEMSEYTSTFFQKLMEESKNAETRAAEANDSQINSIIDFQ 722
Query: 755 KKFEECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQD 814
K +E + + +L+ + +++S R+ +LV +++ +++ + + L + +
Sbjct: 723 KTYEAQSKSDTDKLIADLTNLVSSHIRRQHELVDSRLHNFKDAVSSNKTFLDEHVSAVNN 782
Query: 815 STSFVKAEWMVHMEKTESNYHE--DTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDS 872
T K +W + E+ E D S+ + + +L +LQ + AE + + +S
Sbjct: 783 LTKDAKRKWETFSMQAENEAREGADFSAAKHCRMEL--LLQQSVGHAESAFKHCKITHES 840
Query: 873 LLSLEKRNAGSVDTIVRGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDH 932
L + + V ++VR ++N+ A S+ + +D + DI I+ + +
Sbjct: 841 LKEMTSKQVTDVSSLVRSACDSNEQHDAEVDSARTAAEKDVTKNSDDIIQQIERMSEDEK 900
Query: 933 EACGNLNSMITPCCGDLTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPTRRLFNL 992
+ + + L + + I + A + +YM EP+ +TPT+ +
Sbjct: 901 ASVSKILENVRSHEKTLESFQQDQCCQARCIEDKAQETFQQQYMEYEPTGATPTKNEPEI 960
Query: 993 PSVSSIEELRTPSFEELLKAFWDAKS 1018
P+ ++IE LR E L++ F + S
Sbjct: 961 PTKATIESLRAMPIETLVEEFRENNS 986
>At2g37420 putative kinesin heavy chain
Length = 1022
Score = 834 bits (2155), Expect = 0.0
Identities = 451/1015 (44%), Positives = 657/1015 (64%), Gaps = 48/1015 (4%)
Query: 2 EVQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNE 61
EV ++ G V + PS P T P + R DS SN ++ +KE VNVQV++RC+P++E
Sbjct: 6 EVVSRKSG-VGVIPSPAPFLT--PRLERRRPDSFSNRLDRDNKE--VNVQVILRCKPLSE 60
Query: 62 DEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVY 121
+E + P VISCNE RREV + +IANKQ+DR F FDKVFGP SQQ+ +YDQA++PIV+
Sbjct: 61 EEQKSSVPRVISCNEMRREVNVLHTIANKQVDRLFNFDKVFGPKSQQRSIYDQAIAPIVH 120
Query: 122 EVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEY 181
EVLEG++CT+FAYGQTGTGKTYTMEGG KK G+ P +AGVIPRAV+ IFD LEAQ+A+Y
Sbjct: 121 EVLEGFSCTVFAYGQTGTGKTYTMEGGMRKKGGDLPAEAGVIPRAVRHIFDTLEAQNADY 180
Query: 182 SMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANE 241
SMKVTFLELYNEE+TDLLA +++++ ++K +KPI+LMEDGKG V++RGLEEE+V +AN+
Sbjct: 181 SMKVTFLELYNEEVTDLLAQDDSSRSSEDKQRKPISLMEDGKGSVVLRGLEEEVVYSAND 240
Query: 242 IYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGS 301
IY +LE+GS+KRRTA+TLLNK+SSRSHS+F+IT+HIKE + EE+IKCGKLNLVDLAGS
Sbjct: 241 IYALLERGSSKRRTADTLLNKRSSRSHSVFTITVHIKEESMGDEELIKCGKLNLVDLAGS 300
Query: 302 ENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTK 361
ENI RSGAR+GRAREAGEINKSLLTLGR INALVEHS HVPYRDSKLTRLLRDSLGGKTK
Sbjct: 301 ENILRSGARDGRAREAGEINKSLLTLGRVINALVEHSSHVPYRDSKLTRLLRDSLGGKTK 360
Query: 362 TCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEV 421
TCIIAT+SPS H LEETLSTLDYA+RAKNIKNKPE NQK+ K+ ++KDLY E++R+K++V
Sbjct: 361 TCIIATISPSAHSLEETLSTLDYAYRAKNIKNKPEANQKLSKAVLLKDLYLELERMKEDV 420
Query: 422 YAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAE 481
AAR+KNG+YI +RY EE EKKA E+IE++E + + + N ++L + +L
Sbjct: 421 RAARDKNGVYIAHERYTQEEVEKKARIERIEQLENELNLSESN---FRDLVSRLFILLVR 477
Query: 482 LSAKLE--KTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELR 539
+ K + +++L + + L DL+E + Q + +KEKE ++S + SE L++RA LR
Sbjct: 478 VFLKFQTFMIQRNLHNSNKDLLDLKENYIQVVSKLKEKEVIVSRMKASETSLIDRAKGLR 537
Query: 540 AELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQL 599
+L++A++D+++LF+++++KDK+E +N+ ++ KF SQL Q L+ LHRTV SV Q+QQL
Sbjct: 538 CDLQHASNDINSLFTRLDQKDKLESDNQSMLLKFGSQLDQNLKDLHRTVLGSVSQQQQQL 597
Query: 600 KDMEKDMQSFVSTK-----------SEATEDLRVRVVELKNMYGSGIKALDNLAEELKSN 648
+ ME+ SF++ K ++AT DL R+ + + Y SGI AL L+E L+
Sbjct: 598 RTMEEHTHSFLAHKYDLITLVVDLLTQATRDLESRIGKTSDTYTSGIAALKELSEMLQKK 657
Query: 649 NQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHS 708
E + + A+E A EA ++ D+ N L+ Q+ + A QQ +
Sbjct: 658 ASSDLEKKNTSIVSQIEAVEKFLTTSATEASAVAQDIHNLLNDQKKLLALAARQQEQGLV 717
Query: 709 RAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQL 768
R++ + + +S T F I +E EEKQ
Sbjct: 718 RSMRSAQEISNSTSTIFSNIYN---------------------------QEEAEREEKQA 750
Query: 769 LEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHME 828
L ++ +L+ ++K ++ A +++RE +L + MQ + K E +++
Sbjct: 751 LNDISLILSKLTSKKTAMISDASSNIREHDIQEEKRLYEQMSGMQQVSIGAKEELCDYLK 810
Query: 829 KTESNYHEDTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIV 888
K ++++ E+T + + L+ CL +A W + + +L + ++ +
Sbjct: 811 KEKTHFTENTIASAESITVMDSYLEDCLGRANDSKTLWETTETGIKNLNTKYQQELNVTM 870
Query: 889 RGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGD 948
+ N+ ++ F+S+ S+ + +++++++ SL D E +++ C
Sbjct: 871 EDMAKENEKVQDEFTSTFSSMDANFVSRTNELHAAVNDSLMQDRENKETTEAIVETCMNQ 930
Query: 949 LTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRT 1003
+T L+ H + I A + L+ +Y VD+ TP ++ N+PS+ SIEE+RT
Sbjct: 931 VTLLQENHGQAVSNIRNKAEQSLIKDYQVDQHKNETPKKQSINVPSLDSIEEMRT 985
>At3g44050 kinesin -like protein
Length = 1229
Score = 274 bits (701), Expect = 2e-73
Identities = 249/879 (28%), Positives = 410/879 (46%), Gaps = 84/879 (9%)
Query: 46 KGVNVQVLVRCRPMNEDEMRLH-TPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGP 104
K NVQV++R RP++ E+ + + + G+ A+ I N + F FD V
Sbjct: 90 KDHNVQVIIRTRPLSSSEISVQGNNKCVRQDNGQ----AITWIGNPE--SRFTFDLVADE 143
Query: 105 NSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIP 164
N Q++++ A P+V V+ GYN +FAYGQTG+GKT+TM G + G+ P
Sbjct: 144 NVSQEQMFKVAGVPMVENVVAGYNSCMFAYGQTGSGKTHTMLGDIEGGTRRHSVNCGMTP 203
Query: 165 RAVKQIF-------DILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIA 217
R + +F ++ + + ++ + +FLE+YNE+I DLL P S +
Sbjct: 204 RVFEYLFSRIQKEKEVRKEEKLHFTCRCSFLEIYNEQILDLLDP----------SSYNLQ 253
Query: 218 LMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHI 277
L ED K G+ V L+E V +A ++ + L +G+A R+ A T +N+ SSRSHS+F+ I
Sbjct: 254 LREDHKKGIHVENLKEIEVSSARDVIQQLMQGAANRKVAATNMNRASSRSHSVFTCIIES 313
Query: 278 KECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEH 337
K + +G + +LNLVDLAGSE SGA R +EA INKSL TLG I LV
Sbjct: 314 KWVS-QGVTHHRFARLNLVDLAGSERQKSSGAEGERLKEATNINKSLSTLGLVIMNLVSV 372
Query: 338 SG----HVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKN 393
S HVPYRDSKLT LL+DSLGG +KT IIA +SPS C ETLSTL +A RAK IKN
Sbjct: 373 SNGKSVHVPYRDSKLTFLLQDSLGGNSKTIIIANISPSSSCSLETLSTLKFAQRAKLIKN 432
Query: 394 KPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIER 453
VN+ + L +I +LK+EV R G+ N++ + +M
Sbjct: 433 NAIVNEDASGDVIAMRL--QIQQLKKEVTRLRGMGGVD-------NQDMDTISMGCPASP 483
Query: 454 MELDADSKDKNLVEL---QELYNSQQLLTAELSAKLEKTEK-----SLEETEQTLFDLEE 505
M L D + + L + + + A + A + EK +L + LE+
Sbjct: 484 MSLKWDGFNGSFTPLTTHKRMSKVKDYEVALVGAFRREREKDVALQALTAENEASMKLEK 543
Query: 506 RHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEE 565
+ +K L + +KS + + I + A L+ D L +IE E E
Sbjct: 544 KREDEIRGLKMMLKLRDSAIKSLQGVTSGKIPVEAHLQKEKGD---LMKEIE-----EGE 595
Query: 566 NRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVV 625
+L Q+ Q+ A+ LEAL +MH+ E S + ++ E ++ +
Sbjct: 596 RDILNQQIQALQAKLLEAL----DWKLMHESDSSMVKEDGNISNMFCSNQNQESKKLSSI 651
Query: 626 ELKNMYGSGIKALDNLAEELKSNNQLTYE-DLKSEVAKHSSALEDLFKGIALEADSLLND 684
+ +N + ++A+ N AE L++ D K + K L + +G + + +D
Sbjct: 652 QDENEF-LRMQAIQNRAEMESLQKSLSFSLDEKERLQKLVDNLSNELEGKIRSSGMVGDD 710
Query: 685 LQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMK---FFETIDRHASSLTQIVEE 741
Q + + + ++ EAH A++ ++ + K E ++ Q+ EE
Sbjct: 711 DQMEVKTMVQAIACVSQREAEAHETAIKLSKENDDLRQKIKVLIEDNNKLIELYEQVAEE 770
Query: 742 --------------TQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKLV 787
+ + Q E+ + E+ A E+K+++ + L + +KL+
Sbjct: 771 NSSRAWGKIETDSSSNNADAQNSAEIALEVEKSAAEEQKKMIGNLENQLTEMHDENEKLM 830
Query: 788 QMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKD 847
+ N ++E +L+R L+ D ++A ME + + T + S K
Sbjct: 831 SLYENAMKEK-----DELKR-LLSSPDQKKPIEANSDTEMELCNISSEKSTEDLNSAKLK 884
Query: 848 LAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDT 886
L E+ Q L+ + + + ++++L + K + S +T
Sbjct: 885 L-ELAQEKLSISAKTIGVFSSLEENILDIIKLSKESKET 922
Score = 47.8 bits (112), Expect = 3e-05
Identities = 94/455 (20%), Positives = 178/455 (38%), Gaps = 55/455 (12%)
Query: 383 DYAHRAKNIKNKPEVNQKMMKSA------MIKDLYSEIDRLKQE------VY--AAREKN 428
D + + +N E+ ++ KSA MI +L +++ + E +Y A +EK+
Sbjct: 782 DSSSNNADAQNSAEIALEVEKSAAEEQKKMIGNLENQLTEMHDENEKLMSLYENAMKEKD 841
Query: 429 GIYIPRDRYLNEEAEKKAM-AEKIERMELDADSKDKNLVELQELYNSQQLLTAELS--AK 485
+ R L+ +KK + A MEL S +K+ +L +L +LS AK
Sbjct: 842 EL----KRLLSSPDQKKPIEANSDTEMELCNISSEKSTEDLNSAKLKLELAQEKLSISAK 897
Query: 486 LEKTEKSLEETEQTLFDLEERHRQANATIKEKEF------LISNLLKSEKELVERAIE-L 538
SLEE + L + ++ +KE + +S+ + KE+ E+ + L
Sbjct: 898 TIGVFSSLEENILDIIKLSKESKETEEKVKEHQSELGSIKTVSDQTNARKEVAEKKLAAL 957
Query: 539 RAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQ 598
R L N AS + EE R + F L Q+ E L S H+ +
Sbjct: 958 RCSLSNFASSAVYFQQR-------EERARAHVNSFSGYLNQKNEELDVIRS----HKREI 1006
Query: 599 LKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSG--IKALDNLAEELKSNNQLTYEDL 656
M K QS KS L+++V E + + +DN+ K+ + L ++
Sbjct: 1007 DAAMGKIQQSEAELKSNIVM-LKIKVDEENKRHEEEGVLCTIDNILRTGKATDLLKSQEE 1065
Query: 657 KSEVAKHSSALEDLFKGIALEADSLLN---DLQNSLHKQEANVTAFAHQQREAHSRAVET 713
K+++ + + E D + L+ + E + + + E+ T
Sbjct: 1066 KTKLQSEMKLSREKLASVRKEVDDMTKKSLKLEKEIKTMETEIEKSSKTRTESEMELENT 1125
Query: 714 TRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVA 773
+ I E ++ S + ++ E Q + E + + EE E+ + E++
Sbjct: 1126 IQEKQTIQ----EMEEQGMSEIQNMIIEIH----QLVFESDLRKEEAMIIREELIAEELR 1177
Query: 774 EMLASSNARKKKLVQMAVNDLRESANCRTSKLQRE 808
+N ++ V+ A+ L N + K++ E
Sbjct: 1178 AKDVHTNMIER--VENALKTLENQNNSVSGKIEEE 1210
Score = 36.2 bits (82), Expect = 0.10
Identities = 74/348 (21%), Positives = 140/348 (39%), Gaps = 36/348 (10%)
Query: 651 LTYEDLKSEVAKHSSALEDLF-----KGIALEADSLLNDLQNSLHKQEANVTAFAHQQRE 705
LT S+V + AL F K +AL+A + N+ L K+ + +
Sbjct: 498 LTTHKRMSKVKDYEVALVGAFRREREKDVALQALTAENEASMKLEKKREDEIRGLKMMLK 557
Query: 706 AHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQF-VNDQKLCELEKKFEECTAYE 764
A+++ + V+ + + + L + +EE + + +Q++ L+ K E A +
Sbjct: 558 LRDSAIKSLQGVTSGKIPVEAHLQKEKGDLMKEIEEGERDILNQQIQALQAKLLE--ALD 615
Query: 765 EKQLLEKVAEML-----------ASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQ 813
K + E + M+ ++ N KKL ++ D E + + + E ++Q
Sbjct: 616 WKLMHESDSSMVKEDGNISNMFCSNQNQESKKL--SSIQDENEFLRMQAIQNRAEMESLQ 673
Query: 814 DSTSFVKAEWMVHMEKTESNYHEDT-----SSVESGKKDLAEVLQICLNKAEVGSQQWRN 868
S SF E ++K N + SS G D EV + A V SQ+
Sbjct: 674 KSLSFSLDE-KERLQKLVDNLSNELEGKIRSSGMVGDDDQMEVKTMVQAIACV-SQREAE 731
Query: 869 AQDSLLSLEKRNAG---SVDTIVRGG---MEANQALRARFSSSVSTTLE-DAGIANTDIN 921
A ++ + L K N + ++ +E + + SS +E D+ N D
Sbjct: 732 AHETAIKLSKENDDLRQKIKVLIEDNNKLIELYEQVAEENSSRAWGKIETDSSSNNADAQ 791
Query: 922 SSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEITENAGK 969
+S + +L+++ A MI LTE+ + +++ + ENA K
Sbjct: 792 NSAEIALEVEKSAAEEQKKMIGNLENQLTEMHDEN-EKLMSLYENAMK 838
>At3g17360 kinesin-like protein
Length = 2008
Score = 270 bits (690), Expect = 3e-72
Identities = 245/856 (28%), Positives = 386/856 (44%), Gaps = 144/856 (16%)
Query: 46 KGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPN 105
K NVQVL+R RP+ E N+G A F FD V
Sbjct: 165 KDHNVQVLIRLRPLGTMER---------ANQGYGHPEA-----------RFTFDHVASET 204
Query: 106 SQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPR 165
Q++L+ A P+V L GYN +FAYGQTG+GKTYTM G + G D GV R
Sbjct: 205 ISQEKLFRVAGLPMVENCLSGYNSCVFAYGQTGSGKTYTMMGEISEAEGSLGEDCGVTAR 264
Query: 166 AVKQIFDILEAQSAE-------YSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIAL 218
+ +F ++ + E +S K +FLE+YNE+ITDLL P T + L
Sbjct: 265 IFEYLFSRIKMEEEERRDENLKFSCKCSFLEIYNEQITDLLEPSSTN----------LQL 314
Query: 219 MEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIK 278
ED GV V L E V T +++ K+L +G+ R+ A T +N +SSRSHS+F+ TI
Sbjct: 315 REDLGKGVYVENLVEHNVRTVSDVLKLLLQGATNRKIAATRMNSESSRSHSVFTCTI--- 371
Query: 279 ECTPEGEEMIKC--GKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVE 336
E E + + + +LNLVDLAGSE SGA R +EA INKSL TLG I +LV+
Sbjct: 372 ESLWEKDSLTRSRFARLNLVDLAGSERQKSSGAEGDRLKEAANINKSLSTLGLVIMSLVD 431
Query: 337 HSG----HVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIK 392
+ HVPYRDS+LT LL+DSLGG +KT IIA VSPS+ ETLSTL +A RAK I+
Sbjct: 432 LAHGKHRHVPYRDSRLTFLLQDSLGGNSKTMIIANVSPSLCSTNETLSTLKFAQRAKLIQ 491
Query: 393 NKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAARE---------------KNGIYIPRDRY 437
N +VN+ S + L EI +LK ++ + + + Y +
Sbjct: 492 NNAKVNED--ASGDVTALQQEIRKLKVQLTSLLKNHDSCGALSDCISSLEESRYSGTCKV 549
Query: 438 LNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETE 497
E + K + + + + + N++ + + L+K+E +E +
Sbjct: 550 AGETRQDKCHCQVHNSLRVKVKNMNDNMIGALRREKIAE-------SALQKSEAEIERID 602
Query: 498 QTLFDLEERHRQ------------------ANATIKEKEFLI--SNLLKSEKELVERAIE 537
+ D+EE ++ + ++ KE LI + LK E +L+ +I+
Sbjct: 603 CLVRDMEEDAKRIKIMLNLREEKVGEMEFCTSGSLMTKECLIEENKTLKGEIKLLRDSID 662
Query: 538 LRAELENAASDVSNLFSKIERKDKIEE--ENRVLIQK---FQSQLAQQLEALHRTVSASV 592
EL +A + + L +++R K E E L+ + + QL LEA + S V
Sbjct: 663 KNPELTRSALENTKLREQLQRYQKFYEHGEREALLAEVTGLRDQLLDVLEAKDESFSKHV 722
Query: 593 MHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLT 652
M + + K+ E + +NM S I+ LD + L L
Sbjct: 723 MKENEMEKEFE----------------------DCRNMNSSLIRELDEIQAGL--GRYLN 758
Query: 653 YEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVE 712
++ ++S V S+ + E ++++Q + A +H + V+
Sbjct: 759 FDQIQSNVVASSTRGAE-----QAETMPTISEIQEEV--------AISHSKNYDRGALVK 805
Query: 713 TTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKV 772
T + + ++F L + +EE + +N C+ EK + + +E +E V
Sbjct: 806 TDEGIDRSILQF------KLGKLMKDLEEARTLN----CKYEKDHKSQLSQQED--IEVV 853
Query: 773 AEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTES 832
E + + AR +Q V L+ R L E +++D+ + ++E + E
Sbjct: 854 REQVETETARTILELQEEVIALQSEFQRRICNLTEENQSIKDTITARESEIRALNQDWEK 913
Query: 833 NYHEDTSSVESGKKDL 848
E T+ + +G K +
Sbjct: 914 ATLELTNFIVAGSKSI 929
Score = 43.5 bits (101), Expect = 6e-04
Identities = 49/274 (17%), Positives = 119/274 (42%), Gaps = 17/274 (6%)
Query: 389 KNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMA 448
+++ N+ E + + K L S + +K+ + ++ +D+ ++E ++
Sbjct: 1565 RDLVNRLEKEILHLTTTAEKQLLSAVKSIKENLKKTSDE------KDQIVDEIC---SLN 1615
Query: 449 EKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHR 508
K+E AD K+ VE + + ++ + +++ E S+EE E+T+ LE R
Sbjct: 1616 NKLELAYAIADEKEAIAVEAHQESEASKIYAEQKEEEVKILEISVEELERTINILERRVY 1675
Query: 509 QANATIKEKEFLISNL---LKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEE 565
+ +K +L L++ ++ + R + S I R ++
Sbjct: 1676 DMDEEVKRHRTTQDSLETELQALRQRLFRFENFTGTMVTTNESTEEYKSHISRSTGLQGA 1735
Query: 566 NRVL--IQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVR 623
+ + +QK ++ ++++ L +S ++H E Q ++ M+ K + RVR
Sbjct: 1736 HSQIQVLQKEVAEQTKEIKQLKEYISEILLHSEAQSSAYQEKMK---LEKDQELTMARVR 1792
Query: 624 VVELKNMYGSGIKALDNLAEELKSNNQLTYEDLK 657
V EL+++ K + L + + + +T++ ++
Sbjct: 1793 VEELESLLAVKQKEICTLNTRIAAADSMTHDVIR 1826
Score = 37.7 bits (86), Expect = 0.035
Identities = 40/172 (23%), Positives = 73/172 (42%), Gaps = 24/172 (13%)
Query: 498 QTLFDLEERHRQANATIKEKEFLISNLLKSEKE---LVERAIELRAELENAASDVSNLFS 554
+T FD R + NAT+KE + I L+K+ ++ + E ++ EL S NL
Sbjct: 1217 RTFFD---RFEEVNATMKEADLTICELVKANEKSNSVTEMWLQTHEEL---ISKEKNLMD 1270
Query: 555 KIERKDKI----EEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFV 610
+E+ I EEE +VL+ + + LA + SV E+ ++M++ ++
Sbjct: 1271 DLEQVKSILSACEEEKQVLLNQTHTTLAD--------MENSVSLLEEYFQEMKRGVE--- 1319
Query: 611 STKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAK 662
T R+ EL + + +L+ +A E Y + + K
Sbjct: 1320 ETVEALFSHARLAGKELLQLISNSRPSLEQIASEFMEREFTMYATYQCHIGK 1371
Score = 32.0 bits (71), Expect = 1.9
Identities = 68/360 (18%), Positives = 140/360 (38%), Gaps = 49/360 (13%)
Query: 347 KLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAM 406
KL +L++D +T C S +E + + + + E+ ++++ A+
Sbjct: 818 KLGKLMKDLEEARTLNCKYEKDHKSQLSQQEDIEVVREQVETETARTILELQEEVI--AL 875
Query: 407 IKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLV 466
+ I L +E + ++ R LN++ EK A E + + S
Sbjct: 876 QSEFQRRICNLTEENQSIKDTITARESEIRALNQDWEK-ATLELTNFIVAGSKSIKNAST 934
Query: 467 ELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNL-- 524
+++ + S + A + +EK K+ + E+T+ L++ A + E +++L
Sbjct: 935 QIESIICSFPQVNAWIGDYVEKAAKNCIKKEETILLLQKSLEDARILVAEMNLKLNSLKG 994
Query: 525 ---------LKSEKELVERAIELRAELENAASDVSNLFS----------KIERKDKI--- 562
L E A L +++ + +V L S K ER +
Sbjct: 995 ATIALNEFQLGGNAATTEEAFNLNNDVDRMSDEVDTLESNFKANQYSILKTERHAEAALA 1054
Query: 563 -------EEENRVLIQKFQSQLAQQLEALHRTVSASV---------MHQEQQLKDMEKDM 606
+ +++K Q Q ++ L ++SAS+ + ++ L D
Sbjct: 1055 VTKWLSDSRDQHQMMEKVQDQSVKEFGTL-SSISASLSAEGNADISLSRDGHLSDATYPK 1113
Query: 607 QSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKHSSA 666
+ST S + R + N+ G+ + ++ A+E SNN++T L +AK+ SA
Sbjct: 1114 GDELSTSSSDFSNCRWQHDCALNVKCQGVSSSESDAQE--SNNKITSAAL---IAKNGSA 1168
>At3g54870 kinesin-like protein
Length = 1070
Score = 268 bits (685), Expect = 1e-71
Identities = 233/796 (29%), Positives = 384/796 (47%), Gaps = 116/796 (14%)
Query: 50 VQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQK 109
V+V VR RP N +E+ + E + E+ ++ N ++ FD+VF + QK
Sbjct: 124 VRVSVRVRPRNGEEL-ISDADFADLVELQPEIKRLKLRKNNWNSESYKFDEVFTDTASQK 182
Query: 110 ELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQ 169
+Y+ P+V VL GYN TI AYGQTGTGKTYT+ G I K+ + G++ RA++
Sbjct: 183 RVYEGVAKPVVEGVLSGYNGTIMAYGQTGTGKTYTV--GKIGKDDA--AERGIMVRALED 238
Query: 170 IFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGG-VLV 228
I +L A SA S+++++L+LY E I DLLAPE K I++ ED K G V V
Sbjct: 239 I--LLNASSASISVEISYLQLYMETIQDLLAPE----------KNNISINEDAKTGEVSV 286
Query: 229 RGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEM- 287
G + + ++L+ G R A T +N +SSRSH+I +T++++ E E
Sbjct: 287 PGATVVNIQDLDHFLQVLQVGETNRHAANTKMNTESSRSHAI--LTVYVRRAMNEKTEKA 344
Query: 288 ------------IKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALV 335
++ KL +VDLAGSE I++SG EA IN SL +LG+ INAL
Sbjct: 345 KPESLGDKAIPRVRKSKLLIVDLAGSERINKSGTDGHMIEEAKFINLSLTSLGKCINALA 404
Query: 336 EHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKP 395
E S H+P RDSKLTRLLRDS GG +T +I T+ PS ET ST+ + RA I N
Sbjct: 405 EGSSHIPTRDSKLTRLLRDSFGGSARTSLIITIGPSARYHAETTSTIMFGQRAMKIVNMV 464
Query: 396 EVNQKMMKSAMIKDLYSEIDRLKQEV------------------------YAAREKNGIY 431
++ ++ ++ + L +++D L EV +A EKN
Sbjct: 465 KLKEEFDYESLCRKLETQVDHLTAEVERQNKLRNSEKHELEKRLRECENSFAEAEKNA-- 522
Query: 432 IPRDRYLNEEAEK--KAMAEKIERMELDADS----KDKNLVELQELYNS--QQLLTAELS 483
+ R ++L +E + +M E ++ ++L D DK + +L N+ QQL +
Sbjct: 523 VTRSKFLEKENTRLELSMKELLKDLQLQKDQCDLMHDKAIQLEMKLKNTKQQQLENSAYE 582
Query: 484 AKLEKT----EKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELR 539
AKL T EK + E Q + D + R A + E + ++S KS E + + +
Sbjct: 583 AKLADTSQVYEKKIAELVQRVEDEQARSTNAEHQLTEMKNILSKQQKSIHEQEKGNYQYQ 642
Query: 540 AELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQL 599
EL + +++++ K+E EN +S A+ Q +Q+
Sbjct: 643 RELAETTHTYESKIAELQK--KLEGENA------RSNAAED--------------QLRQM 680
Query: 600 KDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSE 659
K + D Q +S ++E +L++++ EL MY S + L + +L Y+DL +
Sbjct: 681 KRLISDRQ-VISQENEEANELKIKLEELSQMYESTVDELQTV--------KLDYDDLLQQ 731
Query: 660 VAKHSSALEDLFKGIALE------ADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVET 713
K + D+ + + LE +S L+ L+ +L + E V + + + + E+
Sbjct: 732 KEKLGEEVRDMKERLLLEEKQRKQMESELSKLKKNLRESENVVEEKRYMKEDLSKGSAES 791
Query: 714 -TRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKV 772
++ S+ + +++ +++ ++ EE V QK+ +L K + E + KV
Sbjct: 792 GAQTGSQRSQGLKKSLSGQRATMARLCEE---VGIQKILQLIKSED----LEVQIQAVKV 844
Query: 773 AEMLASSNARKKKLVQ 788
LA+ A + K+V+
Sbjct: 845 VANLAAEEANQVKIVE 860
>At1g59540 zcf125 kinesin-like protein
Length = 823
Score = 253 bits (646), Expect = 4e-67
Identities = 190/708 (26%), Positives = 338/708 (46%), Gaps = 55/708 (7%)
Query: 98 FDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFP 157
FD VF +S +Y+ I++ +EG+N T FAYGQT +GKT+TM G
Sbjct: 46 FDHVFDESSTNASVYELLTKDIIHAAVEGFNGTAFAYGQTSSGKTFTMTGSE-------- 97
Query: 158 TDAGVIPRAVKQIFDILEAQS-AEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPI 216
TD G+I R+V+ +F+ + S E+ ++V+++E+YNEEI DLLA E + E ++
Sbjct: 98 TDPGIIRRSVRDVFERIHMISDREFLIRVSYMEIYNEEINDLLAVENQRLQIHEHLER-- 155
Query: 217 ALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIH 276
GV V GL+EEIV A +I K+++ G R ET +N SSRSH+IF + I
Sbjct: 156 --------GVFVAGLKEEIVSDAEQILKLIDSGEVNRHFGETNMNVHSSRSHTIFRMVIE 207
Query: 277 IKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVE 336
+ + I+ LNLVDLAGSE I+++GA R +E INKSL+ LG IN L +
Sbjct: 208 SRGKDNSSSDAIRVSVLNLVDLAGSERIAKTGAGGVRLQEGKYINKSLMILGNVINKLSD 267
Query: 337 HS---GHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKN 393
+ H+PYRDSKLTR+L+ +LGG KTCII T++P H +EE+ TL +A RAK I N
Sbjct: 268 STKLRAHIPYRDSKLTRILQPALGGNAKTCIICTIAPEEHHIEESKGTLQFASRAKRITN 327
Query: 394 KPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIER 453
+VN+ + +A++K EI+ L+ ++ + + + LN + + ER
Sbjct: 328 CAQVNEILTDAALLKRQKLEIEELRMKLQGSHAE----VLEQEILNLSNQMLKYELECER 383
Query: 454 MELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANAT 513
++ + + + E + QQ+ L+ + ++ ++E F + +
Sbjct: 384 LKTQLEEEKRKQKEQENCIKEQQMKIENLNNFVTNSDFKRNQSED--FIISRKTPDGLCN 441
Query: 514 IKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQK- 572
+ + + K + R+ + SD S + + D +E+ + + K
Sbjct: 442 VNDTSDVPGT--PCFKSASRSFVVARSNNYSGLSDFSPMVHSL--GDVADEDTWMKLNKG 497
Query: 573 -----FQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVEL 627
Q Q ++ +S + ++ +D++S + + + L+V+
Sbjct: 498 FVADLDQIQFTPAVKCQPTPLSIATTECPRENHSEVEDLKSRIQLLTNENDSLQVK---- 553
Query: 628 KNMYGSGIKALDNLAEELKSNNQ--LTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDL 685
+ + +NL +E+ Q LT +++ + +++ + +D++K + + SL+ D
Sbjct: 554 ---FNEQVLLSNNLMQEMSELKQETLTVKEIPNRLSESVANCKDVYKDVIVTMKSLITDK 610
Query: 686 QN-------SLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQI 738
++ + ++ A Q +T S+ +ET+ + + T +
Sbjct: 611 ESPTANLLLGTTEITTSLLATLETQFSMIMDGQKTGSSIDHPLSDHWETLRVNLKNTTTL 670
Query: 739 VEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKL 786
+ D+ L K +E A EEK+L ++ + N +K+L
Sbjct: 671 LLSDAQAKDEFL-NSHNKGQETAALEEKKLKSELIIIKERYNELEKEL 717
>At5g47820 kinesin-like protein
Length = 1035
Score = 251 bits (642), Expect = 1e-66
Identities = 205/676 (30%), Positives = 335/676 (49%), Gaps = 77/676 (11%)
Query: 49 NVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQI-DRTFVFDKVFGPN-S 106
+V+V V RP+ DE ++ G+ +V QI +F FD V+G + S
Sbjct: 11 SVKVAVHIRPLIGDERIQGCQDCVTVVTGKPQV---------QIGSHSFTFDHVYGSSGS 61
Query: 107 QQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRA 166
E+Y++ +P+V + +GYN T+ AYGQTG+GKTYTM G G+ + G+IP+
Sbjct: 62 PSTEMYEECAAPLVDGLFQGYNATVLAYGQTGSGKTYTMGTGC----GD-SSQTGIIPQV 116
Query: 167 VKQIFDILEA--QSAEYSMKVTFLELYNEEITDLLAP-----------EETTKFVDEKSK 213
+ +F +E Q E+ + V+F+E++ EE+ DLL P K K
Sbjct: 117 MNALFTKIETLKQQIEFQIHVSFIEIHKEEVQDLLDPCTVNKSDTNNTGHVGKVAHVPGK 176
Query: 214 KPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSI 273
PI + E G + + G E V T E+ L++GS R T T +N QSSRSH+IF+I
Sbjct: 177 PPIQIRETSNGVITLAGSTEVSVSTLKEMAACLDQGSVSRATGSTNMNNQSSRSHAIFTI 236
Query: 274 TIH----IKECTPEG-------EEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINK 322
T+ I +PE +E C KL+LVDLAGSE R+G+ R +E INK
Sbjct: 237 TVEQMRKINTDSPENGAYNGSLKEEYLCAKLHLVDLAGSERAKRTGSDGLRFKEGVHINK 296
Query: 323 SLLTLGRTINALVEHS-----GHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEE 377
LL LG I+AL + HVPYRDSKLTRLL+DSLGG ++T +IA +SP+ EE
Sbjct: 297 GLLALGNVISALGDEKKRKDGAHVPYRDSKLTRLLQDSLGGNSRTVMIACISPADINAEE 356
Query: 378 TLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNG---IYIPR 434
TL+TL YA+RA+NI+NKP VN+ + S M+K + +++ L+ E+ + + +
Sbjct: 357 TLNTLKYANRARNIRNKPVVNRDPVSSEMLK-MRQQVEYLQAELSLRTGGSSCAEVQALK 415
Query: 435 DRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLE 494
+R + E + + ++ + + + +++ + ++ + L+++ S+E
Sbjct: 416 ERIVWLETANEELCRELHEYRSRCPGVEHSEKDFKDI-RADDIVGSVRPDGLKRSLHSIE 474
Query: 495 ETEQTLFDL---EERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELE-----NAA 546
+ + + + R A E + L +++ K EL R E +E++ + A
Sbjct: 475 SSNYPMVEATTGDSREIDEEAKEWEHKLLQNSMDKELYELNRRLEEKESEMKLFDGYDPA 534
Query: 547 SDVSNLFSKI-----ERKDKIEEENRVL-----------IQKFQSQLAQQLEALHRTVSA 590
+ + KI E++ EE NR+L QK Q AQ L+AL +
Sbjct: 535 ALKQHFGKKIAEVEDEKRSVQEERNRLLAEIENLASDGQAQKLQDVHAQNLKALEAQILD 594
Query: 591 SVMHQEQQLKDMEKDMQSFVSTK--SEATEDLRVRVVELKN-MYGSGIKALDNLAEELKS 647
QE Q++ +++ +S + + + + ++ + V+L++ M + A K
Sbjct: 595 LKKKQESQVQLLKQKQKSDDAARRLQDEIQSIKAQKVQLQHRMKQEAEQFRQWKASREKE 654
Query: 648 NNQLTYEDLKSEVAKH 663
QL E KSE +H
Sbjct: 655 LLQLRKEGRKSEYERH 670
Score = 37.0 bits (84), Expect = 0.059
Identities = 89/437 (20%), Positives = 163/437 (36%), Gaps = 74/437 (16%)
Query: 459 DSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKE 518
DSK L++ NS+ ++ A +S + + EET TL N + ++
Sbjct: 324 DSKLTRLLQDSLGGNSRTVMIACISP----ADINAEETLNTLKYANRARNIRNKPVVNRD 379
Query: 519 FLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLA 578
+ S +LK +++ EL L S + + + ER +E N L ++
Sbjct: 380 PVSSEMLKMRQQVEYLQAEL--SLRTGGSSCAEVQALKERIVWLETANEELCRELHE--- 434
Query: 579 QQLEALHRTVSASVMHQEQQLKDMEKD-----------MQSFVSTKS------EATEDLR 621
+R+ V H E+ KD+ D +S S +S EAT
Sbjct: 435 ------YRSRCPGVEHSEKDFKDIRADDIVGSVRPDGLKRSLHSIESSNYPMVEATTGDS 488
Query: 622 VRVVE---------LKNMYGSGIKALDNLAEELKSNNQLT--YED--LKSEVAKHSSALE 668
+ E L+N + L+ EE +S +L Y+ LK K + +E
Sbjct: 489 REIDEEAKEWEHKLLQNSMDKELYELNRRLEEKESEMKLFDGYDPAALKQHFGKKIAEVE 548
Query: 669 DLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQR-EAHSRAVETTRSVSKITMKFFET 727
D + + E + LL +++N +A H Q +A + + + ++ +
Sbjct: 549 DEKRSVQEERNRLLAEIENLASDGQAQKLQDVHAQNLKALEAQILDLKKKQESQVQLLKQ 608
Query: 728 IDRHASSLTQIVEETQFVNDQK------LCELEKKFEECTAYEEKQLLEKVAE------- 774
+ + ++ +E Q + QK + + ++F + A EK+LL+ E
Sbjct: 609 KQKSDDAARRLQDEIQSIKAQKVQLQHRMKQEAEQFRQWKASREKELLQLRKEGRKSEYE 668
Query: 775 --MLASSNARKKKLVQ-------MAVNDLRESANCRTSKLQREAL------TMQDSTSFV 819
L + N R+K ++Q MA L+E R S + + T +
Sbjct: 669 RHKLQALNQRQKMVLQRKTEEAAMATKRLKELLEARKSSPREHSAGTNGFGTNGQTNEKS 728
Query: 820 KAEWMVHMEKTESNYHE 836
W+ H + N HE
Sbjct: 729 LQRWLDHELEVMVNVHE 745
>At3g50240 kinesin-related protein (kicp-02)
Length = 1051
Score = 247 bits (631), Expect = 2e-65
Identities = 235/847 (27%), Positives = 396/847 (46%), Gaps = 106/847 (12%)
Query: 32 ADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQ 91
+ S+S+S + V+V V RP+ DE+ +G RE +V + +
Sbjct: 8 SSSSSSSPPSSLSSESCCVKVAVNVRPLIGDEV----------TQGCRECVSVSPVTPQV 57
Query: 92 IDRT--FVFDKVFGPNSQQKEL-YDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGG 148
T F FD V+G N L +++ V+P+V + GYN T+ AYGQTG+GKTYTM G
Sbjct: 58 QMGTHPFTFDHVYGSNGSPSSLMFEECVAPLVDGLFHGYNATVLAYGQTGSGKTYTMGTG 117
Query: 149 AIKKNGEFPTDAGVIPRAVKQIFDILEAQSAE--YSMKVTFLELYNEEITDLLAPE---- 202
K+G T G+IP+ + +F+ +++ + + + V+F+E+ EE+ DLL
Sbjct: 118 I--KDG---TKNGLIPQVMSALFNKIDSVKHQMGFQLHVSFIEILKEEVLDLLDSSVPFN 172
Query: 203 ---ETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETL 259
T SK P+ + E G + + G E + T E+ LE+GS R T T
Sbjct: 173 RLANGTPGKVVLSKSPVQIRESPNGVITLSGATEVPIATKEEMASCLEQGSLTRATGSTN 232
Query: 260 LNKQSSRSHSIFSITIH----------IKECTPE--GEEMIKCGKLNLVDLAGSENISRS 307
+N +SSRSH+IF+IT+ +K+ E GEE C KL+LVDLAGSE R+
Sbjct: 233 MNNESSRSHAIFTITLEQMRKISSISVVKDTVDEDMGEEYC-CAKLHLVDLAGSERAKRT 291
Query: 308 GAREGRAREAGEINKSLLTLGRTINALVEHS-----GHVPYRDSKLTRLLRDSLGGKTKT 362
G+ R +E IN+ LL LG I+AL + HVPYRDSKLTRLL+DSLGG +KT
Sbjct: 292 GSGGVRLKEGIHINRGLLALGNVISALGDEKRRKEGAHVPYRDSKLTRLLQDSLGGNSKT 351
Query: 363 CIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVY 422
+IA +SP+ EETL+TL YA+RA+NI+NKP N+ ++ S M K + E+ L+ +
Sbjct: 352 VMIACISPADINAEETLNTLKYANRARNIQNKPVANKDLICSEMQK-MRQELQYLQATLC 410
Query: 423 A--AREKNGIYIPRDRYL-----NEEAEKKAMAEKIERMELD-----------ADSKDKN 464
A A + + R++ + NEE ++ + +R+ LD SKD
Sbjct: 411 ARGATSSEEVQVMREKIMKLESANEELSRELHIYRSKRVTLDYCNIDAQEDGVIFSKDDG 470
Query: 465 LVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFD-------------LEERHRQAN 511
L E +S ++ S + + + EE E L LEE+ +
Sbjct: 471 LKRGFESMDSDYEMSEATSGGISEDIGAAEEWEHALRQNSMGKELNELSKRLEEKESEMR 530
Query: 512 ATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRV-LI 570
E + + K EL + ++ E + ++V L + +R+ ++ +N +
Sbjct: 531 VCGIGTETIRQHFEKKMMELEKEKRTVQDERDMLLAEVEELAASSDRQAQVARDNHAHKL 590
Query: 571 QKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEK----DMQSFVSTKSEATEDLRVRVVE 626
+ ++Q+ + V V+ Q+Q+ +D K ++Q + K + + ++ +
Sbjct: 591 KALETQILNLKKKQENQV--EVLKQKQKSEDAAKRLKTEIQCIKAQKVQLQQKMKQEAEQ 648
Query: 627 LKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEV--AKHSSALEDLFKGIALEADSLLND 684
+ S K L L +E + +E LK E + L+ + A+ A L +
Sbjct: 649 FRQWKASQEKELLQLKKE---GRKTEHERLKLEALNRRQKMVLQRKTEEAAM-ATKRLKE 704
Query: 685 LQNSLHKQEANVTAFAHQQ---REAHSRA--------VETTRSVSKITMKFFETIDRHAS 733
L + +++ A+ Q R+ + ++ +E V ++ ++ + I A+
Sbjct: 705 LLEARKSSPHDISVIANGQPPSRQTNEKSLRKWLDNELEVMAKVHQVRFQYEKQIQVRAA 764
Query: 734 ---SLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEML-ASSNARKKKLVQM 789
LT + +E +F ++ E +F + + + + ML SSNA Q+
Sbjct: 765 LAVELTSLRQEMEFPSNSHQ-EKNGQFRFLSPNTRLERIASLESMLDVSSNALTAMGSQL 823
Query: 790 AVNDLRE 796
+ + RE
Sbjct: 824 SEAEERE 830
>At2g21380 putative kinesin heavy chain
Length = 1058
Score = 234 bits (596), Expect = 3e-61
Identities = 235/846 (27%), Positives = 389/846 (45%), Gaps = 88/846 (10%)
Query: 49 NVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRT-FVFDKVFGPNSQ 107
++ V VR RPM+E E + +V + A + + N+ T + FDKVFGP S
Sbjct: 104 SISVTVRFRPMSEREYQRGDEIVWYPD-------ADKMVRNEYNPLTAYAFDKVFGPQST 156
Query: 108 QKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAV 167
E+YD A P+V +EG N T+FAYG T +GKT+TM G + +FP G+IP A+
Sbjct: 157 TPEVYDVAAKPVVKAAMEGVNGTVFAYGVTSSGKTHTMHG-----DQDFP---GIIPLAI 208
Query: 168 KQIFDIL-EAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGV 226
K +F I+ E E+ ++V++LE+YNE I DLL P + E S+ G
Sbjct: 209 KDVFSIIQETTGREFLLRVSYLEIYNEVINDLLDPTGQNLRIREDSQ-----------GT 257
Query: 227 LVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEE 286
V G++EE+V + + G R N SSRSH+IF++ I E + G++
Sbjct: 258 YVEGIKEEVVLSPGHALSFIAAGEEHRHVGSNNFNLMSSRSHTIFTLMI---ESSAHGDQ 314
Query: 287 M--IKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVE-HSGHVPY 343
+ +LNL+DLAGSE+ S++ R +E INKSLLTLG I L E + HVP+
Sbjct: 315 YDGVIFSQLNLIDLAGSES-SKTETTGLRRKEGAYINKSLLTLGTVIGKLTEGKTTHVPF 373
Query: 344 RDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMK 403
RDSKLTRLL+ SL G +I TV+P+ EET +TL +A RAK I+ N+ + +
Sbjct: 374 RDSKLTRLLQSSLSGHGHVSLICTVTPASSSTEETHNTLKFASRAKRIEINASRNKIIDE 433
Query: 404 SAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIE-RMELDADSKD 462
++IK EI LK E+ R + + + L+ + + + K++ R+E + ++K
Sbjct: 434 KSLIKKYQKEISTLKVELDQLRRGVLVGVSHEELLSLKQQLQEGQVKMQSRLEEEEEAKA 493
Query: 463 KNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEK---EF 519
+ +Q+L L T+ S+ L D R +A +K
Sbjct: 494 ALMSRIQKL----------TKLILVSTKNSI---PGYLGDTPAHSRSISAGKDDKLDSLL 540
Query: 520 LISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKI--EEENRVLIQKFQ--- 574
L S+ L S + A + R + S + S+ E + +E +L+++ +
Sbjct: 541 LDSDNLASPSSTLSLASDARRSSSKFKDENSPVGSRAELTQGVMTPDEMDLLVEQVKMLA 600
Query: 575 SQLAQQLEALHRTVSASVMHQEQ---QLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMY 631
++A L R V S+ E Q++++E D+Q K + L R+ E
Sbjct: 601 GEIAFGTSTLKRLVDQSMNDPENSKTQIQNLENDIQE----KQRQMKSLEQRITESGEAS 656
Query: 632 GSGIKALDNLAEELKSNNQLTYEDLKSE-VAKHSSALEDLFKGIALEADSLLNDLQNSLH 690
+ +++ + ++ Q + + E ++ + L++ + E N+L +H
Sbjct: 657 IANASSIEMQEKVMRLMTQCNEKSFELEIISADNRILQEQLQTKCTEN----NELHEKVH 712
Query: 691 KQEANVTAFAHQQREAHSRAVETTRSVSKITMKF-FETIDRHASSL--TQIVEETQ--FV 745
E +++ Q+ V T V ++ K + I+ L Q VEE V
Sbjct: 713 LLEQRLSS---QKATLSCCDVVTEEYVDELKKKVQSQEIENEKLKLEHVQSVEEKSGLRV 769
Query: 746 NDQKLCE---LEKKFEECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRT 802
+QKL E K+ A E K L ++V ++ + +K+LV A DL +A R
Sbjct: 770 QNQKLAEEASYAKELASAAAIELKNLADEVTKLSLQNAKLEKELV--AARDLAAAAQKRN 827
Query: 803 SKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVE-SGKKDLAEVLQICLNKAEV 861
+ A + KA N +++ ++E +K VL+ L + E
Sbjct: 828 NNSMNSAANRNGTRPGRKAR-----ISDSWNLNQENLTMELQARKQREAVLEAALAEKEY 882
Query: 862 GSQQWR 867
+++R
Sbjct: 883 IEEEFR 888
Score = 42.0 bits (97), Expect = 0.002
Identities = 109/566 (19%), Positives = 214/566 (37%), Gaps = 102/566 (18%)
Query: 311 EGRAREAGEINK-SLLTLGRTINALVEHSGHVPY--------RDSKLTRLLRDS--LGGK 359
E +A I K + L L T N++ + G P +D KL LL DS L
Sbjct: 490 EAKAALMSRIQKLTKLILVSTKNSIPGYLGDTPAHSRSISAGKDDKLDSLLLDSDNLASP 549
Query: 360 TKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQ 419
+ T +A+ + S+ + + ++ E+ Q +M + L ++ L
Sbjct: 550 SSTLSLAS--------DARRSSSKFKDENSPVGSRAELTQGVMTPDEMDLLVEQVKMLAG 601
Query: 420 EVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLT 479
E+ + D+ +N+ K +I+ +E D K + + L++ +
Sbjct: 602 EIAFGTSTLKRLV--DQSMNDPENSKT---QIQNLENDIQEKQRQMKSLEQRITESGEAS 656
Query: 480 AELSAKLEKTEKSLE---ETEQTLFDLE---------ERHRQANAT----IKEKEFLISN 523
++ +E EK + + + F+LE + Q T + EK L+
Sbjct: 657 IANASSIEMQEKVMRLMTQCNEKSFELEIISADNRILQEQLQTKCTENNELHEKVHLLEQ 716
Query: 524 LLKSEKELV--------ERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQK--- 572
L S+K + E EL+ ++++ ++ N K+E +EE++ + +Q
Sbjct: 717 RLSSQKATLSCCDVVTEEYVDELKKKVQS--QEIENEKLKLEHVQSVEEKSGLRVQNQKL 774
Query: 573 -----FQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVEL 627
+ +LA + ++ V Q +EK++ + + A + +
Sbjct: 775 AEEASYAKELASAAAIELKNLADEVTKLSLQNAKLEKELVAARDLAAAAQKRNNNSMNSA 834
Query: 628 KNMYGS--GIKAL---------DNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIAL 676
N G+ G KA +NL EL++ Q E + +E+ F+ A
Sbjct: 835 ANRNGTRPGRKARISDSWNLNQENLTMELQARKQR--EAVLEAALAEKEYIEEEFRKKAE 892
Query: 677 EADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLT 736
EA L+N L AN+ + ++A+S A+ +S
Sbjct: 893 EAKRREEALENDL----ANMWVLVAKLKKANSGALSIQKS-------------------- 928
Query: 737 QIVEETQFVNDQKLCELEKKFEECTAYEEKQLL---EKVAEMLASSNARKKKLVQMAVND 793
+E + + ++ EL+ K E+ +E+QL+ E+V A +++ LV
Sbjct: 929 ---DEAEPAKEDEVTELDNKNEQNAILKERQLVNGHEEVIVAKAEETPKEEPLVARLKAR 985
Query: 794 LRESANCRTSKLQREALTMQDSTSFV 819
++E + K Q A D+ S +
Sbjct: 986 MQEMKE-KEMKSQAAAAANADANSHI 1010
>At3g12020 unknown protein
Length = 956
Score = 231 bits (590), Expect = 1e-60
Identities = 216/795 (27%), Positives = 379/795 (47%), Gaps = 99/795 (12%)
Query: 49 NVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRT--FVFDKVFGPNS 106
NV V VR RP++ E+R V A ++I + + T + +D+VFGP +
Sbjct: 69 NVTVTVRFRPLSPREIRQGEEVAW--------YADGETIVRNEHNPTIAYAYDRVFGPTT 120
Query: 107 QQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRA 166
+ +YD A +V +EG N TIFAYG T +GKT+TM G G+IP A
Sbjct: 121 TTRNVYDIAAHHVVNGAMEGINGTIFAYGVTSSGKTHTMHGDQ--------RSPGIIPLA 172
Query: 167 VKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGV 226
VK F L ++++++E+YNE + DLL P + + + ED K G
Sbjct: 173 VKDAFKFL--------LRISYMEIYNEVVNDLLNP----------AGHNLRIRED-KQGT 213
Query: 227 LVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGE- 285
V G++EE+V + ++ G +R T N SSRSH+IF++TI E +P G+
Sbjct: 214 FVEGIKEEVVLSPAHALSLIAAGEEQRHVGSTNFNLLSSRSHTIFTLTI---ESSPLGDK 270
Query: 286 ---EMIKCGKLNLVDLAGSEN--ISRSGAREGRAREAGEINKSLLTLGRTINALVE-HSG 339
E + +LNLVDLAGSE+ + SG R +E INKSLLTLG I+ L + +
Sbjct: 271 SKGEAVHLSQLNLVDLAGSESSKVETSGVRR---KEGSYINKSLLTLGTVISKLTDVRAS 327
Query: 340 HVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQ 399
HVPYRDSKLTR+L+ SL G + +I TV+P+ EET +TL +AHRAK+I+ + E N+
Sbjct: 328 HVPYRDSKLTRILQSSLSGHDRVSLICTVTPASSSSEETHNTLKFAHRAKHIEIQAEQNK 387
Query: 400 KMMKSAMIKDLYSEIDRLKQEVYAARE-----------------------KNGIYIPRDR 436
+ + ++IK EI +LK+E+ ++ ++G + R
Sbjct: 388 IIDEKSLIKKYQREIRQLKEELEQLKQEIVPVPQLKDIGADDIVLLKQKLEDGQVKLQSR 447
Query: 437 YLNEEAEKKAMAEKIERME--LDADSKDKNLVELQELYNSQQLLT---AELSAKLEKTEK 491
EE K A+ +I+R+ + +K+ L +N ++ + EL+ K
Sbjct: 448 LEEEEEAKAALLSRIQRLTKLILVSTKNPQASRLPHRFNPRRRHSFGEEELAYLPYKRRD 507
Query: 492 SLEETEQTLF-DLEERHR-QANATIKEKEFL---ISNLLKSEKELVERAIELRAELENAA 546
+++ + L+ +E H + NA +EK+ + N LK +K + ++ + +
Sbjct: 508 MMDDEQLDLYVSVEGNHEIRDNAYREEKKTRKHGLLNWLKPKKRDHSSSASDQSSVVKSN 567
Query: 547 SDVSNL---FSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDME 603
S S S + + ++ E + ++ Q S+ + EAL S+ M + M
Sbjct: 568 STPSTPQGGGSHLHTESRLSEGSPLMEQ--LSEPREDREALED--SSHEMEIPETSNKMS 623
Query: 604 KDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKH 663
++ K +E+ +++ LK M K+ N EE+ ++ +D+K++ +
Sbjct: 624 DELDLLREQKKILSEEAALQLSSLKRMSDEAAKSPQN--EEINEEIKVLNDDIKAKNDQI 681
Query: 664 SSALEDLFKGIALEADSL-LNDLQNSLH--KQEANVTAFAHQQREAHSRAVETTRSVSKI 720
++ + + ++L +D+ ++ + + N +F + + A +R ++ T +
Sbjct: 682 ATLERQIMDFVMTSHEALDKSDIMQAVAELRDQLNEKSFELEVKAADNRIIQQTLNEKTC 741
Query: 721 TMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSN 780
+ + + A+ Q+ E + K+ EL++ +E + E K+ LE LA +
Sbjct: 742 ECEVLQ--EEVANLKQQLSEALELAQGTKIKELKQDAKELS--ESKEQLELRNRKLAEES 797
Query: 781 ARKKKLVQMAVNDLR 795
+ K L A +L+
Sbjct: 798 SYAKGLASAAAVELK 812
>At1g18370 AtNACK1 kinesin-like protein (AtNACK1)
Length = 974
Score = 230 bits (587), Expect = 3e-60
Identities = 216/789 (27%), Positives = 356/789 (44%), Gaps = 101/789 (12%)
Query: 50 VQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQK 109
+ V VR RPMN+ E+ V C V+ Q +F FDKVFGP S +
Sbjct: 32 IVVTVRLRPMNKRELLAKDQVAWECVNDHTIVSKPQVQERLHHQSSFTFDKVFGPESLTE 91
Query: 110 ELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQ 169
+Y+ V + L G N TIFAYGQT +GKTYTM G V +AV
Sbjct: 92 NVYEDGVKNVALSALMGINATIFAYGQTSSGKTYTMRG--------------VTEKAVND 137
Query: 170 IFD-ILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLV 228
I++ I++ ++++K++ LE+YNE + DLL + S + + L++D + G +V
Sbjct: 138 IYNHIIKTPERDFTIKISGLEIYNENVRDLL---------NSDSGRALKLLDDPEKGTVV 188
Query: 229 RGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMI 288
L EE N + ++ A+R+ ET LN SSRSH I +TI + E + +
Sbjct: 189 EKLVEETANNDNHLRHLISICEAQRQVGETALNDTSSRSHQIIRLTI--QSTHRENSDCV 246
Query: 289 KC--GKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALV--EHSGHVPYR 344
+ LN VDLAGSE S+S A R RE IN SL+TL I L + SGH+PYR
Sbjct: 247 RSYMASLNFVDLAGSERASQSQADGTRLREGCHINLSLMTLTTVIRKLSVGKRSGHIPYR 306
Query: 345 DSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKS 404
DSKLTR+L+ SLGG +T II T+SP++ +E++ +TL +A+RAK + N VN +
Sbjct: 307 DSKLTRILQHSLGGNARTAIICTLSPALAHVEQSRNTLYFANRAKEVTNNAHVNMVVSDK 366
Query: 405 AMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKN 464
++K L E+ RL+ E R G +D KI++ME++
Sbjct: 367 QLVKHLQKEVARLEAE----RRTPGPSTEKD-------------FKIQQMEME------- 402
Query: 465 LVELQELYNSQQLLTAELSAKL---EKTEKSLEETEQTLFDLEERHRQANATIKEKEFLI 521
+ EL+ + Q+ EL KL ++ K L E + + + A E
Sbjct: 403 IGELRRQRDDAQIQLEELRQKLQGDQQQNKGLNPFESPDPPVRKCLSYSVAVTPSSE--- 459
Query: 522 SNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQL 581
N + E + ++ + +++ L +I + + ++E+ K L +++
Sbjct: 460 -NKTLNRNERARKTTMRQSMIRQSSTAPFTLMHEIRKLEHLQEQLGEEATKALEVLQKEV 518
Query: 582 EALHR----TVSASVMHQEQQLKDME--------KDMQSFVSTKSEATEDLRVRVVELKN 629
A HR + ++ + ++++M K++ ++ + +L+ + L +
Sbjct: 519 -ACHRLGNQDAAQTIAKLQAEIREMRTVKPSAMLKEVGDVIAPNKSVSANLKEEITRLHS 577
Query: 630 MYGSGI-----------KALDNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEA 678
GS I K++D L L SN E K++ H S + L A
Sbjct: 578 Q-GSTIANLEEQLESVQKSIDKLVMSLPSNISAGDETPKTKNHHHQSKKKKLLPLTPSSA 636
Query: 679 DSLLNDLQN-----SLHKQEANVTAFAHQQREAHS---RAVETTRSVSKIT-MKFFETID 729
+ N L++ S +Q + A +E +S R T + K T K E+ D
Sbjct: 637 SNRQNFLKSPCSPLSASRQVLDCDAENKAPQENNSSAARGATTPQGSEKETPQKGEESGD 696
Query: 730 RHASSLTQIVEETQFVNDQKLCEL-----EKKFEECTAYEEKQLLEKVAEMLASSNARKK 784
+ T + VN +K+ ++ E+ AY +L E+VA++
Sbjct: 697 VSSREGTPGYRRSSSVNMKKMQQMFQNAAEENVRSIRAY-VTELKERVAKLQYQKQLLVC 755
Query: 785 KLVQMAVND 793
+++++ ND
Sbjct: 756 QVLELEAND 764
>At1g12430 kinesin-like protein
Length = 919
Score = 229 bits (583), Expect = 8e-60
Identities = 182/617 (29%), Positives = 291/617 (46%), Gaps = 73/617 (11%)
Query: 50 VQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQK 109
V+V VR RP N +E+ C E + E+ ++ N TF FD+V + QK
Sbjct: 71 VRVAVRLRPRNGEELIADADFA-DCVELQPELKRLKLRKNNWDTDTFEFDEVLTEYASQK 129
Query: 110 ELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQ 169
+Y+ P+V VL+GYN TI AYGQTGTGKTYT+ + E D G++ RA++
Sbjct: 130 RVYEVVAKPVVEGVLDGYNGTIMAYGQTGTGKTYTLG----QLGEEDVADRGIMVRAMED 185
Query: 170 IFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGG-VLV 228
I + E S+ V++L+LY E + DLL P S IA++ED K G V +
Sbjct: 186 I--LAEVSLETDSISVSYLQLYMETVQDLLDP----------SNDNIAIVEDPKNGDVSL 233
Query: 229 RGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEG---- 284
G + ++L+ G A R A T LN +SSRSH+I + + T +G
Sbjct: 234 PGATLVEIRDQQSFLELLQLGEAHRFAANTKLNTESSRSHAILMVNVRRSMKTRDGLSSE 293
Query: 285 ------------EEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTIN 332
+++ GKL +VDLAGSE I++SG+ EA IN SL LG+ IN
Sbjct: 294 SNGNSHMTKSLKPPVVRKGKLVVVDLAGSERINKSGSEGHTLEEAKSINLSLSALGKCIN 353
Query: 333 ALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIK 392
AL E+S HVP+RDSKLTRLLRDS GG +T ++ T+ PS ET ST+ + RA ++
Sbjct: 354 ALAENSSHVPFRDSKLTRLLRDSFGGTARTSLVITIGPSPRHRGETTSTIMFGQRAMKVE 413
Query: 393 NKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIE 452
N ++ ++ ++ + L ++D L +E E ++KA ++IE
Sbjct: 414 NMVKIKEEFDYKSLSRRLEVQLDNLIEE-------------------NERQQKAFVDEIE 454
Query: 453 RMEL-------DADSKDKNLVELQEL--YNSQQLLTAELSAKLEKTEKSLEET-----EQ 498
R+ + +A+ + N +E ++L N +L K +K L E+
Sbjct: 455 RITVEAHNQISEAEKRYANALEDEKLRYQNDYMESIKKLEENWSKNQKKLAAERLALGEK 514
Query: 499 TLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELE-NAASDVSNLFSKIE 557
D+ ++ A E+ + LL+ E + ++ AE E N N F K+E
Sbjct: 515 NGLDITSNGNRSIAPALEEVSELKKLLQKEAQ-----SKMAAEEEVNRLKHQLNEFKKVE 569
Query: 558 RKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEAT 617
E + + ++Q ++LE T+ + ++ + ++++ S K+
Sbjct: 570 ASGNSEIMRLHKMLENETQQKEKLEGEIATLHSQLLQLSLTADETRRNLEQHGSEKTSGA 629
Query: 618 EDLRVRVVELKNMYGSG 634
D + + L + G
Sbjct: 630 RDSLMSQLRLPQIQDPG 646
>At3g10180 putative kinesin-like centromere protein
Length = 459
Score = 227 bits (578), Expect = 3e-59
Identities = 146/404 (36%), Positives = 222/404 (54%), Gaps = 43/404 (10%)
Query: 97 VFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEF 156
+ D++F + + ++Y+ IV + G+N T+FAYGQT +GKT+TM G
Sbjct: 43 ITDRIFREDCKTVQVYEARTKEIVSAAVRGFNGTVFAYGQTNSGKTHTMRGS-------- 94
Query: 157 PTDAGVIPRAVKQIFD-ILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKP 215
P + GVIP AV +FD I + S E+ +++++LE+YNE+I DLLAPE + E +K
Sbjct: 95 PIEPGVIPLAVHDLFDTIYQDASREFLLRMSYLEIYNEDINDLLAPEHRKLQIHENLEK- 153
Query: 216 IALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITI 275
G+ V GL EEIV + ++ +++E G + R ET +N SSRSH+IF + I
Sbjct: 154 ---------GIFVAGLREEIVASPQQVLEMMEFGESHRHIGETNMNLYSSRSHTIFRMII 204
Query: 276 HIKE-CTPEGE----EMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRT 330
++ EG + ++ LNLVDLAGSE +++GA R +E INKSL+TLG
Sbjct: 205 ESRQKMQDEGVGNSCDAVRVSVLNLVDLAGSERAAKTGAEGVRLKEGSHINKSLMTLGTV 264
Query: 331 INALVE----HSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAH 386
I L E GHVPYRDSKLTR+L+ +LGG T II ++ + +ET S+L +A
Sbjct: 265 IKKLSEGVETQGGHVPYRDSKLTRILQPALGGNANTAIICNITLAPIHADETKSSLQFAS 324
Query: 387 RAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNG-------------IYIP 433
RA + N VN+ + +A++K EI+ L+ ++ + + +
Sbjct: 325 RALRVTNCAHVNEILTDAALLKRQKKEIEELRSKLKTSHSDHSEEEILNLRNTLLKSELE 384
Query: 434 RDRY-LNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQ 476
R+R L E EKKA A++ ER+ + K KNL + L N +
Sbjct: 385 RERIALELEEEKKAQAQR-ERVLQEQAKKIKNLSSMVLLSNRDE 427
>At3g49650 kinesin-like protein
Length = 813
Score = 224 bits (571), Expect = 2e-58
Identities = 166/530 (31%), Positives = 271/530 (50%), Gaps = 55/530 (10%)
Query: 45 EKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAA--------VQSIANKQIDRTF 96
+K + V V+CRP+ E E ++ N + V + I N+ ++ +
Sbjct: 10 KKTTTLTVAVKCRPLMEKERGRD---IVRVNNSKEVVVLDPDLSKDYLDRIQNRTKEKKY 66
Query: 97 VFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEF 156
FD FGP S K +Y +++S ++ V+ G N T+FAYG TG+GKTYTM G
Sbjct: 67 CFDHAFGPESTNKNVY-RSMSSVISSVVHGLNATVFAYGSTGSGKTYTMVGTR------- 118
Query: 157 PTDAGVIPRAVKQIFDILEAQ--SAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKK 214
+D G++ ++ IFD++++ S E+ + ++LE+YNE I DLL EKS
Sbjct: 119 -SDPGLMVLSLNTIFDMIKSDKSSDEFEVTCSYLEVYNEVIYDLL----------EKSSG 167
Query: 215 PIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSIT 274
+ L ED + G++V GL V +A+ I ++L G+++R+T T +N SSRSH++ I
Sbjct: 168 HLELREDPEQGIVVAGLRSIKVHSADRILELLNLGNSRRKTESTEMNGTSSRSHAVLEIA 227
Query: 275 IHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINAL 334
+ ++ + + + GKL LVDLAGSE + + + R+ IN+SLL L INAL
Sbjct: 228 VKRRQ---KNQNQVMRGKLALVDLAGSERAAETNNGGQKLRDGANINRSLLALANCINAL 284
Query: 335 VEHS----GHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKN 390
+ +VPYR+SKLTR+L+D L G ++T ++AT+SP+ T++TL YA RAK
Sbjct: 285 GKQHKKGLAYVPYRNSKLTRILKDGLSGNSQTVMVATISPADSQYHHTVNTLKYADRAKE 344
Query: 391 IKNKPEVNQKMMKS------AMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAE- 443
IK + N + + MI +L SE+ +LK ++ A +E P +R + E
Sbjct: 345 IKTHIQKNIGTIDTHMSDYQRMIDNLQSEVSQLKTQL-AEKESQLSIKPFERGVERELSW 403
Query: 444 KKAMAEKIERMELDADSKDKNLVELQE--LYNSQQLLTAELSAKLEKTEKSLEETEQTLF 501
++ +I D + K L EL+E L N +L + + + TEK + E
Sbjct: 404 LDGLSHQISENVQDRINLQKALFELEETNLRNRTELQHLDDAIAKQATEKDVVEA----- 458
Query: 502 DLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSN 551
L R + I++ + N + +E + EL+ L A ++ N
Sbjct: 459 -LSSRRQVILDNIRDNDEAGVNYQRDIEENEKHRCELQDMLNEAINNNGN 507
>At3g43210 kinesin -like protein
Length = 932
Score = 223 bits (569), Expect = 3e-58
Identities = 189/655 (28%), Positives = 309/655 (46%), Gaps = 66/655 (10%)
Query: 25 PARDLRSADSNSNSH----NKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRRE 80
P R S SN + +K +EK + V VR RP+N E + + C + E
Sbjct: 3 PPRTPLSKIDKSNPYTPCGSKVTEEK---ILVTVRMRPLNWREHAKYDLIAWECPDD--E 57
Query: 81 VAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTG 140
++ + + FDKVF P +E+Y+ + L G N TIFAYGQT +G
Sbjct: 58 TIVFKNPNPDKAPTKYSFDKVFEPTCATQEVYEGGSRDVALSALAGTNATIFAYGQTSSG 117
Query: 141 KTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFD-ILEAQSAEYSMKVTFLELYNEEITDLL 199
KT+TM G V VK I++ I + Q + +KV+ LE+YNE + DLL
Sbjct: 118 KTFTMRG--------------VTESVVKDIYEHIRKTQERSFVLKVSALEIYNETVVDLL 163
Query: 200 APEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETL 259
+ P+ L++D + G +V L EE+V + + ++ +R+ ET
Sbjct: 164 ----------NRDTGPLRLLDDPEKGTIVENLVEEVVESRQHLQHLISICEDQRQVGETA 213
Query: 260 LNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGE 319
LN +SSRSH I +TIH G LNLVDLAGSE ++ A R +E
Sbjct: 214 LNDKSSRSHQIIRLTIHSSLREIAGCVQSFMATLNLVDLAGSERAFQTNADGLRLKEGSH 273
Query: 320 INKSLLTLGRTINALVE--HSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEE 377
IN+SLLTL I L HVPYRDSKLTR+L++SLGG +T II T+SP++ +E+
Sbjct: 274 INRSLLTLTTVIRKLSSGRKRDHVPYRDSKLTRILQNSLGGNARTAIICTISPALSHVEQ 333
Query: 378 TLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKN-----GIYI 432
T TL +A AK + N +VN + + ++K L ++ +L+ E+ + + + I
Sbjct: 334 TKKTLSFAMSAKEVTNCAKVNMVVSEKKLLKHLQQKVAKLESELRSPEPSSSTCLKSLLI 393
Query: 433 PRDRYLNE-EAEKKAMAEK--IERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKT 489
++ + + E+E K + + I + ELD + K K E Q+ E++
Sbjct: 394 EKEMKIQQMESEMKELKRQRDIAQSELDLERKAKERKGSSECEPFSQVARCLSYHTKEES 453
Query: 490 --EKSLEETEQTLFDLEERHRQANATIKEKEFLISN---LLKSEKELVERAIE----LRA 540
KS+ + +T D + + + + T + L+ L K +K+L E A + +
Sbjct: 454 IPSKSVPSSRRTARDRRKDNVRQSLTSADPTALVQEIRLLEKHQKKLGEEANQALDLIHK 513
Query: 541 EL-------ENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVM 593
E+ + AA V+ + S+I +++ N + + A E ++R S +
Sbjct: 514 EVTSHKLGDQQAAEKVAKMLSEIR---DMQKSNLLTEEIVVGDKANLKEEINRLNSQEIA 570
Query: 594 HQEQQLKDMEKDMQSFVST--KSEATEDLRVRVVELKNMYGSGIKALDNLAEELK 646
E++L+ ++ + VS+ E T D R + V+ K + G+ NL ++
Sbjct: 571 ALEKKLECVQNTIDMLVSSFQTDEQTPDFRTQ-VKKKRLLPFGLSNSPNLQHMIR 624
Score = 33.9 bits (76), Expect = 0.50
Identities = 50/238 (21%), Positives = 104/238 (43%), Gaps = 16/238 (6%)
Query: 523 NLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLE 582
N++ SEK+L++ + A+LE+ S + IE+E + IQ+ +S++ +
Sbjct: 354 NMVVSEKKLLKHLQQKVAKLESELRSPEPSSSTCLKSLLIEKEMK--IQQMESEMKELKR 411
Query: 583 ALHRTVSASVMHQEQQLKDME--KDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDN 640
R ++ S + E++ K+ + + + F + + + K++ S A D
Sbjct: 412 --QRDIAQSELDLERKAKERKGSSECEPFSQVARCLSYHTKEESIPSKSVPSSRRTARDR 469
Query: 641 LAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFA 700
+ ++ + LT D + + + LE K + EA+ L+ +HK+ +
Sbjct: 470 RKDNVRQS--LTSAD-PTALVQEIRLLEKHQKKLGEEANQALD----LIHKEVTSHKLGD 522
Query: 701 HQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFE 758
Q E ++ + R + K + E + ++L EE +N Q++ LEKK E
Sbjct: 523 QQAAEKVAKMLSEIRDMQKSNLLTEEIVVGDKANLK---EEINRLNSQEIAALEKKLE 577
>At1g72250 kinesin, putative
Length = 1195
Score = 223 bits (567), Expect = 6e-58
Identities = 174/536 (32%), Positives = 278/536 (51%), Gaps = 55/536 (10%)
Query: 39 HNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVF 98
+NK + KG N++V RCRP+N +E + I + V S N ++F F
Sbjct: 482 YNKILELKG-NIRVFCRCRPLNFEETEAGVSMGIDVESTKNGEVIVMS--NGFPKKSFKF 538
Query: 99 DKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPT 158
D VFGPN+ Q ++++ +P V++GYN IFAYGQTGTGKT+TMEG
Sbjct: 539 DSVFGPNASQADVFEDT-APFATSVIDGYNVCIFAYGQTGTGKTFTMEGTQ--------H 589
Query: 159 DAGVIPRAVKQIFDILEAQSAEYS--MKVTFLELYNEEITDLLAPEETTKFVDEKSKKPI 216
D GV R ++ +F I++A+ Y+ + V+ LE+YNE+I DLL P + + K
Sbjct: 590 DRGVNYRTLENLFRIIKAREHRYNYEISVSVLEVYNEQIRDLLVPASQSA----SAPKRF 645
Query: 217 ALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIH 276
+ + +G V GL E V + E++ +L+ GS R +T N+ SSRSH I + +
Sbjct: 646 EIRQLSEGNHHVPGLVEAPVKSIEEVWDVLKTGSNARAVGKTTANEHSSRSHCIHCVMV- 704
Query: 277 IKECTPEGEEMI--KC--GKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTIN 332
+GE ++ +C KL LVDLAGSE ++++ + R +E INKSL LG I
Sbjct: 705 ------KGENLLNGECTKSKLWLVDLAGSERVAKTEVQGERLKETQNINKSLSALGDVIF 758
Query: 333 ALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIK 392
AL S H+P+R+SKLT LL+DSLGG +KT + +SP+ + ETL +L++A R + I+
Sbjct: 759 ALANKSSHIPFRNSKLTHLLQDSLGGDSKTLMFVQISPNENDQSETLCSLNFASRVRGIE 818
Query: 393 NKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIE 452
P +K + + + +++ KQ++ E+ I + EA+ K
Sbjct: 819 LGPA--KKQLDNTELLKYKQMVEKWKQDMKGKDEQ--IRKMEETMYGLEAKIK------- 867
Query: 453 RMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANA 512
+ D+K+K L + + SQ L+ +L+ + T+ + ++T+Q D N
Sbjct: 868 ----ERDTKNKTLQDKVKELESQLLVERKLARQHVDTKIAEQQTKQQTED-------ENN 916
Query: 513 TIKEKEFLISNLLKSEKELVERAIELRAELENAAS-DVSNLFSKIER-KDKIEEEN 566
T K L + KE+V + + LE+ S D++ L S + + D E+EN
Sbjct: 917 TSKRPPLTNILLGSASKEMVN--LTRPSLLESTTSYDLAPLPSGVPKYNDLSEKEN 970
>At3g19050 hypothetical protein
Length = 539
Score = 222 bits (565), Expect = 1e-57
Identities = 144/364 (39%), Positives = 200/364 (54%), Gaps = 30/364 (8%)
Query: 41 KYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDK 100
K D ++ +L+R RP+N E ++ E + VA + + F FD
Sbjct: 183 KEDPSFWMDHNILIRVRPLNSMERSINGYNRCLKQESSQCVAWIGPPETR-----FQFDH 237
Query: 101 VFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDA 160
V Q+ L+ A P+V L GYN IFAYGQTG+GKTYTM G + +
Sbjct: 238 VACETIDQETLFRVAGLPMVENCLSGYNSCIFAYGQTGSGKTYTMLGEVGDLEFKPSPNR 297
Query: 161 GVIPRAVKQIFDILEAQSA-------EYSMKVTFLELYNEEITDLLAPEETTKFVDEKSK 213
G++PR + +F ++A+ +Y+ K +FLE+YNE+ITDLL P T
Sbjct: 298 GMMPRIFEFLFARIQAEEESRRDERLKYNCKCSFLEIYNEQITDLLEPSSTN-------- 349
Query: 214 KPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSI 273
++D K GV V L E V + +I ++ +GS RR T +N++SSRSHS+F+
Sbjct: 350 -----LQDIKSGVYVENLTECEVQSVQDILGLITQGSLNRRVGATNMNRESSRSHSVFTC 404
Query: 274 TIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINA 333
I + + ++ +LNLVDLAGSE SGA R +EA INKSL TLG I
Sbjct: 405 VIESR-WEKDSTANMRFARLNLVDLAGSERQKTSGAEGDRLKEAASINKSLSTLGHVIMV 463
Query: 334 LVEHSG----HVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAK 389
LV+ + H+PYRDS+LT LL+DSLGG +KT IIA SPS+ C ETL+TL +A RAK
Sbjct: 464 LVDVANGKPRHIPYRDSRLTFLLQDSLGGNSKTMIIANASPSVSCAAETLNTLKFAQRAK 523
Query: 390 NIKN 393
I+N
Sbjct: 524 LIQN 527
>At1g01950 unknown protein
Length = 893
Score = 218 bits (554), Expect = 2e-56
Identities = 180/595 (30%), Positives = 278/595 (46%), Gaps = 76/595 (12%)
Query: 16 SQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCN 75
S+ P S P R S+S S D V+V VR RP N DE C
Sbjct: 33 SRIPSSAPAPRR------SSSASIGAADNGVPGRVRVAVRLRPRNADESVADADFA-DCV 85
Query: 76 EGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYG 135
E + E+ ++ N T+ FD+V + QK +Y+ P+V EGYN T+ AYG
Sbjct: 86 ELQPELKRLKLRKNNWDTETYEFDEVLTEAASQKRVYEVVAKPVV----EGYNGTVMAYG 141
Query: 136 QTGTGKTYTMEGGAIKKNGEFPTDA-GVIPRAVKQIFDILEAQSAEY-SMKVTFLELYNE 193
QTGTGKT+T+ + G+ T A G++ R+++ DI+ S + S+ V++L+LY E
Sbjct: 142 QTGTGKTFTLG-----RLGDEDTAARGIMVRSME---DIIGGTSLDTDSISVSYLQLYME 193
Query: 194 EITDLLAPEETTKFVDEKSKKPIALMEDGK-GGVLVRGLEEEIVCTANEIYKILEKGSAK 252
I DLL P IA++ED + G V + G + ++L+ G
Sbjct: 194 TIQDLLDPTNDN----------IAIVEDPRTGDVSLPGATHVEIRNQQNFLELLQLGETH 243
Query: 253 RRTAETLLNKQSSRSHSIFSITIHIKECTPEGE------------------EMIKCGKLN 294
R A T LN +SSRSH+I + +H+K E E +++ KL
Sbjct: 244 RVAANTKLNTESSRSHAI--LMVHVKRSVVENEFPVSNEMESSSHFVRPSKPLVRRSKLV 301
Query: 295 LVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRD 354
LVDLAGSE + +SG+ EA IN SL LG+ INA+ E+S HVP RDSKLTRLLRD
Sbjct: 302 LVDLAGSERVHKSGSEGHMLEEAKSINLSLSALGKCINAIAENSPHVPLRDSKLTRLLRD 361
Query: 355 SLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEI 414
S GG +T +I T+ PS ET ST+ + RA ++N ++ ++ ++ K L ++
Sbjct: 362 SFGGTARTSLIVTIGPSPRHRGETTSTILFGQRAMKVENMLKIKEEFDYKSLSKKLEVQL 421
Query: 415 DRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNS 474
D++ E E + KA + +ER+ A +N + E +
Sbjct: 422 DKVIAE-------------------NERQLKAFDDDVERINRQA----QNRISEVEKNFA 458
Query: 475 QQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVER 534
+ L +L ++E E S+++ E+ L + H + + ++ KE +E
Sbjct: 459 EALEKEKLKCQMEYME-SVKKLEEKLISNQRNHENGKRNGEVNGVVTASEFTRLKESLEN 517
Query: 535 AIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVS 589
++LR E S V + + R + E+ +QK A Q + L V+
Sbjct: 518 EMKLRKSAEEEVSKVKSQSTLKTRSGEGEDAGITRLQKLLEDEALQKKKLEEEVT 572
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.127 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,217,796
Number of Sequences: 26719
Number of extensions: 904235
Number of successful extensions: 4530
Number of sequences better than 10.0: 416
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 310
Number of HSP's that attempted gapping in prelim test: 3314
Number of HSP's gapped (non-prelim): 816
length of query: 1049
length of database: 11,318,596
effective HSP length: 109
effective length of query: 940
effective length of database: 8,406,225
effective search space: 7901851500
effective search space used: 7901851500
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC149490.9


