

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.13 + phase: 0
(227 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
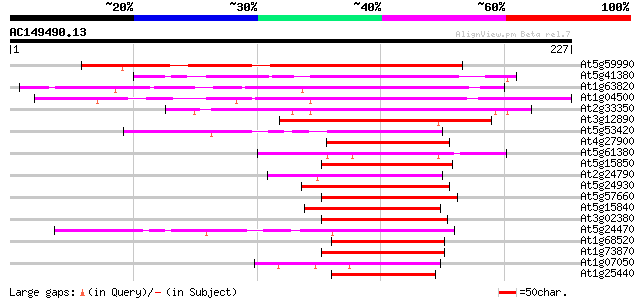
Score E
Sequences producing significant alignments: (bits) Value
At5g59990 putative protein 137 5e-33
At5g41380 putative protein 108 3e-24
At1g63820 hypothetical protein 107 7e-24
At1g04500 zinc finger CONSTANS-related like protein 97 9e-21
At2g33350 hypothetical protein 92 2e-19
At3g12890 unknown protein 89 1e-18
At5g53420 unknown protein 73 1e-13
At4g27900 unknown protein 69 3e-12
At5g61380 pseudo-response regulator 1 62 2e-10
At5g15850 CONSTANS-like 1 60 9e-10
At2g24790 CONSTANS-like B-box zinc finger protein 60 1e-09
At5g24930 unknown protein 59 2e-09
At5g57660 CONSTANS-like B-box zinc finger protein-like 54 5e-08
At5g15840 CONSTANS 54 5e-08
At3g02380 unknown protein 54 7e-08
At5g24470 pseudo-response regulator 5 (APRR5) 51 4e-07
At1g68520 putative B-box zinc finger protein 51 6e-07
At1g73870 unknown protein 50 7e-07
At1g07050 unknown protein 50 7e-07
At1g25440 unknown protein 49 2e-06
>At5g59990 putative protein
Length = 241
Score = 137 bits (345), Expect = 5e-33
Identities = 81/163 (49%), Positives = 102/163 (61%), Gaps = 23/163 (14%)
Query: 30 SGYSSYGGSPSPSTP---------ILMQRSISSHSLQYNNGTHHYPLSAFFADLLDSDDA 80
SG + GGS S +P ++MQRS+SSH+ N T + D ++ DD
Sbjct: 67 SGMNGGGGSSSCDSPSSMGSGGESLVMQRSVSSHNGFSGNYTAAH-------DFVNDDDG 119
Query: 81 PVRKVCSTGDLQRINGMQHNHHHHVDSPLSSESSMIIEGMSRVCPYSPEEKKVRIERYRI 140
PVR+ S GDL R + + S + SES+ IIEGMS+ YSPEEKK +IE+YR
Sbjct: 120 PVRRALSAGDLPRSSRRE-------SSTVWSESNAIIEGMSKAYKYSPEEKKEKIEKYRS 172
Query: 141 KRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKKPIVE 183
KRN RNFNK+IKY CRKTLAD RPRIRGRFARNDEI ++ V+
Sbjct: 173 KRNLRNFNKRIKYECRKTLADSRPRIRGRFARNDEISQQEQVD 215
>At5g41380 putative protein
Length = 315
Score = 108 bits (269), Expect = 3e-24
Identities = 71/165 (43%), Positives = 92/165 (55%), Gaps = 36/165 (21%)
Query: 51 ISSHSLQYNNGTHHYPLSAFFADLLDSDDAPVRKVCSTGDLQRINGMQHNHHHHVDSPLS 110
+ S +L YNN + P +AFF+ + +R+V STGDLQ MQ + + P S
Sbjct: 151 MDSSNLSYNNLSS--PENAFFS-------SQMRRVYSTGDLQNNMEMQRSSENST-MPFS 200
Query: 111 SESSMIIEGMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRF 170
E + +V YS EE+K +I +YR KRNQRNF K IKY CRKTLAD RPRIRGRF
Sbjct: 201 EEQNF------KVGRYSAEERKEKISKYRAKRNQRNFTKTIKYACRKTLADSRPRIRGRF 254
Query: 171 ARNDEIDKKPIVEWSHIGGGEEEDEEDEKW----------NNIFN 205
ARNDE+ + P + ED++ E W +NIFN
Sbjct: 255 ARNDEVVEIPNI----------EDDDSELWVSHIQQLGFFSNIFN 289
>At1g63820 hypothetical protein
Length = 265
Score = 107 bits (266), Expect = 7e-24
Identities = 75/205 (36%), Positives = 107/205 (51%), Gaps = 24/205 (11%)
Query: 5 NSIPDFTELDSFQDTLSQLNNSELSSGYSSYGGSPSP----STPILMQRSISSHSLQYNN 60
+S P ++L + TL+ N+ SG+ ++ + ++P S + Q N
Sbjct: 68 SSSPPISQLQTL--TLTHTNSFPNFSGFENFDTVKTEQLLFNSPFDAPFMEDSSNFQ-NQ 124
Query: 61 GTHHYPLSAFFADLLDSDDAPVRKVCSTGDLQRINGMQHNHHHHVDSPLSSESSMII--- 117
+ P +FF+D + R+V STGDLQ + G SPL++ESS
Sbjct: 125 NLLNSPEDSFFSDHM-------RRVYSTGDLQNL-GRDFTGQRSYSSPLAAESSPTTVFS 176
Query: 118 --EGMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDE 175
E RV YS EE+K +I +YR KR QRNF K IKY CRKTLAD RPR+RGRFARNDE
Sbjct: 177 GDEQSLRVGRYSSEERKEKISKYRAKRTQRNFTKTIKYACRKTLADNRPRVRGRFARNDE 236
Query: 176 IDKKPIVEWSHIGGGEEEDEEDEKW 200
+ + P + S ++ +D+ W
Sbjct: 237 VFENPKIASSF----TRQENDDDLW 257
>At1g04500 zinc finger CONSTANS-related like protein
Length = 386
Score = 96.7 bits (239), Expect = 9e-21
Identities = 73/228 (32%), Positives = 111/228 (48%), Gaps = 36/228 (15%)
Query: 11 TELDSFQDTLSQLNNSELSSGYSS----YGGSPSPSTPILMQRSISSHSLQYNNGTHHYP 66
T L ++ + N+ L SG+ S G PS LM+ +Q +NG P
Sbjct: 178 TGLPTYMTVTGNMMNTGLGSGFYSGNIHLGSDFKPSHDQLME-------IQADNGGLFCP 230
Query: 67 LSAFFADLLDSDDAPVRKVCSTGD--LQRINGMQHNHHHHVDSPLSSESSMIIEGM---- 120
P++ + + GD LQ ++G++ N +H V P+ + I G+
Sbjct: 231 -------------DPIKPIFNPGDHHLQGLDGVE-NQNHMVAQPVLPQLGTEITGLDDPS 276
Query: 121 -SRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKK 179
++V S E++K +I RY KRN+RNF+KKIKY CRKTLAD RPR+RGRFA+NDE +
Sbjct: 277 FNKVGKLSAEQRKEKIHRYMKKRNERNFSKKIKYACRKTLADSRPRVRGRFAKNDEFGEP 336
Query: 180 PIVEWSHIGGGEEEDEEDEKWNNIFNSIVAANLVHDEFQGSSSFGLLY 227
S ED++D+ +V ++ + G +SF Y
Sbjct: 337 NRQACS----SHHEDDDDDVGVKEEEQLVDSSDIFSHISGVNSFKCNY 380
>At2g33350 hypothetical protein
Length = 440
Score = 92.4 bits (228), Expect = 2e-19
Identities = 65/167 (38%), Positives = 94/167 (55%), Gaps = 23/167 (13%)
Query: 64 HYPLSAFFAD-----LLDSDDAPVRKVCSTGDLQRINGMQHNHHHHVDSPLSSES--SMI 116
H PL F AD DS ++++ + DLQ+ G + H+ +P + + +
Sbjct: 266 HDPLMDFQADNGGFFCPDS----IKRMFNPEDLQKALGGGAENQSHLVTPQAHPALGPVD 321
Query: 117 IEGM-----SRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFA 171
I G+ ++V SPE++K +I RY KRN+RNFNKKIKY CRKTLAD RPR+RGRFA
Sbjct: 322 INGLEDSTLNKVGKLSPEQRKEKIRRYMKKRNERNFNKKIKYACRKTLADSRPRVRGRFA 381
Query: 172 RNDEIDKKPIVEWSHIGGGEEEDE---EDEKW----NNIFNSIVAAN 211
+NDE + +S E+ED+ +DE+ ++IF I AN
Sbjct: 382 KNDEFGEPNRQAFSSHHDDEDEDDMGVKDEEQLVDSSDIFAHISGAN 428
>At3g12890 unknown protein
Length = 251
Score = 89.4 bits (220), Expect = 1e-18
Identities = 47/87 (54%), Positives = 62/87 (71%), Gaps = 1/87 (1%)
Query: 110 SSESSMIIEGMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGR 169
+SE + E ++V YS EE+K RI RY K+NQRNFNK IKYVCRKTLADRR R+RGR
Sbjct: 118 NSEGGIQAEPNTKVGRYSVEERKDRIMRYLKKKNQRNFNKTIKYVCRKTLADRRVRVRGR 177
Query: 170 FAR-NDEIDKKPIVEWSHIGGGEEEDE 195
FAR ND +++P + +H E+E++
Sbjct: 178 FARNNDTCEEQPHMSKNHNNHSEKEED 204
>At5g53420 unknown protein
Length = 264
Score = 73.2 bits (178), Expect = 1e-13
Identities = 45/130 (34%), Positives = 73/130 (55%), Gaps = 17/130 (13%)
Query: 47 MQRSISSHSLQYNNGTHHYPLSAFFADLLDSDDA-PVRKVCSTGDLQRINGMQHNHHHHV 105
MQ+S+SS +L + +H + + D D +R+ S GD+Q++ V
Sbjct: 149 MQKSVSSGNLSSMDWSHAQQETVMIQNFPDFDFGYGMRRAFSEGDIQKLGAGL------V 202
Query: 106 DSPLSSESSMIIEGMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPR 165
SPL +I+ S E+++ ++ RY+ K+++RNF +KIKY CRK LAD +PR
Sbjct: 203 QSPLDR---IIVSCTS-------EDRREKLSRYKNKKSRRNFGRKIKYACRKALADSQPR 252
Query: 166 IRGRFARNDE 175
+RGRFA+ +E
Sbjct: 253 VRGRFAKTEE 262
>At4g27900 unknown protein
Length = 261
Score = 68.6 bits (166), Expect = 3e-12
Identities = 28/50 (56%), Positives = 41/50 (82%)
Query: 129 EEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDK 178
E+++ ++ RYR K+++RNF +KIKY CRK LAD +PRIRGRFA+ +E+ K
Sbjct: 212 EDRREKLSRYRNKKSKRNFGRKIKYACRKALADSQPRIRGRFAKTEEMQK 261
>At5g61380 pseudo-response regulator 1
Length = 618
Score = 62.0 bits (149), Expect = 2e-10
Identities = 40/122 (32%), Positives = 66/122 (53%), Gaps = 23/122 (18%)
Query: 101 HHHHVDSPLSSESSMIIEGMSRVCPYS------PEEKKVRIER----------YRIKRNQ 144
+HH +++ L + G + +S P +VR+ + +R KRNQ
Sbjct: 487 YHHPMNTSLQHSQMSLQNGQMSMVHHSWSPAGNPPSNEVRVNKLDRREEALLKFRRKRNQ 546
Query: 145 RNFNKKIKYVCRKTLADRRPRIRGRFAR-----NDEIDKKPIVEWSHIGGGEEEDEEDEK 199
R F+KKI+YV RK LA+RRPR++G+F R N +++ +P + + EEE+EE+E+
Sbjct: 547 RCFDKKIRYVNRKRLAERRPRVKGQFVRKMNGVNVDLNGQP--DSADYDDEEEEEEEEEE 604
Query: 200 WN 201
N
Sbjct: 605 EN 606
>At5g15850 CONSTANS-like 1
Length = 355
Score = 60.1 bits (144), Expect = 9e-10
Identities = 24/53 (45%), Positives = 38/53 (71%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKK 179
SP +++ R+ RYR K+ R F K I+Y RK A++RPRI+GRFA+ ++D++
Sbjct: 282 SPRDREARVLRYREKKKMRKFEKTIRYASRKAYAEKRPRIKGRFAKKKDVDEE 334
>At2g24790 CONSTANS-like B-box zinc finger protein
Length = 294
Score = 59.7 bits (143), Expect = 1e-09
Identities = 31/75 (41%), Positives = 42/75 (55%), Gaps = 4/75 (5%)
Query: 105 VDSPLSSESSMIIEGMSRV----CPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLA 160
+D PL ES + M+ SP E++ R+ RYR KR R F K I+Y RK A
Sbjct: 199 IDVPLVPESGGVTAEMTNTETPAVQLSPAEREARVLRYREKRKNRKFEKTIRYASRKAYA 258
Query: 161 DRRPRIRGRFARNDE 175
+ RPRI+GRFA+ +
Sbjct: 259 EMRPRIKGRFAKRTD 273
>At5g24930 unknown protein
Length = 362
Score = 58.9 bits (141), Expect = 2e-09
Identities = 26/60 (43%), Positives = 37/60 (61%)
Query: 119 GMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDK 178
G R P + E++ R+ RYR KR R F K I+Y RK A+ RPRI+GRFA+ + ++
Sbjct: 283 GTQRAVPLTSAEREARVMRYREKRKNRKFEKTIRYASRKAYAEMRPRIKGRFAKRTDTNE 342
>At5g57660 CONSTANS-like B-box zinc finger protein-like
Length = 355
Score = 54.3 bits (129), Expect = 5e-08
Identities = 25/55 (45%), Positives = 34/55 (61%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKKPI 181
S +++ R+ RYR KR R F K I+Y RK A+ RPRI+GRFA+ E + I
Sbjct: 281 SSMDREARVLRYREKRKNRKFEKTIRYASRKAYAESRPRIKGRFAKRTETENDDI 335
>At5g15840 CONSTANS
Length = 373
Score = 54.3 bits (129), Expect = 5e-08
Identities = 25/55 (45%), Positives = 34/55 (61%)
Query: 120 MSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARND 174
M V SP +++ R+ RYR KR R F K I+Y RK A+ RPR+ GRFA+ +
Sbjct: 295 MITVTQLSPMDREARVLRYREKRKTRKFEKTIRYASRKAYAEIRPRVNGRFAKRE 349
>At3g02380 unknown protein
Length = 347
Score = 53.9 bits (128), Expect = 7e-08
Identities = 24/51 (47%), Positives = 35/51 (68%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEID 177
+P E++ R+ RYR K+ R F+K I+Y RK A+ RPRI+GRFA+ E +
Sbjct: 274 TPMEREARVLRYREKKKTRKFDKTIRYASRKAYAEIRPRIKGRFAKRIETE 324
>At5g24470 pseudo-response regulator 5 (APRR5)
Length = 667
Score = 51.2 bits (121), Expect = 4e-07
Identities = 41/167 (24%), Positives = 81/167 (47%), Gaps = 18/167 (10%)
Query: 19 TLSQLNNSELSSGYSSYGGSPSPSTPILMQRSISSHSLQYNNGTHHYPLSAFFADLLDSD 78
T + +NN + ++Y + +P++ S+S H +Y++ H P ++ L D D
Sbjct: 514 TPTPINNIQFRDPNTAYTSAMAPASLSPSPSSVSPH--EYSSMFH--PFNSKPEGLQDRD 569
Query: 79 ---DAPVRKVCSTGDLQRINGMQHNHHHHVDSPLSSESSMIIEGMSRVCPYSPE--EKKV 133
D R+ S+ G +H+D + ++ +G S + +++
Sbjct: 570 CSMDVDERRYVSSATEHSAIG------NHIDQLIEKKNE---DGYSLSVGKIQQSLQREA 620
Query: 134 RIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKKP 180
+ ++R+KR R + KK++Y RK LA++RPRI+G+F R + + P
Sbjct: 621 ALTKFRMKRKDRCYEKKVRYESRKKLAEQRPRIKGQFVRQVQSTQAP 667
>At1g68520 putative B-box zinc finger protein
Length = 406
Score = 50.8 bits (120), Expect = 6e-07
Identities = 22/46 (47%), Positives = 32/46 (68%)
Query: 131 KKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEI 176
++ R+ RYR KR R F+KKI+Y RK A++RPR++GRF + I
Sbjct: 357 REARVSRYREKRRTRLFSKKIRYEVRKLNAEKRPRMKGRFVKRSSI 402
>At1g73870 unknown protein
Length = 392
Score = 50.4 bits (119), Expect = 7e-07
Identities = 23/50 (46%), Positives = 34/50 (68%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEI 176
S E++ R+ RY+ KR R F+KKI+Y RK A++RPRI+GRF + +
Sbjct: 341 SDGEREARVLRYKEKRRTRLFSKKIRYEVRKLNAEQRPRIKGRFVKRTSL 390
>At1g07050 unknown protein
Length = 195
Score = 50.4 bits (119), Expect = 7e-07
Identities = 31/86 (36%), Positives = 48/86 (55%), Gaps = 11/86 (12%)
Query: 100 NHHHHVDS------PLSSESSMIIEGMSR-VCPYSPEEKKVRIE----RYRIKRNQRNFN 148
NH +D+ PL +++S + + R P E++ +R E RY+ KR R F+
Sbjct: 109 NHQEVIDAWSDHQKPLWTDTSTLDNSVYRGEVPVIEEKRNMRREASVLRYKEKRQSRLFS 168
Query: 149 KKIKYVCRKTLADRRPRIRGRFARND 174
KKI+Y RK AD+RPR +GRF + +
Sbjct: 169 KKIRYQVRKLNADKRPRFKGRFVKRE 194
>At1g25440 unknown protein
Length = 417
Score = 49.3 bits (116), Expect = 2e-06
Identities = 21/42 (50%), Positives = 31/42 (73%)
Query: 131 KKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFAR 172
++ R+ RYR KR R F+KKI+Y RK A++RPR++GRF +
Sbjct: 361 REARVSRYREKRRTRLFSKKIRYEVRKLNAEKRPRMKGRFVK 402
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,779,552
Number of Sequences: 26719
Number of extensions: 267195
Number of successful extensions: 1085
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1017
Number of HSP's gapped (non-prelim): 76
length of query: 227
length of database: 11,318,596
effective HSP length: 96
effective length of query: 131
effective length of database: 8,753,572
effective search space: 1146717932
effective search space used: 1146717932
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149490.13


