

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.2 - phase: 1
(317 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
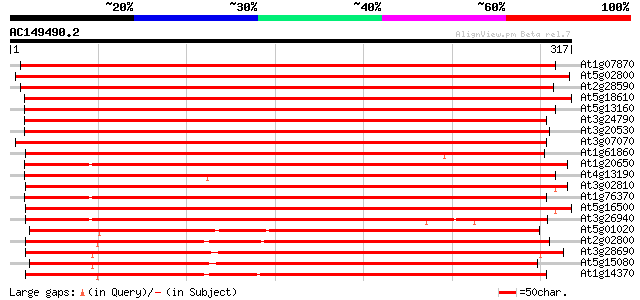
Score E
Sequences producing significant alignments: (bits) Value
At1g07870 putative protein kinase (At1g07870) 496 e-141
At5g02800 protein kinase - like 489 e-138
At2g28590 putative protein kinase 484 e-137
At5g18610 protein kinase -like protein 468 e-132
At5g13160 protein kinase-like 439 e-124
At3g24790 protein kinase, putative 410 e-115
At3g20530 protein kinase, putative 408 e-114
At3g07070 putative protein kinase 397 e-111
At1g61860 hypothetical protein 393 e-110
At1g20650 unknown protein 389 e-108
At4g13190 putative protein 379 e-105
At3g02810 putative protein kinase 373 e-104
At1g76370 putative protein kinase 370 e-103
At5g16500 protein kinase-like protein 360 e-100
At3g26940 protein kinase, putative 337 4e-93
At5g01020 protein kinase -like protein 312 1e-85
At2g02800 protein kinase like protein 311 2e-85
At3g28690 protein kinase like protein 308 2e-84
At5g15080 serine/threonine specific protein kinase -like 308 3e-84
At1g14370 putative protein 307 4e-84
>At1g07870 putative protein kinase (At1g07870)
Length = 423
Score = 496 bits (1276), Expect = e-141
Identities = 239/302 (79%), Positives = 267/302 (88%)
Query: 7 KVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREF 66
K A+ FTF ELA AT +FR DCF+GEGGFGKV+KG I+K++Q VAIKQLD NG+QG REF
Sbjct: 86 KKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREF 145
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRM 126
VEVLTLSLA+HPNLVKL+GFCAEG+QRLLVYEYMP GSLE+HLH LP GKKPLDWNTRM
Sbjct: 146 VVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRM 205
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+IAAG A+GLEYLHD M PPVIYRDLKCSNILLG+DY PKLSDFGLAKVGP GD THVST
Sbjct: 206 KIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST 265
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RVMGTYGYCAPDYAMTGQLT KSDIYS GV LLELITGRKA D +K K+QNLV WA PL
Sbjct: 266 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPL 325
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKY 306
FK++R F KMVDPLL+GQYP RGLYQALA++AMCV+EQ +MRPV++DVV AL+F+AS KY
Sbjct: 326 FKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKY 385
Query: 307 DP 308
DP
Sbjct: 386 DP 387
>At5g02800 protein kinase - like
Length = 395
Score = 489 bits (1258), Expect = e-138
Identities = 230/313 (73%), Positives = 268/313 (85%)
Query: 4 TDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGT 63
+D VA+ FTF ELA AT++FR +C +GEGGFG+VYKGY+ +Q AIKQLD NGLQG
Sbjct: 53 SDHIVAQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTSQTAAIKQLDHNGLQGN 112
Query: 64 REFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWN 123
REF VEVL LSL HPNLV L+G+CA+G+QRLLVYEYMPLGSLE+HLHD+ PGK+PLDWN
Sbjct: 113 REFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPGKQPLDWN 172
Query: 124 TRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTH 183
TRM+IAAG AKGLEYLHD+ PPVIYRDLKCSNILL DDY PKLSDFGLAK+GP+GD +H
Sbjct: 173 TRMKIAAGAAKGLEYLHDKTMPPVIYRDLKCSNILLDDDYFPKLSDFGLAKLGPVGDKSH 232
Query: 184 VSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWA 243
VSTRVMGTYGYCAP+YAMTGQLT KSD+YS GV LLE+ITGRKA D S+ EQNLVAWA
Sbjct: 233 VSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDSSRSTGEQNLVAWA 292
Query: 244 YPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
PLFK++RKFS+M DP+L+GQYP RGLYQALAVAAMCV+EQ ++RP+IADVV AL ++AS
Sbjct: 293 RPLFKDRRKFSQMADPMLQGQYPPRGLYQALAVAAMCVQEQPNLRPLIADVVTALSYLAS 352
Query: 304 HKYDPQVHPIQSS 316
K+DP P+Q S
Sbjct: 353 QKFDPLAQPVQGS 365
>At2g28590 putative protein kinase
Length = 424
Score = 484 bits (1245), Expect = e-137
Identities = 233/301 (77%), Positives = 261/301 (86%)
Query: 7 KVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREF 66
K A+ FTF+EL+ +T +F+ DCF+GEGGFGKVYKG+I+KINQ VAIKQLD NG QG REF
Sbjct: 81 KKAQTFTFEELSVSTGNFKSDCFLGEGGFGKVYKGFIEKINQVVAIKQLDRNGAQGIREF 140
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRM 126
VEVLTLSLA+HPNLVKL+GFCAEG QRLLVYEYMPLGSL+NHLHDLP GK PL WNTRM
Sbjct: 141 VVEVLTLSLADHPNLVKLIGFCAEGVQRLLVYEYMPLGSLDNHLHDLPSGKNPLAWNTRM 200
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+IAAG A+GLEYLHD MKPPVIYRDLKCSNIL+ + YH KLSDFGLAKVGP G THVST
Sbjct: 201 KIAAGAARGLEYLHDTMKPPVIYRDLKCSNILIDEGYHAKLSDFGLAKVGPRGSETHVST 260
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RVMGTYGYCAPDYA+TGQLT KSD+YS GV LLELITGRKA+D ++ Q+LV WA PL
Sbjct: 261 RVMGTYGYCAPDYALTGQLTFKSDVYSFGVVLLELITGRKAYDNTRTRNHQSLVEWANPL 320
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKY 306
FK+++ F KMVDPLLEG YP RGLYQALA+AAMCV+EQ SMRPVIADVV ALD +AS KY
Sbjct: 321 FKDRKNFKKMVDPLLEGDYPVRGLYQALAIAAMCVQEQPSMRPVIADVVMALDHLASSKY 380
Query: 307 D 307
D
Sbjct: 381 D 381
>At5g18610 protein kinase -like protein
Length = 513
Score = 468 bits (1203), Expect = e-132
Identities = 224/309 (72%), Positives = 259/309 (83%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF ELAAATK+FR +C +GEGGFG+VYKG ++ Q VA+KQLD NGLQG REF V
Sbjct: 68 AQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLETTGQIVAVKQLDRNGLQGNREFLV 127
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV L+G+CA+G+QRLLVYEYMPLGSLE+HLHDLPP K+PLDW+TRM I
Sbjct: 128 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDLPPDKEPLDWSTRMTI 187
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLEYLHD+ PPVIYRDLK SNILLGD YHPKLSDFGLAK+GP+GD THVSTRV
Sbjct: 188 AAGAAKGLEYLHDKANPPVIYRDLKSSNILLGDGYHPKLSDFGLAKLGPVGDKTHVSTRV 247
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D ++ E NLVAWA PLFK
Sbjct: 248 MGTYGYCAPEYAMTGQLTLKSDVYSFGVVFLELITGRKAIDNARAPGEHNLVAWARPLFK 307
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RKF KM DP L+G+YP RGLYQALAVAAMC++EQ++ RP+I DVV AL ++AS +DP
Sbjct: 308 DRRKFPKMADPSLQGRYPMRGLYQALAVAAMCLQEQAATRPLIGDVVTALTYLASQTFDP 367
Query: 309 QVHPIQSSR 317
Q+SR
Sbjct: 368 NAPSGQNSR 376
>At5g13160 protein kinase-like
Length = 456
Score = 439 bits (1129), Expect = e-124
Identities = 209/300 (69%), Positives = 245/300 (81%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A F F ELAAAT +F D F+GEGGFG+VYKG + Q VA+KQLD NGLQG REF V
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHDLPP K+ LDWN RM+I
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLE+LHD+ PPVIYRD K SNILL + +HPKLSDFGLAK+GP GD +HVSTRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D P EQNLVAWA PLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RKF K+ DP L+G++P R LYQALAVA+MC++EQ++ RP+IADVV AL ++A+ YDP
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
>At3g24790 protein kinase, putative
Length = 379
Score = 410 bits (1053), Expect = e-115
Identities = 192/295 (65%), Positives = 239/295 (80%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+IFTF ELA ATK+FR +C +GEGGFG+VYKG ++ Q VA+KQLD NGLQG REF V
Sbjct: 48 ARIFTFRELATATKNFRQECLIGEGGFGRVYKGKLENPAQVVAVKQLDRNGLQGQREFLV 107
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL H NLV L+G+CA+G+QRLLVYEYMPLGSLE+HL DL PG+KPLDWNTR++I
Sbjct: 108 EVLMLSLLHHRNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLLDLEPGQKPLDWNTRIKI 167
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A G AKG+EYLHDE PPVIYRDLK SNILL +Y KLSDFGLAK+GP+GD HVS+RV
Sbjct: 168 ALGAAKGIEYLHDEADPPVIYRDLKSSNILLDPEYVAKLSDFGLAKLGPVGDTLHVSSRV 227
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+Y TG LT+KSD+YS GV LLELI+GR+ D +P+ EQNLV WA P+F+
Sbjct: 228 MGTYGYCAPEYQRTGYLTNKSDVYSFGVVLLELISGRRVIDTMRPSHEQNLVTWALPIFR 287
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
+ ++ ++ DPLL G YP + L QA+AVAAMC+ E+ ++RP+++DV+ AL F+ +
Sbjct: 288 DPTRYWQLADPLLRGDYPEKSLNQAIAVAAMCLHEEPTVRPLMSDVITALSFLGA 342
>At3g20530 protein kinase, putative
Length = 386
Score = 408 bits (1049), Expect = e-114
Identities = 197/298 (66%), Positives = 242/298 (81%), Gaps = 1/298 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A IFTF EL ATK+F D +GEGGFG+VYKG I+ Q VA+KQLD NG QG REF V
Sbjct: 67 AHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNREFLV 126
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKK-PLDWNTRMR 127
EV+ LSL H NLV L+G+CA+G+QR+LVYEYM GSLE+HL +L KK PLDW+TRM+
Sbjct: 127 EVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWDTRMK 186
Query: 128 IAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTR 187
+AAG A+GLEYLH+ PPVIYRD K SNILL ++++PKLSDFGLAKVGP G THVSTR
Sbjct: 187 VAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGETHVSTR 246
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
VMGTYGYCAP+YA+TGQLT KSD+YS GV LE+ITGR+ D +KP +EQNLV WA PLF
Sbjct: 247 VMGTYGYCAPEYALTGQLTVKSDVYSFGVVFLEMITGRRVIDTTKPTEEQNLVTWASPLF 306
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHK 305
K++RKF+ M DPLLEG+YP +GLYQALAVAAMC++E+++ RP+++DVV AL+++A K
Sbjct: 307 KDRRKFTLMADPLLEGKYPIKGLYQALAVAAMCLQEEAATRPMMSDVVTALEYLAVTK 364
>At3g07070 putative protein kinase
Length = 414
Score = 397 bits (1021), Expect = e-111
Identities = 187/300 (62%), Positives = 239/300 (79%)
Query: 4 TDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGT 63
T+ A+ F+F ELA ATK+FR +C +GEGGFG+VYKG ++K VA+KQLD NGLQG
Sbjct: 59 TNNIAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLDRNGLQGN 118
Query: 64 REFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWN 123
+EF VEVL LSL H +LV L+G+CA+G+QRLLVYEYM GSLE+HL DL P + PLDW+
Sbjct: 119 KEFIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHLLDLTPDQIPLDWD 178
Query: 124 TRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTH 183
TR+RIA G A GLEYLHD+ PPVIYRDLK +NILL +++ KLSDFGLAK+GP+GD H
Sbjct: 179 TRIRIALGAAMGLEYLHDKANPPVIYRDLKAANILLDGEFNAKLSDFGLAKLGPVGDKQH 238
Query: 184 VSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWA 243
VS+RVMGTYGYCAP+Y TGQLT+KSD+YS GV LLELITGR+ D ++P EQNLV WA
Sbjct: 239 VSSRVMGTYGYCAPEYQRTGQLTTKSDVYSFGVVLLELITGRRVIDTTRPKDEQNLVTWA 298
Query: 244 YPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
P+FKE +F ++ DP LEG +P + L QA+AVAAMC++E++++RP+++DVV AL F+ +
Sbjct: 299 QPVFKEPSRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPLMSDVVTALGFLGT 358
>At1g61860 hypothetical protein
Length = 420
Score = 393 bits (1010), Expect = e-110
Identities = 189/324 (58%), Positives = 238/324 (73%), Gaps = 31/324 (9%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
+IF F EL AAT +F +DC +GEGGFG+VYKG++ +NQ VA+K+LD NGLQGTREF E
Sbjct: 71 RIFKFKELIAATDNFSMDCMIGEGGFGRVYKGFLTSLNQVVAVKRLDRNGLQGTREFFAE 130
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
V+ LSLA+HPNLV L+G+C E EQR+LVYE+MP GSLE+HL DLP G LDW TRMRI
Sbjct: 131 VMVLSLAQHPNLVNLIGYCVEDEQRVLVYEFMPNGSLEDHLFDLPEGSPSLDWFTRMRIV 190
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVM 189
G AKGLEYLHD PPVIYRD K SNILL D++ KLSDFGLA++GP HVSTRVM
Sbjct: 191 HGAAKGLEYLHDYADPPVIYRDFKASNILLQSDFNSKLSDFGLARLGPTEGKDHVSTRVM 250
Query: 190 GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAY----- 244
GTYGYCAP+YAMTGQLT+KSD+YS GV LLE+I+GR+A D +P +EQNL++W +
Sbjct: 251 GTYGYCAPEYAMTGQLTAKSDVYSFGVVLLEIISGRRAIDGDRPTEEQNLISWVFHQARV 310
Query: 245 --------------------------PLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAA 278
PL K++R F+++VDP L+G YP +GL+QALA+AA
Sbjct: 311 LLTYLCCCLRRKKPMKVFFFLVWQAEPLLKDRRMFAQIVDPNLDGNYPVKGLHQALAIAA 370
Query: 279 MCVEEQSSMRPVIADVVAALDFIA 302
MC++E++ RP++ DVV AL+F+A
Sbjct: 371 MCLQEEAETRPLMGDVVTALEFLA 394
>At1g20650 unknown protein
Length = 381
Score = 389 bits (998), Expect = e-108
Identities = 189/307 (61%), Positives = 233/307 (75%), Gaps = 1/307 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF ELAAAT++FR +GEGGFG+VYKG + Q VAIKQL+P+GLQG REF V
Sbjct: 63 ARSFTFKELAAATRNFREVNLLGEGGFGRVYKGRLDS-GQVVAIKQLNPDGLQGNREFIV 121
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV L+G+C G+QRLLVYEYMP+GSLE+HL DL ++PL WNTRM+I
Sbjct: 122 EVLMLSLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDLESNQEPLSWNTRMKI 181
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A G A+G+EYLH PPVIYRDLK +NILL ++ PKLSDFGLAK+GP+GD THVSTRV
Sbjct: 182 AVGAARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRTHVSTRV 241
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAM+G+LT KSDIY GV LLELITGRKA D + EQNLV W+ P K
Sbjct: 242 MGTYGYCAPEYAMSGKLTVKSDIYCFGVVLLELITGRKAIDLGQKQGEQNLVTWSRPYLK 301
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
+Q+KF +VDP L G+YP R L A+A+ AMC+ E++ RP I D+V AL+++A+
Sbjct: 302 DQKKFGHLVDPSLRGKYPRRCLNYAIAIIAMCLNEEAHYRPFIGDIVVALEYLAAQSRSH 361
Query: 309 QVHPIQS 315
+ + S
Sbjct: 362 EARNVSS 368
>At4g13190 putative protein
Length = 405
Score = 379 bits (972), Expect = e-105
Identities = 182/316 (57%), Positives = 235/316 (73%), Gaps = 16/316 (5%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
AK F F ELA AT SFR + +GEGGFG+VYKG ++K Q VA+KQLD NGLQG REF V
Sbjct: 56 AKSFKFRELATATNSFRQEFLIGEGGFGRVYKGKMEKTGQVVAVKQLDRNGLQGNREFLV 115
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHL----------------HD 112
E+ LSL HPNL L+G+C +G+QRLLV+E+MPLGSLE+HL D
Sbjct: 116 EIFRLSLLHHPNLANLIGYCLDGDQRLLVHEFMPLGSLEDHLLEFCTIVMELFNYLIEPD 175
Query: 113 LPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGL 172
+ G++PLDWN+R+RIA G AKGLEYLH++ PPVIYRD K SNILL D+ KLSDFGL
Sbjct: 176 VVVGQQPLDWNSRIRIALGAAKGLEYLHEKANPPVIYRDFKSSNILLNVDFDAKLSDFGL 235
Query: 173 AKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSK 232
AK+G +GD +VS+RV+GTYGYCAP+Y TGQLT KSD+YS GV LLELITG++ D ++
Sbjct: 236 AKLGSVGDTQNVSSRVVGTYGYCAPEYHKTGQLTVKSDVYSFGVVLLELITGKRVIDTTR 295
Query: 233 PAKEQNLVAWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIA 292
P EQNLV WA P+F+E +F ++ DPLL+G++P + L QA+A+AAMC++E+ +RP+I+
Sbjct: 296 PCHEQNLVTWAQPIFREPNRFPELADPLLQGEFPEKSLNQAVAIAAMCLQEEPIVRPLIS 355
Query: 293 DVVAALDFIASHKYDP 308
DVV AL F+++ P
Sbjct: 356 DVVTALSFMSTETGSP 371
>At3g02810 putative protein kinase
Length = 558
Score = 373 bits (957), Expect = e-104
Identities = 182/310 (58%), Positives = 236/310 (75%), Gaps = 4/310 (1%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
KIFTF ELA ATK+FR +C +GEGGFG+VYKG +K Q VA+KQLD +GL G +EF E
Sbjct: 50 KIFTFRELATATKNFRQECLLGEGGFGRVYKGTLKSTGQVVAVKQLDKHGLHGNKEFQAE 109
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
VL+L +HPNLVKL+G+CA+G+QRLLVY+Y+ GSL++HLH+ P+DW TRM+IA
Sbjct: 110 VLSLGQLDHPNLVKLIGYCADGDQRLLVYDYISGGSLQDHLHEPKADSDPMDWTTRMQIA 169
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGP-IGD-MTHVSTR 187
A+GL+YLHD+ PPVIYRDLK SNILL DD+ PKLSDFGL K+GP GD M +S+R
Sbjct: 170 YAAAQGLDYLHDKANPPVIYRDLKASNILLDDDFSPKLSDFGLHKLGPGTGDKMMALSSR 229
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
VMGTYGY AP+Y G LT KSD+YS GV LLELITGR+A D ++P EQNLV+WA P+F
Sbjct: 230 VMGTYGYSAPEYTRGGNLTLKSDVYSFGVVLLELITGRRALDTTRPNDEQNLVSWAQPIF 289
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYD 307
++ +++ M DP+LE ++ RGL QA+A+A+MCV+E++S RP+I+DV+ AL F++ D
Sbjct: 290 RDPKRYPDMADPVLENKFSERGLNQAVAIASMCVQEEASARPLISDVMVALSFLSMPTED 349
Query: 308 --PQVHPIQS 315
P PI S
Sbjct: 350 GIPTTVPILS 359
>At1g76370 putative protein kinase
Length = 381
Score = 370 bits (949), Expect = e-103
Identities = 183/295 (62%), Positives = 226/295 (76%), Gaps = 1/295 (0%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF ELAAATK+FR +G+GGFG VYKG + Q VAIKQL+P+G QG +EF V
Sbjct: 60 ARSFTFKELAAATKNFREGNIIGKGGFGSVYKGRLDS-GQVVAIKQLNPDGHQGNQEFIV 118
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EV LS+ HPNLV L+G+C G QRLLVYEYMP+GSLE+HL DL P + PL W TRM+I
Sbjct: 119 EVCMLSVFHHPNLVTLIGYCTSGAQRLLVYEYMPMGSLEDHLFDLEPDQTPLSWYTRMKI 178
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A G A+G+EYLH ++ P VIYRDLK +NILL ++ KLSDFGLAKVGP+G+ THVSTRV
Sbjct: 179 AVGAARGIEYLHCKISPSVIYRDLKSANILLDKEFSVKLSDFGLAKVGPVGNRTHVSTRV 238
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAM+G+LT KSDIYS GV LLELI+GRKA D SKP EQ LVAWA P K
Sbjct: 239 MGTYGYCAPEYAMSGRLTIKSDIYSFGVVLLELISGRKAIDLSKPNGEQYLVAWARPYLK 298
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIAS 303
+ +KF +VDPLL G++ R L A+++ MC+ ++++ RP I DVV A ++IAS
Sbjct: 299 DPKKFGLLVDPLLRGKFSKRCLNYAISITEMCLNDEANHRPKIGDVVVAFEYIAS 353
>At5g16500 protein kinase-like protein
Length = 636
Score = 360 bits (924), Expect = e-100
Identities = 174/311 (55%), Positives = 232/311 (73%), Gaps = 3/311 (0%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
K F F ELA ATK+FR +C +GEGGFG+VYKG ++ Q VA+KQLD +GL G +EF E
Sbjct: 60 KTFNFRELATATKNFRQECLLGEGGFGRVYKGTLQSTGQLVAVKQLDKHGLHGNKEFLAE 119
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
VL+L+ EHPNLVKL+G+CA+G+QRLLV+EY+ GSL++HL++ PG+KP+DW TRM+IA
Sbjct: 120 VLSLAKLEHPNLVKLIGYCADGDQRLLVFEYVSGGSLQDHLYEQKPGQKPMDWITRMKIA 179
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGP-IGDMTHVSTRV 188
G A+GL+YLHD++ P VIYRDLK SNILL +++PKL DFGL + P GD +S+RV
Sbjct: 180 FGAAQGLDYLHDKVTPAVIYRDLKASNILLDAEFYPKLCDFGLHNLEPGTGDSLFLSSRV 239
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
M TYGY AP+Y LT KSD+YS GV LLELITGR+A D +KP EQNLVAWA P+FK
Sbjct: 240 MDTYGYSAPEYTRGDDLTVKSDVYSFGVVLLELITGRRAIDTTKPNDEQNLVAWAQPIFK 299
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYD- 307
+ +++ M DPLL + RGL QA+A+ +MC++E+ + RP+I+DV+ AL F++ D
Sbjct: 300 DPKRYPDMADPLLRKNFSERGLNQAVAITSMCLQEEPTARPLISDVMVALSFLSMSTEDG 359
Query: 308 -PQVHPIQSSR 317
P P++S R
Sbjct: 360 IPATVPMESFR 370
>At3g26940 protein kinase, putative
Length = 432
Score = 337 bits (865), Expect = 4e-93
Identities = 176/300 (58%), Positives = 219/300 (72%), Gaps = 7/300 (2%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
+IF++ ELA AT SFR + +G GGFG VYKG + Q +A+K LD +G+QG +EF VE
Sbjct: 60 QIFSYRELAIATNSFRNESLIGRGGFGTVYKGRLST-GQNIAVKMLDQSGIQGDKEFLVE 118
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
VL LSL H NLV L G+CAEG+QRL+VYEYMPLGS+E+HL+DL G++ LDW TRM+IA
Sbjct: 119 VLMLSLLHHRNLVHLFGYCAEGDQRLVVYEYMPLGSVEDHLYDLSEGQEALDWKTRMKIA 178
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVM 189
G AKGL +LH+E +PPVIYRDLK SNILL DY PKLSDFGLAK GP DM+HVSTRVM
Sbjct: 179 LGAAKGLAFLHNEAQPPVIYRDLKTSNILLDHDYKPKLSDFGLAKFGPSDDMSHVSTRVM 238
Query: 190 GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPA---KEQNLVAWAYPL 246
GT+GYCAP+YA TG+LT KSDIYS GV LLELI+GRKA PS + + LV WA PL
Sbjct: 239 GTHGYCAPEYANTGKLTLKSDIYSFGVVLLELISGRKALMPSSECVGNQSRYLVHWARPL 298
Query: 247 FKEQRKFSKMVDPLL--EGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASH 304
F R ++VDP L +G + LY+ + VA +C+ E+++ RP I+ VV L +I H
Sbjct: 299 FLNGR-IRQIVDPRLARKGGFSNILLYRGIEVAFLCLAEEANARPSISQVVECLKYIIDH 357
>At5g01020 protein kinase -like protein
Length = 410
Score = 312 bits (800), Expect = 1e-85
Identities = 163/294 (55%), Positives = 200/294 (67%), Gaps = 9/294 (3%)
Query: 12 FTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQF------VAIKQLDPNGLQGTRE 65
FT EL TKSFR D +GEGGFG VYKGYI + VA+K L+ GLQG RE
Sbjct: 57 FTLFELETITKSFRPDYILGEGGFGTVYKGYIDDNLRVGLKSLPVAVKVLNKEGLQGHRE 116
Query: 66 FAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTR 125
+ EV L HPNLVKL+G+C E + RLLVYE+M GSLENHL PL W+ R
Sbjct: 117 WLTEVNFLGQLRHPNLVKLIGYCCEDDHRLLVYEFMLRGSLENHLFRKTTA--PLSWSRR 174
Query: 126 MRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVS 185
M IA G AKGL +LH+ +P VIYRD K SNILL DY KLSDFGLAK GP GD THVS
Sbjct: 175 MMIALGAAKGLAFLHNAERP-VIYRDFKTSNILLDSDYTAKLSDFGLAKAGPQGDETHVS 233
Query: 186 TRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYP 245
TRVMGTYGY AP+Y MTG LT++SD+YS GV LLE++TGRK+ D ++P+KEQNLV WA P
Sbjct: 234 TRVMGTYGYAAPEYVMTGHLTARSDVYSFGVVLLEMLTGRKSVDKTRPSKEQNLVDWARP 293
Query: 246 LFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
++RK +++DP LE QY R +A ++A C+ + RP+++DVV L+
Sbjct: 294 KLNDKRKLLQIIDPRLENQYSVRAAQKACSLAYYCLSQNPKARPLMSDVVETLE 347
>At2g02800 protein kinase like protein
Length = 426
Score = 311 bits (798), Expect = 2e-85
Identities = 161/305 (52%), Positives = 208/305 (67%), Gaps = 12/305 (3%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQ---------FVAIKQLDPNGL 60
K FTF+EL AT++FR D +GEGGFG V+KG+I VA+K+L G
Sbjct: 69 KAFTFNELKNATRNFRPDSLLGEGGFGYVFKGWIDGTTLTASKPGSGIVVAVKKLKTEGY 128
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG +E+ EV L HPNLVKL+G+C EGE RLLVYE+MP GSLENHL G +PL
Sbjct: 129 QGHKEWLTEVNYLGQLSHPNLVKLVGYCVEGENRLLVYEFMPKGSLENHL--FRRGAQPL 186
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
W RM++A G AKGL +LHD K VIYRD K +NILL +++ KLSDFGLAK GP GD
Sbjct: 187 TWAIRMKVAIGAAKGLTFLHDA-KSQVIYRDFKAANILLDAEFNSKLSDFGLAKAGPTGD 245
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
THVST+VMGT+GY AP+Y TG+LT+KSD+YS GV LLEL++GR+A D SK EQ+LV
Sbjct: 246 KTHVSTQVMGTHGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDKSKVGMEQSLV 305
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
WA P ++RK +++D L GQYP +G Y A ++A C+ + +RP +++V+A LD
Sbjct: 306 DWATPYLGDKRKLFRIMDTRLGGQYPQKGAYTAASLALQCLNPDAKLRPKMSEVLAKLDQ 365
Query: 301 IASHK 305
+ S K
Sbjct: 366 LESTK 370
>At3g28690 protein kinase like protein
Length = 376
Score = 308 bits (790), Expect = 2e-84
Identities = 160/320 (50%), Positives = 214/320 (66%), Gaps = 19/320 (5%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKK---------INQFVAIKQLDPNGL 60
+IF F++L AT++FR + +GEGGFG V+KG+I++ VA+K L+P+GL
Sbjct: 12 RIFMFNDLKLATRNFRPESLLGEGGFGCVFKGWIEENGTAPVKPGTGLTVAVKTLNPDGL 71
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG +E+ E+ L HP+LVKL+G+C E +QRLLVYE+MP GSLENHL PL
Sbjct: 72 QGHKEWLAEINFLGNLVHPSLVKLVGYCMEEDQRLLVYEFMPRGSLENHLFRRT---LPL 128
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
W+ RM+IA G AKGL +LH+E + PVIYRD K SNILL +Y+ KLSDFGLAK P
Sbjct: 129 PWSVRMKIALGAAKGLAFLHEEAEKPVIYRDFKTSNILLDGEYNAKLSDFGLAKDAPDEK 188
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
+HVSTRVMGTYGY AP+Y MTG LT+KSD+YS GV LLE++TGR++ D S+P EQNLV
Sbjct: 189 KSHVSTRVMGTYGYAAPEYVMTGHLTTKSDVYSFGVVLLEILTGRRSVDKSRPNGEQNLV 248
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL-- 298
W P ++++F +++DP LEG Y +G +A VAA C+ S RP +++VV AL
Sbjct: 249 EWVRPHLLDKKRFYRLLDPRLEGHYSIKGAQKATQVAAQCLNRDSKARPKMSEVVEALKP 308
Query: 299 -----DFIASHKYDPQVHPI 313
DF +S + P+
Sbjct: 309 LPNLKDFASSSSSFQTMQPV 328
>At5g15080 serine/threonine specific protein kinase -like
Length = 493
Score = 308 bits (789), Expect = 3e-84
Identities = 156/296 (52%), Positives = 204/296 (68%), Gaps = 12/296 (4%)
Query: 12 FTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKK---------INQFVAIKQLDPNGLQG 62
FTF++L +T++FR + +GEGGFG V+KG+I++ VA+K L+P+GLQG
Sbjct: 130 FTFNDLKLSTRNFRPESLLGEGGFGCVFKGWIEENGTAPVKPGTGLTVAVKTLNPDGLQG 189
Query: 63 TREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDW 122
+E+ E+ L HPNLVKL+G+C E +QRLLVYE+MP GSLENHL PL W
Sbjct: 190 HKEWLAEINFLGNLLHPNLVKLVGYCIEDDQRLLVYEFMPRGSLENHLFRR---SLPLPW 246
Query: 123 NTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMT 182
+ RM+IA G AKGL +LH+E PVIYRD K SNILL DY+ KLSDFGLAK P T
Sbjct: 247 SIRMKIALGAAKGLSFLHEEALKPVIYRDFKTSNILLDADYNAKLSDFGLAKDAPDEGKT 306
Query: 183 HVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAW 242
HVSTRVMGTYGY AP+Y MTG LTSKSD+YS GV LLE++TGR++ D ++P E NLV W
Sbjct: 307 HVSTRVMGTYGYAAPEYVMTGHLTSKSDVYSFGVVLLEMLTGRRSMDKNRPNGEHNLVEW 366
Query: 243 AYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
A P ++R+F +++DP LEG + +G + +AA C+ +RP ++DVV AL
Sbjct: 367 ARPHLLDKRRFYRLLDPRLEGHFSIKGAQKVTQLAAQCLSRDPKIRPKMSDVVEAL 422
>At1g14370 putative protein
Length = 426
Score = 307 bits (787), Expect = 4e-84
Identities = 159/303 (52%), Positives = 206/303 (67%), Gaps = 12/303 (3%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQ---------FVAIKQLDPNGL 60
K FTF+EL ATK+FR D +GEGGFG V+KG+I + + VA+KQL P G
Sbjct: 72 KAFTFNELKNATKNFRQDNLLGEGGFGCVFKGWIDQTSLTASRPGSGIVVAVKQLKPEGF 131
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG +E+ EV L HPNLV L+G+CAEGE RLLVYE+MP GSLENHL G +PL
Sbjct: 132 QGHKEWLTEVNYLGQLSHPNLVLLVGYCAEGENRLLVYEFMPKGSLENHL--FRRGAQPL 189
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
W RM++A G AKGL +LH E K VIYRD K +NILL D++ KLSDFGLAK GP GD
Sbjct: 190 TWAIRMKVAVGAAKGLTFLH-EAKSQVIYRDFKAANILLDADFNAKLSDFGLAKAGPTGD 248
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
THVST+V+GT+GY AP+Y TG+LT+KSD+YS GV LLELI+GR+A D S E +LV
Sbjct: 249 NTHVSTKVIGTHGYAAPEYVATGRLTAKSDVYSFGVVLLELISGRRAMDNSNGGNEYSLV 308
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
WA P ++RK +++D L GQYP +G + A +A C+ + +RP +++V+ L+
Sbjct: 309 DWATPYLGDKRKLFRIMDTKLGGQYPQKGAFTAANLALQCLNPDAKLRPKMSEVLVTLEQ 368
Query: 301 IAS 303
+ S
Sbjct: 369 LES 371
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,130,924
Number of Sequences: 26719
Number of extensions: 300149
Number of successful extensions: 3941
Number of sequences better than 10.0: 971
Number of HSP's better than 10.0 without gapping: 881
Number of HSP's successfully gapped in prelim test: 90
Number of HSP's that attempted gapping in prelim test: 626
Number of HSP's gapped (non-prelim): 1056
length of query: 317
length of database: 11,318,596
effective HSP length: 99
effective length of query: 218
effective length of database: 8,673,415
effective search space: 1890804470
effective search space used: 1890804470
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149490.2


