

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.3 + phase: 0
(620 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
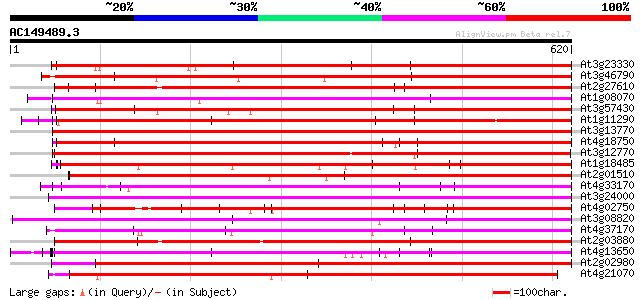
Score E
Sequences producing significant alignments: (bits) Value
At3g23330 hypothetical protein 489 e-138
At3g46790 putative protein 478 e-135
At2g27610 putative selenium-binding protein 475 e-134
At1g08070 unknown protein 474 e-134
At3g57430 putative protein 462 e-130
At1g11290 hypothetical protein 462 e-130
At3g13770 hypothetical protein 456 e-128
At4g18750 putative protein 455 e-128
At3g12770 unknown protein 450 e-127
At1g18485 hypothetical protein 449 e-126
At2g01510 hypothetical protein 444 e-125
At4g33170 putative protein 437 e-123
At3g24000 hypothetical protein 437 e-123
At4g02750 hypothetical protein 437 e-122
At3g08820 unknown protein 435 e-122
At4g37170 putative protein 434 e-122
At2g03880 putative selenium-binding protein 434 e-122
At4g13650 unknown protein 433 e-121
At2g02980 unknown protein 431 e-121
At4g21070 putative protein (fragment) 430 e-120
>At3g23330 hypothetical protein
Length = 715
Score = 489 bits (1260), Expect = e-138
Identities = 247/611 (40%), Positives = 374/611 (60%), Gaps = 37/611 (6%)
Query: 47 HYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYA------------- 93
H + S+L+SC L G+ +H LG+ + L+++YA
Sbjct: 105 HNVFPSVLKSCTMMMDLRFGESVHGFIVRLGMDCDLYTGNALMNMYAKLLGMGSKISVGN 164
Query: 94 --------VSNS---------------LLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHD 130
SNS + + R +F+ +P++++ +N +I GYA +G ++
Sbjct: 165 VFDEMPQRTSNSGDEDVKAETCIMPFGIDSVRRVFEVMPRKDVVSYNTIIAGYAQSGMYE 224
Query: 131 NAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALI 190
+A+ + +M L+PD+FTL VL S + +G+ IH YVI+ G + D+++G++L+
Sbjct: 225 DALRMVREMGTTDLKPDSFTLSSVLPIFSEYVDVIKGKEIHGYVIRKGIDSDVYIGSSLV 284
Query: 191 DMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEA 250
DMYAK + D+ RVF ++ RD + WNS++A Y QNG +E++ L R+M V+P
Sbjct: 285 DMYAKSARIEDSERVFSRLYCRDGISWNSLVAGYVQNGRYNEALRLFRQMVTAKVKPGAV 344
Query: 251 TLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERL 310
+VI + A +A L G+++HG+ R GF SN + +AL+DMY+KCG++K A +F+R+
Sbjct: 345 AFSSVIPACAHLATLHLGKQLHGYVLRGGFGSNIFIASALVDMYSKCGNIKAARKIFDRM 404
Query: 311 REKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGR 369
VSW AII G+A+HG A+ LF++M+++ +P+ + FV VL ACS L+DE
Sbjct: 405 NVLDEVSWTAIIMGHALHGHGHEAVSLFEEMKRQGVKPNQVAFVAVLTACSHVGLVDEAW 464
Query: 370 ALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKI 429
+N M + YG+ ++HY + DLLG G+L+EAY+ I M V+P VW LL+SC +
Sbjct: 465 GYFNSMTKVYGLNQELEHYAAVADLLGRAGKLEEAYNFISKMCVEPTGSVWSTLLSSCSV 524
Query: 430 HGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACS 489
H N+ELAE EK+ ++ ++ G YV++ NMYA +G+W+ + KLR M K ++K ACS
Sbjct: 525 HKNLELAEKVAEKIFTVDSENMGAYVLMCNMYASNGRWKEMAKLRLRMRKKGLRKKPACS 584
Query: 490 WIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTGSVFHDVEEDEKTSMV 549
WIE+KNK + F++GD SH + D I LK + M + GY DT V HDV+E+ K ++
Sbjct: 585 WIEMKNKTHGFVSGDRSHPSMDKINEFLKAVMEQMEKEGYVADTSGVLHDVDEEHKRELL 644
Query: 550 CSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIMEREITVRDVNRYHSF 609
HSERLA+AFG+I+T PGT + +TKN+RIC DCHVAIKFISKI EREI VRD +R+H F
Sbjct: 645 FGHSERLAVAFGIINTEPGTTIRVTKNIRICTDCHVAIKFISKITEREIIVRDNSRFHHF 704
Query: 610 KHGMCSCGDHW 620
G CSCGD+W
Sbjct: 705 NRGNCSCGDYW 715
Score = 166 bits (420), Expect = 4e-41
Identities = 113/429 (26%), Positives = 208/429 (48%), Gaps = 40/429 (9%)
Query: 52 SLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQ 111
+L+++ K+ + KQLHAQF + + A+ ++ +Y L A LF +
Sbjct: 10 TLIKNPTRIKSKSQAKQLHAQFIRTQ-SLSHTSASIVISIYTNLKLLHEALLLFKTLKSP 68
Query: 112 NLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIH 171
+ W +IR + A+ + +M G PD+ P VLK+C+ + + G S+H
Sbjct: 69 PVLAWKSVIRCFTDQSLFSKALASFVEMRASGRCPDHNVFPSVLKSCTMMMDLRFGESVH 128
Query: 172 EYVIKSGWERDLFVGAALIDMYAKC-------------------------------GCVM 200
++++ G + DL+ G AL++MYAK C+M
Sbjct: 129 GFIVRLGMDCDLYTGNALMNMYAKLLGMGSKISVGNVFDEMPQRTSNSGDEDVKAETCIM 188
Query: 201 DAG-----RVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTV 255
G RVF+ + +D V +N+++A YAQ+G ++++ + REM ++P TL +V
Sbjct: 189 PFGIDSVRRVFEVMPRKDVVSYNTIIAGYAQSGMYEDALRMVREMGTTDLKPDSFTLSSV 248
Query: 256 ISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRV 315
+ ++ + G+EIHG+ R G S+ + ++L+DMYAK ++ + +F RL +
Sbjct: 249 LPIFSEYVDVIKGKEIHGYVIRKGIDSDVYIGSSLVDMYAKSARIEDSERVFSRLYCRDG 308
Query: 316 VSWNAIITGYAMHGLAVGALDLFDKM-RKEDRPDHITFVGVLAACSRGRLLDEGRALYNL 374
+SWN+++ GY +G AL LF +M + +P + F V+ AC+ L G+ L+
Sbjct: 309 ISWNSLVAGYVQNGRYNEALRLFRQMVTAKVKPGAVAFSSVIPACAHLATLHLGKQLHGY 368
Query: 375 MVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVE 434
++R G + + ++D+ CG + A + M+V D W A++ +HG+
Sbjct: 369 VLRG-GFGSNIFIASALVDMYSKCGNIKAARKIFDRMNVL-DEVSWTAIIMGHALHGHGH 426
Query: 435 LAELALEKL 443
A E++
Sbjct: 427 EAVSLFEEM 435
Score = 46.2 bits (108), Expect = 5e-05
Identities = 34/131 (25%), Positives = 62/131 (46%), Gaps = 2/131 (1%)
Query: 248 TEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALF 307
++A + T+I + + +++H R S+ + +I +Y + AL LF
Sbjct: 4 SKALIKTLIKNPTRIKSKSQAKQLHAQFIRTQSLSHTSA-SIVISIYTNLKLLHEALLLF 62
Query: 308 ERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDR-PDHITFVGVLAACSRGRLLD 366
+ L+ V++W ++I + L AL F +MR R PDH F VL +C+ L
Sbjct: 63 KTLKSPPVLAWKSVIRCFTDQSLFSKALASFVEMRASGRCPDHNVFPSVLKSCTMMMDLR 122
Query: 367 EGRALYNLMVR 377
G +++ +VR
Sbjct: 123 FGESVHGFIVR 133
>At3g46790 putative protein
Length = 657
Score = 478 bits (1231), Expect = e-135
Identities = 244/592 (41%), Positives = 373/592 (62%), Gaps = 11/592 (1%)
Query: 36 SVDSFPPQPTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVS 95
S +S P Q T Y L+ C +L+ ++H G + LATKL+ +Y+
Sbjct: 70 SQESSPSQQT----YELLILCCGHRSSLSDALRVHRHILDNGSDQDPFLATKLIGMYSDL 125
Query: 96 NSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVL 155
S+ AR +FDK K+ +++WN L R G + + LY KM G+ D FT +VL
Sbjct: 126 GSVDYARKVFDKTRKRTIYVWNALFRALTLAGHGEEVLGLYWKMNRIGVESDRFTYTYVL 185
Query: 156 KACSA----LSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVV 211
KAC A ++ + +G+ IH ++ + G+ +++ L+DMYA+ GCV A VF + V
Sbjct: 186 KACVASECTVNHLMKGKEIHAHLTRRGYSSHVYIMTTLVDMYARFGCVDYASYVFGGMPV 245
Query: 212 RDAVLWNSMLAAYAQNGHPDESISLCREMA--ANGVRPTEATLVTVISSSADVACLPYGR 269
R+ V W++M+A YA+NG E++ REM P T+V+V+ + A +A L G+
Sbjct: 246 RNVVSWSAMIACYAKNGKAFEALRTFREMMRETKDSSPNSVTMVSVLQACASLAALEQGK 305
Query: 270 EIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHG 329
IHG+ R G S V +AL+ MY +CG ++V +F+R+ ++ VVSWN++I+ Y +HG
Sbjct: 306 LIHGYILRRGLDSILPVISALVTMYGRCGKLEVGQRVFDRMHDRDVVSWNSLISSYGVHG 365
Query: 330 LAVGALDLFDKMRKEDR-PDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHY 388
A+ +F++M P +TFV VL ACS L++EG+ L+ M RD+GI P ++HY
Sbjct: 366 YGKKAIQIFEEMLANGASPTPVTFVSVLGACSHEGLVEEGKRLFETMWRDHGIKPQIEHY 425
Query: 389 TCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEP 448
CM+DLLG +LDEA ++++M +P VWG+LL SC+IHGNVELAE A +L LEP
Sbjct: 426 ACMVDLLGRANRLDEAAKMVQDMRTEPGPKVWGSLLGSCRIHGNVELAERASRRLFALEP 485
Query: 449 DDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHS 508
++GNYV+LA++YA++ W+ V+++++++ + ++K W+EV+ K+Y+F++ D +
Sbjct: 486 KNAGNYVLLADIYAEAQMWDEVKRVKKLLEHRGLQKLPGRCWMEVRRKMYSFVSVDEFNP 545
Query: 509 NSDAIYAELKRLEGLMHEAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPG 568
+ I+A L +L M E GY P T V +++E +EK +V HSE+LA+AFGLI+TS G
Sbjct: 546 LMEQIHAFLVKLAEDMKEKGYIPQTKGVLYELETEEKERIVLGHSEKLALAFGLINTSKG 605
Query: 569 TRLLITKNLRICEDCHVAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
+ ITKNLR+CEDCH+ KFISK ME+EI VRDVNR+H FK+G+CSCGD+W
Sbjct: 606 EPIRITKNLRLCEDCHLFTKFISKFMEKEILVRDVNRFHRFKNGVCSCGDYW 657
Score = 147 bits (371), Expect = 2e-35
Identities = 98/335 (29%), Positives = 167/335 (49%), Gaps = 13/335 (3%)
Query: 117 NVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIK 176
N LI+ G AI ++L P T ++ C S++ + +H +++
Sbjct: 50 NQLIQSLCKEGKLKQAI----RVLSQESSPSQQTYELLILCCGHRSSLSDALRVHRHILD 105
Query: 177 SGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISL 236
+G ++D F+ LI MY+ G V A +VFDK R +WN++ A GH +E + L
Sbjct: 106 NGSDQDPFLATKLIGMYSDLGSVDYARKVFDKTRKRTIYVWNALFRALTLAGHGEEVLGL 165
Query: 237 CREMAANGVRPTEAT----LVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALID 292
+M GV T L ++S V L G+EIH R G+ S+ + T L+D
Sbjct: 166 YWKMNRIGVESDRFTYTYVLKACVASECTVNHLMKGKEIHAHLTRRGYSSHVYIMTTLVD 225
Query: 293 MYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDR---PDH 349
MYA+ G V A +F + + VVSW+A+I YA +G A AL F +M +E + P+
Sbjct: 226 MYARFGCVDYASYVFGGMPVRNVVSWSAMIACYAKNGKAFEALRTFREMMRETKDSSPNS 285
Query: 350 ITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIR 409
+T V VL AC+ L++G+ ++ ++R G+ + + ++ + G CG+L+ +
Sbjct: 286 VTMVSVLQACASLAALEQGKLIHGYILR-RGLDSILPVISALVTMYGRCGKLEVGQRVFD 344
Query: 410 NMSVKPDSGVWGALLNSCKIHGNVELAELALEKLI 444
M + D W +L++S +HG + A E+++
Sbjct: 345 RMHDR-DVVSWNSLISSYGVHGYGKKAIQIFEEML 378
>At2g27610 putative selenium-binding protein
Length = 868
Score = 475 bits (1222), Expect = e-134
Identities = 242/575 (42%), Positives = 363/575 (63%), Gaps = 8/575 (1%)
Query: 50 YTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIP 109
+ S+++ C + K L +QLH G ++Q++ T L+ Y+ ++L+A LF +I
Sbjct: 298 FASVIKLCANLKELRFTEQLHCSVVKYGFLFDQNIRTALMVAYSKCTAMLDALRLFKEIG 357
Query: 110 -KQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGR 168
N+ W +I G+ N + A+ L+ +M G+RP+ FT +L A +S
Sbjct: 358 CVGNVVSWTAMISGFLQNDGKEEAVDLFSEMKRKGVRPNEFTYSVILTALPVISP----S 413
Query: 169 SIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNG 228
+H V+K+ +ER VG AL+D Y K G V +A +VF I +D V W++MLA YAQ G
Sbjct: 414 EVHAQVVKTNYERSSTVGTALLDAYVKLGKVEEAAKVFSGIDDKDIVAWSAMLAGYAQTG 473
Query: 229 HPDESISLCREMAANGVRPTEATLVTVIS-SSADVACLPYGREIHGFGWRHGFQSNDKVK 287
+ +I + E+ G++P E T ++++ +A A + G++ HGF + S+ V
Sbjct: 474 ETEAAIKMFGELTKGGIKPNEFTFSSILNVCAATNASMGQGKQFHGFAIKSRLDSSLCVS 533
Query: 288 TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-R 346
+AL+ MYAK G+++ A +F+R REK +VSWN++I+GYA HG A+ ALD+F +M+K +
Sbjct: 534 SALLTMYAKKGNIESAEEVFKRQREKDLVSWNSMISGYAQHGQAMKALDVFKEMKKRKVK 593
Query: 347 PDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYD 406
D +TF+GV AAC+ L++EG +++MVRD I PT +H +CM+DL GQL++A
Sbjct: 594 MDGVTFIGVFAACTHAGLVEEGEKYFDIMVRDCKIAPTKEHNSCMVDLYSRAGQLEKAMK 653
Query: 407 LIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGK 466
+I NM S +W +L +C++H EL LA EK+I ++P+DS YV+L+NMYA+SG
Sbjct: 654 VIENMPNPAGSTIWRTILAACRVHKKTELGRLAAEKIIAMKPEDSAAYVLLSNMYAESGD 713
Query: 467 WEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHE 526
W+ K+R++M ++ +KK SWIEVKNK Y+FLAGD SH D IY +L+ L + +
Sbjct: 714 WQERAKVRKLMNERNVKKEPGYSWIEVKNKTYSFLAGDRSHPLKDQIYMKLEDLSTRLKD 773
Query: 527 AGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVA 586
GY PDT V D++++ K +++ HSERLAIAFGLI+T G+ LLI KNLR+C DCH+
Sbjct: 774 LGYEPDTSYVLQDIDDEHKEAVLAQHSERLAIAFGLIATPKGSPLLIIKNLRVCGDCHLV 833
Query: 587 IKFISKIMEREITVRDVNRYHSF-KHGMCSCGDHW 620
IK I+KI EREI VRD NR+H F G+CSCGD W
Sbjct: 834 IKLIAKIEEREIVVRDSNRFHHFSSDGVCSCGDFW 868
Score = 182 bits (463), Expect = 4e-46
Identities = 115/373 (30%), Positives = 195/373 (51%), Gaps = 8/373 (2%)
Query: 66 GKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAW 125
G+QLH Q G + + T LV Y ++ + R +FD++ ++N+ W LI GYA
Sbjct: 112 GRQLHCQCIKFGFLDDVSVGTSLVDTYMKGSNFKDGRKVFDEMKERNVVTWTTLISGYAR 171
Query: 126 NGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFV 185
N +D + L+ +M + G +P++FT L + G G +H V+K+G ++ + V
Sbjct: 172 NSMNDEVLTLFMRMQNEGTQPNSFTFAAALGVLAEEGVGGRGLQVHTVVVKNGLDKTIPV 231
Query: 186 GAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGV 245
+LI++Y KCG V A +FDK V+ V WNSM++ YA NG E++ + M N V
Sbjct: 232 SNSLINLYLKCGNVRKARILFDKTEVKSVVTWNSMISGYAANGLDLEALGMFYSMRLNYV 291
Query: 246 RPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALA 305
R +E++ +VI A++ L + ++H ++GF + ++TAL+ Y+KC ++ AL
Sbjct: 292 RLSESSFASVIKLCANLKELRFTEQLHCSVVKYGFLFDQNIRTALMVAYSKCTAMLDALR 351
Query: 306 LFERLR-EKRVVSWNAIITGYAMHGLAVGALDLFDKM-RKEDRPDHITFVGVLAACSRGR 363
LF+ + VVSW A+I+G+ + A+DLF +M RK RP+ T+ +L A
Sbjct: 352 LFKEIGCVGNVVSWTAMISGFLQNDGKEEAVDLFSEMKRKGVRPNEFTYSVILTALP--- 408
Query: 364 LLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGAL 423
++ ++ +Y + TV T ++D G+++EA + + K D W A+
Sbjct: 409 VISPSEVHAQVVKTNYERSSTVG--TALLDAYVKLGKVEEAAKVFSGIDDK-DIVAWSAM 465
Query: 424 LNSCKIHGNVELA 436
L G E A
Sbjct: 466 LAGYAQTGETEAA 478
Score = 145 bits (365), Expect = 9e-35
Identities = 97/332 (29%), Positives = 163/332 (48%), Gaps = 2/332 (0%)
Query: 95 SNSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFV 154
S+ L NA NLFDK P ++ + L+ G++ +G A L+ + G+ D V
Sbjct: 40 SSRLYNAHNLFDKSPGRDRESYISLLFGFSRDGRTQEAKRLFLNIHRLGMEMDCSIFSSV 99
Query: 155 LKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDA 214
LK + L GR +H IK G+ D+ VG +L+D Y K D +VFD++ R+
Sbjct: 100 LKVSATLCDELFGRQLHCQCIKFGFLDDVSVGTSLVDTYMKGSNFKDGRKVFDEMKERNV 159
Query: 215 VLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGF 274
V W ++++ YA+N DE ++L M G +P T + A+ G ++H
Sbjct: 160 VTWTTLISGYARNSMNDEVLTLFMRMQNEGTQPNSFTFAAALGVLAEEGVGGRGLQVHTV 219
Query: 275 GWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGA 334
++G V +LI++Y KCG+V+ A LF++ K VV+WN++I+GYA +GL + A
Sbjct: 220 VVKNGLDKTIPVSNSLINLYLKCGNVRKARILFDKTEVKSVVTWNSMISGYAANGLDLEA 279
Query: 335 LDLFDKMR-KEDRPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMID 393
L +F MR R +F V+ C+ + L L+ +V+ YG T ++
Sbjct: 280 LGMFYSMRLNYVRLSESSFASVIKLCANLKELRFTEQLHCSVVK-YGFLFDQNIRTALMV 338
Query: 394 LLGHCGQLDEAYDLIRNMSVKPDSGVWGALLN 425
C + +A L + + + W A+++
Sbjct: 339 AYSKCTAMLDALRLFKEIGCVGNVVSWTAMIS 370
>At1g08070 unknown protein
Length = 741
Score = 474 bits (1221), Expect = e-134
Identities = 241/607 (39%), Positives = 366/607 (59%), Gaps = 34/607 (5%)
Query: 48 YGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLL-------- 99
Y + +L+SC SKA G+Q+H LG + + T L+ +Y + L
Sbjct: 135 YTFPFVLKSCAKSKAFKEGQQIHGHVLKLGCDLDLYVHTSLISMYVQNGRLEDAHKVFDK 194
Query: 100 -----------------------NARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILY 136
NA+ LFD+IP +++ WN +I GYA G + A+ L+
Sbjct: 195 SPHRDVVSYTALIKGYASRGYIENAQKLFDEIPVKDVVSWNAMISGYAETGNYKEALELF 254
Query: 137 HKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKC 196
M+ +RPD T+ V+ AC+ +I GR +H ++ G+ +L + ALID+Y+KC
Sbjct: 255 KDMMKTNVRPDESTMVTVVSACAQSGSIELGRQVHLWIDDHGFGSNLKIVNALIDLYSKC 314
Query: 197 GCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVI 256
G + A +F+++ +D + WN+++ Y E++ L +EM +G P + T+++++
Sbjct: 315 GELETACGLFERLPYKDVISWNTLIGGYTHMNLYKEALLLFQEMLRSGETPNDVTMLSIL 374
Query: 257 SSSADVACLPYGREIHGFGWRH--GFQSNDKVKTALIDMYAKCGSVKVALALFERLREKR 314
+ A + + GR IH + + G + ++T+LIDMYAKCG ++ A +F + K
Sbjct: 375 PACAHLGAIDIGRWIHVYIDKRLKGVTNASSLRTSLIDMYAKCGDIEAAHQVFNSILHKS 434
Query: 315 VVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYN 373
+ SWNA+I G+AMHG A + DLF +MRK +PD ITFVG+L+ACS +LD GR ++
Sbjct: 435 LSSWNAMIFGFAMHGRADASFDLFSRMRKIGIQPDDITFVGLLSACSHSGMLDLGRHIFR 494
Query: 374 LMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNV 433
M +DY +TP ++HY CMIDLLGH G EA ++I M ++PD +W +LL +CK+HGNV
Sbjct: 495 TMTQDYKMTPKLEHYGCMIDLLGHSGLFKEAEEMINMMEMEPDGVIWCSLLKACKMHGNV 554
Query: 434 ELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACSWIEV 493
EL E E LI++EP++ G+YV+L+N+YA +G+W V K R ++ DK +KK CS IE+
Sbjct: 555 ELGESFAENLIKIEPENPGSYVLLSNIYASAGRWNEVAKTRALLNDKGMKKVPGCSSIEI 614
Query: 494 KNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTGSVFHDVEEDEKTSMVCSHS 553
+ V+ F+ GD H + IY L+ +E L+ +AG+ PDT V ++EE+ K + HS
Sbjct: 615 DSVVHEFIIGDKFHPRNREIYGMLEEMEVLLEKAGFVPDTSEVLQEMEEEWKEGALRHHS 674
Query: 554 ERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIMEREITVRDVNRYHSFKHGM 613
E+LAIAFGLIST PGT+L I KNLR+C +CH A K ISKI +REI RD R+H F+ G+
Sbjct: 675 EKLAIAFGLISTKPGTKLTIVKNLRVCRNCHEATKLISKIYKREIIARDRTRFHHFRDGV 734
Query: 614 CSCGDHW 620
CSC D+W
Sbjct: 735 CSCNDYW 741
Score = 243 bits (620), Expect = 2e-64
Identities = 159/484 (32%), Positives = 251/484 (51%), Gaps = 48/484 (9%)
Query: 20 PFSQSFYHSLATHQTASVDSFPPQPTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIA 79
P S +H L + DS P+ SLL +C K L + +HAQ +G+
Sbjct: 11 PSSSYPFHFLPSSSDPPYDSIRNHPSL-----SLLHNC---KTLQSLRIIHAQMIKIGLH 62
Query: 80 YNQDLATKLVHLYAVS---NSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILY 136
+KL+ +S L A ++F I + NL +WN + RG+A + +A+ LY
Sbjct: 63 NTNYALSKLIEFCILSPHFEGLPYAISVFKTIQEPNLLIWNTMFRGHALSSDPVSALKLY 122
Query: 137 HKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKC 196
M+ GL P+++T PFVLK+C+ A EG+ IH +V+K G + DL+V +LI MY +
Sbjct: 123 VCMISLGLLPNSYTFPFVLKSCAKSKAFKEGQQIHGHVLKLGCDLDLYVHTSLISMYVQN 182
Query: 197 GCVMDAGRVFDK-------------------------------IVVRDAVLWNSMLAAYA 225
G + DA +VFDK I V+D V WN+M++ YA
Sbjct: 183 GRLEDAHKVFDKSPHRDVVSYTALIKGYASRGYIENAQKLFDEIPVKDVVSWNAMISGYA 242
Query: 226 QNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDK 285
+ G+ E++ L ++M VRP E+T+VTV+S+ A + GR++H + HGF SN K
Sbjct: 243 ETGNYKEALELFKDMMKTNVRPDESTMVTVVSACAQSGSIELGRQVHLWIDDHGFGSNLK 302
Query: 286 VKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKM-RKE 344
+ ALID+Y+KCG ++ A LFERL K V+SWN +I GY L AL LF +M R
Sbjct: 303 IVNALIDLYSKCGELETACGLFERLPYKDVISWNTLIGGYTHMNLYKEALLLFQEMLRSG 362
Query: 345 DRPDHITFVGVLAACSRGRLLDEGRALY-NLMVRDYGITPTVQHYTCMIDLLGHCGQLDE 403
+ P+ +T + +L AC+ +D GR ++ + R G+T T +ID+ CG ++
Sbjct: 363 ETPNDVTMLSILPACAHLGAIDIGRWIHVYIDKRLKGVTNASSLRTSLIDMYAKCGDIEA 422
Query: 404 AYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKL--IELEPDDSGNYVILANMY 461
A+ + ++ K S W A++ +HG + + ++ I ++PDD +V L +
Sbjct: 423 AHQVFNSILHKSLSS-WNAMIFGFAMHGRADASFDLFSRMRKIGIQPDDI-TFVGLLSAC 480
Query: 462 AQSG 465
+ SG
Sbjct: 481 SHSG 484
>At3g57430 putative protein
Length = 803
Score = 462 bits (1190), Expect = e-130
Identities = 230/585 (39%), Positives = 368/585 (62%), Gaps = 15/585 (2%)
Query: 51 TSLLQSCIDSKALNPGKQLHAQFYHLG-IAYNQDLATKLVHLYAVSNSLLNARNLFDKIP 109
+S+L +C + L GK+LHA G + N + + LV +Y +L+ R +FD +
Sbjct: 219 SSVLPACSHLEMLRTGKELHAYALKNGSLDENSFVGSALVDMYCNCKQVLSGRRVFDGMF 278
Query: 110 KQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDY-GLRPDNFTLPFVLKACSALSAIGEGR 168
+ + LWN +I GY+ N A++L+ M + GL ++ T+ V+ AC A
Sbjct: 279 DRKIGLWNAMIAGYSQNEHDKEALLLFIGMEESAGLLANSTTMAGVVPACVRSGAFSRKE 338
Query: 169 SIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNG 228
+IH +V+K G +RD FV L+DMY++ G + A R+F K+ RD V WN+M+ Y +
Sbjct: 339 AIHGFVVKRGLDRDRFVQNTLMDMYSRLGKIDIAMRIFGKMEDRDLVTWNTMITGYVFSE 398
Query: 229 HPDESISLCREM-----------AANGVRPTEATLVTVISSSADVACLPYGREIHGFGWR 277
H ++++ L +M + ++P TL+T++ S A ++ L G+EIH + +
Sbjct: 399 HHEDALLLLHKMQNLERKVSKGASRVSLKPNSITLMTILPSCAALSALAKGKEIHAYAIK 458
Query: 278 HGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDL 337
+ ++ V +AL+DMYAKCG ++++ +F+++ +K V++WN II Y MHG A+DL
Sbjct: 459 NNLATDVAVGSALVDMYAKCGCLQMSRKVFDQIPQKNVITWNVIIMAYGMHGNGQEAIDL 518
Query: 338 FDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLG 396
M + +P+ +TF+ V AACS ++DEG ++ +M DYG+ P+ HY C++DLLG
Sbjct: 519 LRMMMVQGVKPNEVTFISVFAACSHSGMVDEGLRIFYVMKPDYGVEPSSDHYACVVDLLG 578
Query: 397 HCGQLDEAYDLIRNMSVKPD-SGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYV 455
G++ EAY L+ M + +G W +LL + +IH N+E+ E+A + LI+LEP+ + +YV
Sbjct: 579 RAGRIKEAYQLMNMMPRDFNKAGAWSSLLGASRIHNNLEIGEIAAQNLIQLEPNVASHYV 638
Query: 456 ILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYA 515
+LAN+Y+ +G W+ ++R+ M ++ ++K CSWIE ++V+ F+AGD SH S+ +
Sbjct: 639 LLANIYSSAGLWDKATEVRRNMKEQGVRKEPGCSWIEHGDEVHKFVAGDSSHPQSEKLSG 698
Query: 516 ELKRLEGLMHEAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITK 575
L+ L M + GY PDT V H+VEEDEK ++C HSE+LAIAFG+++TSPGT + + K
Sbjct: 699 YLETLWERMRKEGYVPDTSCVLHNVEEDEKEILLCGHSEKLAIAFGILNTSPGTIIRVAK 758
Query: 576 NLRICEDCHVAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
NLR+C DCH+A KFISKI++REI +RDV R+H FK+G CSCGD+W
Sbjct: 759 NLRVCNDCHLATKFISKIVDREIILRDVRRFHRFKNGTCSCGDYW 803
Score = 165 bits (418), Expect = 6e-41
Identities = 115/408 (28%), Positives = 195/408 (47%), Gaps = 10/408 (2%)
Query: 47 HYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQ-DLATKLVHLYAVSNSLLNARNLF 105
+Y + +LL++ D + + GKQ+HA Y G + +A LV+LY +F
Sbjct: 10 NYAFPALLKAVADLQDMELGKQIHAHVYKFGYGVDSVTVANTLVNLYRKCGDFGAVYKVF 69
Query: 106 DKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSAL---S 162
D+I ++N WN LI + A+ + MLD + P +FTL V+ ACS L
Sbjct: 70 DRISERNQVSWNSLISSLCSFEKWEMALEAFRCMLDENVEPSSFTLVSVVTACSNLPMPE 129
Query: 163 AIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLA 222
+ G+ +H Y ++ G E + F+ L+ MY K G + + + RD V WN++L+
Sbjct: 130 GLMMGKQVHAYGLRKG-ELNSFIINTLVAMYGKLGKLASSKVLLGSFGGRDLVTWNTVLS 188
Query: 223 AYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHG-FQ 281
+ QN E++ REM GV P E T+ +V+ + + + L G+E+H + ++G
Sbjct: 189 SLCQNEQLLEALEYLREMVLEGVEPDEFTISSVLPACSHLEMLRTGKELHAYALKNGSLD 248
Query: 282 SNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKM 341
N V +AL+DMY C V +F+ + ++++ WNA+I GY+ + AL LF M
Sbjct: 249 ENSFVGSALVDMYCNCKQVLSGRRVFDGMFDRKIGLWNAMIAGYSQNEHDKEALLLFIGM 308
Query: 342 RKED--RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCG 399
+ + T GV+ AC R A++ +V+ G+ ++D+ G
Sbjct: 309 EESAGLLANSTTMAGVVPACVRSGAFSRKEAIHGFVVK-RGLDRDRFVQNTLMDMYSRLG 367
Query: 400 QLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELE 447
++D A + M + D W ++ + E A L L K+ LE
Sbjct: 368 KIDIAMRIFGKMEDR-DLVTWNTMITGYVFSEHHEDALLLLHKMQNLE 414
Score = 121 bits (304), Expect = 1e-27
Identities = 78/291 (26%), Positives = 147/291 (49%), Gaps = 7/291 (2%)
Query: 139 MLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERD-LFVGAALIDMYAKCG 197
M+ G++PDN+ P +LKA + L + G+ IH +V K G+ D + V L+++Y KCG
Sbjct: 1 MIVLGIKPDNYAFPALLKAVADLQDMELGKQIHAHVYKFGYGVDSVTVANTLVNLYRKCG 60
Query: 198 CVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVIS 257
+VFD+I R+ V WNS++++ + ++ R M V P+ TLV+V++
Sbjct: 61 DFGAVYKVFDRISERNQVSWNSLISSLCSFEKWEMALEAFRCMLDENVEPSSFTLVSVVT 120
Query: 258 SSADVAC---LPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKR 314
+ +++ L G+++H +G R G + N + L+ MY K G + + L +
Sbjct: 121 ACSNLPMPEGLMMGKQVHAYGLRKG-ELNSFIINTLVAMYGKLGKLASSKVLLGSFGGRD 179
Query: 315 VVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYN 373
+V+WN +++ + + AL+ +M E PD T VL ACS +L G+ L+
Sbjct: 180 LVTWNTVLSSLCQNEQLLEALEYLREMVLEGVEPDEFTISSVLPACSHLEMLRTGKELHA 239
Query: 374 LMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALL 424
+++ + + ++D+ +C Q+ + M + G+W A++
Sbjct: 240 YALKNGSLDENSFVGSALVDMYCNCKQVLSGRRVFDGMFDR-KIGLWNAMI 289
>At1g11290 hypothetical protein
Length = 809
Score = 462 bits (1190), Expect = e-130
Identities = 232/570 (40%), Positives = 360/570 (62%), Gaps = 2/570 (0%)
Query: 52 SLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQ 111
S+L + + ++ GK++H G +++T LV +YA SL AR LFD + ++
Sbjct: 241 SVLPAVSALRLISVGKEIHGYAMRSGFDSLVNISTALVDMYAKCGSLETARQLFDGMLER 300
Query: 112 NLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIH 171
N+ WN +I Y N A++++ KMLD G++P + ++ L AC+ L + GR IH
Sbjct: 301 NVVSWNSMIDAYVQNENPKEAMLIFQKMLDEGVKPTDVSVMGALHACADLGDLERGRFIH 360
Query: 172 EYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPD 231
+ ++ G +R++ V +LI MY KC V A +F K+ R V WN+M+ +AQNG P
Sbjct: 361 KLSVELGLDRNVSVVNSLISMYCKCKEVDTAASMFGKLQSRTLVSWNAMILGFAQNGRPI 420
Query: 232 ESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALI 291
++++ +M + V+P T V+VI++ A+++ + + IHG R N V TAL+
Sbjct: 421 DALNYFSQMRSRTVKPDTFTYVSVITAIAELSITHHAKWIHGVVMRSCLDKNVFVTTALV 480
Query: 292 DMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHI 350
DMYAKCG++ +A +F+ + E+ V +WNA+I GY HG AL+LF++M+K +P+ +
Sbjct: 481 DMYAKCGAIMIARLIFDMMSERHVTTWNAMIDGYGTHGFGKAALELFEEMQKGTIKPNGV 540
Query: 351 TFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRN 410
TF+ V++ACS L++ G + +M +Y I ++ HY M+DLLG G+L+EA+D I
Sbjct: 541 TFLSVISACSHSGLVEAGLKCFYMMKENYSIELSMDHYGAMVDLLGRAGRLNEAWDFIMQ 600
Query: 411 MSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGV 470
M VKP V+GA+L +C+IH NV AE A E+L EL PDD G +V+LAN+Y + WE V
Sbjct: 601 MPVKPAVNVYGAMLGACQIHKNVNFAEKAAERLFELNPDDGGYHVLLANIYRAASMWEKV 660
Query: 471 EKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYA 530
++R M+ + ++K CS +E+KN+V++F +G +H +S IYA L++L + EAGY
Sbjct: 661 GQVRVSMLRQGLRKTPGCSMVEIKNEVHSFFSGSTAHPDSKKIYAFLEKLICHIKEAGYV 720
Query: 531 PDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFI 590
PDT V VE D K ++ +HSE+LAI+FGL++T+ GT + + KNLR+C DCH A K+I
Sbjct: 721 PDTNLVL-GVENDVKEQLLSTHSEKLAISFGLLNTTAGTTIHVRKNLRVCADCHNATKYI 779
Query: 591 SKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
S + REI VRD+ R+H FK+G CSCGD+W
Sbjct: 780 SLVTGREIVVRDMQRFHHFKNGACSCGDYW 809
Score = 235 bits (599), Expect = 6e-62
Identities = 132/401 (32%), Positives = 221/401 (54%), Gaps = 6/401 (1%)
Query: 48 YGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDK 107
Y +T LL+ C D L GK++H G + + T L ++YA + AR +FD+
Sbjct: 136 YNFTYLLKVCGDEAELRVGKEIHGLLVKSGFSLDLFAMTGLENMYAKCRQVNEARKVFDR 195
Query: 108 IPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEG 167
+P+++L WN ++ GY+ NG A+ + M + L+P T+ VL A SAL I G
Sbjct: 196 MPERDLVSWNTIVAGYSQNGMARMALEMVKSMCEENLKPSFITIVSVLPAVSALRLISVG 255
Query: 168 RSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQN 227
+ IH Y ++SG++ + + AL+DMYAKCG + A ++FD ++ R+ V WNSM+ AY QN
Sbjct: 256 KEIHGYAMRSGFDSLVNISTALVDMYAKCGSLETARQLFDGMLERNVVSWNSMIDAYVQN 315
Query: 228 GHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVK 287
+P E++ + ++M GV+PT+ +++ + + AD+ L GR IH G N V
Sbjct: 316 ENPKEAMLIFQKMLDEGVKPTDVSVMGALHACADLGDLERGRFIHKLSVELGLDRNVSVV 375
Query: 288 TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-R 346
+LI MY KC V A ++F +L+ + +VSWNA+I G+A +G + AL+ F +MR +
Sbjct: 376 NSLISMYCKCKEVDTAASMFGKLQSRTLVSWNAMILGFAQNGRPIDALNYFSQMRSRTVK 435
Query: 347 PDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYD 406
PD T+V V+ A + + + ++ +++R + V T ++D+ CG + A
Sbjct: 436 PDTFTYVSVITAIAELSITHHAKWIHGVVMRSC-LDKNVFVTTALVDMYAKCGAIMIA-R 493
Query: 407 LIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELE 447
LI +M + W A+++ HG + ALE E++
Sbjct: 494 LIFDMMSERHVTTWNAMIDGYGTHG---FGKAALELFEEMQ 531
Score = 197 bits (500), Expect = 2e-50
Identities = 119/389 (30%), Positives = 203/389 (51%), Gaps = 21/389 (5%)
Query: 33 QTASVDSFPPQPTTHYGYTS----------------LLQSCIDSKALNPGKQLHAQFYHL 76
Q ++V P P+ H + S LL+ C K L +Q+ +
Sbjct: 7 QFSTVPQIPNPPSRHRHFLSERNYIPANVYEHPAALLLERCSSLKEL---RQILPLVFKN 63
Query: 77 GIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILY 136
G+ TKLV L+ S+ A +F+ I + L++ +++G+A D A+ +
Sbjct: 64 GLYQEHFFQTKLVSLFCRYGSVDEAARVFEPIDSKLNVLYHTMLKGFAKVSDLDKALQFF 123
Query: 137 HKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKC 196
+M + P + ++LK C + + G+ IH ++KSG+ DLF L +MYAKC
Sbjct: 124 VRMRYDDVEPVVYNFTYLLKVCGDEAELRVGKEIHGLLVKSGFSLDLFAMTGLENMYAKC 183
Query: 197 GCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVI 256
V +A +VFD++ RD V WN+++A Y+QNG ++ + + M ++P+ T+V+V+
Sbjct: 184 RQVNEARKVFDRMPERDLVSWNTIVAGYSQNGMARMALEMVKSMCEENLKPSFITIVSVL 243
Query: 257 SSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVV 316
+ + + + G+EIHG+ R GF S + TAL+DMYAKCGS++ A LF+ + E+ VV
Sbjct: 244 PAVSALRLISVGKEIHGYAMRSGFDSLVNISTALVDMYAKCGSLETARQLFDGMLERNVV 303
Query: 317 SWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLM 375
SWN++I Y + A+ +F KM E +P ++ +G L AC+ L+ GR ++ L
Sbjct: 304 SWNSMIDAYVQNENPKEAMLIFQKMLDEGVKPTDVSVMGALHACADLGDLERGRFIHKLS 363
Query: 376 VRDYGITPTVQHYTCMIDLLGHCGQLDEA 404
V + G+ V +I + C ++D A
Sbjct: 364 V-ELGLDRNVSVVNSLISMYCKCKEVDTA 391
Score = 65.1 bits (157), Expect = 1e-10
Identities = 51/213 (23%), Positives = 98/213 (45%), Gaps = 12/213 (5%)
Query: 14 FIFNLFPFSQSFYHSLATHQTASVDSFPPQPTTHYGYTSLLQSCIDSKALNPGKQLHAQF 73
F N P Y S +T D+F Y S++ + + + K +H
Sbjct: 413 FAQNGRPIDALNYFSQMRSRTVKPDTFT--------YVSVITAIAELSITHHAKWIHGVV 464
Query: 74 YHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAI 133
+ N + T LV +YA +++ AR +FD + ++++ WN +I GY +G A+
Sbjct: 465 MRSCLDKNVFVTTALVDMYAKCGAIMIARLIFDMMSERHVTTWNAMIDGYGTHGFGKAAL 524
Query: 134 ILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVG--AALID 191
L+ +M ++P+ T V+ ACS + G Y++K + +L + A++D
Sbjct: 525 ELFEEMQKGTIKPNGVTFLSVISACSHSGLVEAGLKCF-YMMKENYSIELSMDHYGAMVD 583
Query: 192 MYAKCGCVMDAGRVFDKIVVRDAV-LWNSMLAA 223
+ + G + +A ++ V+ AV ++ +ML A
Sbjct: 584 LLGRAGRLNEAWDFIMQMPVKPAVNVYGAMLGA 616
>At3g13770 hypothetical protein
Length = 628
Score = 456 bits (1172), Expect = e-128
Identities = 215/576 (37%), Positives = 358/576 (61%), Gaps = 3/576 (0%)
Query: 48 YGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDK 107
+GY +LL +C+D +AL G+++HA L T+L+ Y + L +AR + D+
Sbjct: 53 HGYDALLNACLDKRALRDGQRVHAHMIKTRYLPATYLRTRLLIFYGKCDCLEDARKVLDE 112
Query: 108 IPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEG 167
+P++N+ W +I Y+ G A+ ++ +M+ +P+ FT VL +C S +G G
Sbjct: 113 MPEKNVVSWTAMISRYSQTGHSSEALTVFAEMMRSDGKPNEFTFATVLTSCIRASGLGLG 172
Query: 168 RSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQN 227
+ IH ++K ++ +FVG++L+DMYAK G + +A +F+ + RD V +++A YAQ
Sbjct: 173 KQIHGLIVKWNYDSHIFVGSSLLDMYAKAGQIKEAREIFECLPERDVVSCTAIIAGYAQL 232
Query: 228 GHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVK 287
G +E++ + + + G+ P T +++++ + +A L +G++ H R ++
Sbjct: 233 GLDEEALEMFHRLHSEGMSPNYVTYASLLTALSGLALLDHGKQAHCHVLRRELPFYAVLQ 292
Query: 288 TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDR- 346
+LIDMY+KCG++ A LF+ + E+ +SWNA++ GY+ HGL L+LF MR E R
Sbjct: 293 NSLIDMYSKCGNLSYARRLFDNMPERTAISWNAMLVGYSKHGLGREVLELFRLMRDEKRV 352
Query: 347 -PDHITFVGVLAACSRGRLLDEGRALYNLMVR-DYGITPTVQHYTCMIDLLGHCGQLDEA 404
PD +T + VL+ CS GR+ D G +++ MV +YG P +HY C++D+LG G++DEA
Sbjct: 353 KPDAVTLLAVLSGCSHGRMEDTGLNIFDGMVAGEYGTKPGTEHYGCIVDMLGRAGRIDEA 412
Query: 405 YDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQS 464
++ I+ M KP +GV G+LL +C++H +V++ E +LIE+EP+++GNYVIL+N+YA +
Sbjct: 413 FEFIKRMPSKPTAGVLGSLLGACRVHLSVDIGESVGRRLIEIEPENAGNYVILSNLYASA 472
Query: 465 GKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLM 524
G+W V +R +M+ K + K SWI+ + ++ F A D +H + + A++K + M
Sbjct: 473 GRWADVNNVRAMMMQKAVTKEPGRSWIQHEQTLHYFHANDRTHPRREEVLAKMKEISIKM 532
Query: 525 HEAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCH 584
+AGY PD V +DV+E++K M+ HSE+LA+ FGLI+T G + + KNLRIC DCH
Sbjct: 533 KQAGYVPDLSCVLYDVDEEQKEKMLLGHSEKLALTFGLIATGEGIPIRVFKNLRICVDCH 592
Query: 585 VAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
K SK+ ERE+++RD NR+H G+CSCGD+W
Sbjct: 593 NFAKIFSKVFEREVSLRDKNRFHQIVDGICSCGDYW 628
>At4g18750 putative protein
Length = 871
Score = 455 bits (1170), Expect = e-128
Identities = 225/571 (39%), Positives = 356/571 (61%), Gaps = 2/571 (0%)
Query: 52 SLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQ 111
S+ C DS+ ++ G+ +H+ + L+ +Y+ L +A+ +F ++ +
Sbjct: 301 SVFAGCADSRLISLGRAVHSIGVKACFSREDRFCNTLLDMYSKCGDLDSAKAVFREMSDR 360
Query: 112 NLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIH 171
++ + +I GYA G A+ L+ +M + G+ PD +T+ VL C+ + EG+ +H
Sbjct: 361 SVVSYTSMIAGYAREGLAGEAVKLFEEMEEEGISPDVYTVTAVLNCCARYRLLDEGKRVH 420
Query: 172 EYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPD 231
E++ ++ D+FV AL+DMYAKCG + +A VF ++ V+D + WN+++ Y++N + +
Sbjct: 421 EWIKENDLGFDIFVSNALMDMYAKCGSMQEAELVFSEMRVKDIISWNTIIGGYSKNCYAN 480
Query: 232 ESISLCREMAANG-VRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTAL 290
E++SL + P E T+ V+ + A ++ GREIHG+ R+G+ S+ V +L
Sbjct: 481 EALSLFNLLLEEKRFSPDERTVACVLPACASLSAFDKGREIHGYIMRNGYFSDRHVANSL 540
Query: 291 IDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDH 349
+DMYAKCG++ +A LF+ + K +VSW +I GY MHG A+ LF++MR+ D
Sbjct: 541 VDMYAKCGALLLAHMLFDDIASKDLVSWTVMIAGYGMHGFGKEAIALFNQMRQAGIEADE 600
Query: 350 ITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIR 409
I+FV +L ACS L+DEG +N+M + I PTV+HY C++D+L G L +AY I
Sbjct: 601 ISFVSLLYACSHSGLVDEGWRFFNIMRHECKIEPTVEHYACIVDMLARTGDLIKAYRFIE 660
Query: 410 NMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEG 469
NM + PD+ +WGALL C+IH +V+LAE EK+ ELEP+++G YV++AN+YA++ KWE
Sbjct: 661 NMPIPPDATIWGALLCGCRIHHDVKLAEKVAEKVFELEPENTGYYVLMANIYAEAEKWEQ 720
Query: 470 VEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGY 529
V++LR+ + + ++KN CSWIE+K +V F+AGD S+ ++ I A L+++ M E GY
Sbjct: 721 VKRLRKRIGQRGLRKNPGCSWIEIKGRVNIFVAGDSSNPETENIEAFLRKVRARMIEEGY 780
Query: 530 APDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKF 589
+P T D EE EK +C HSE+LA+A G+IS+ G + +TKNLR+C DCH KF
Sbjct: 781 SPLTKYALIDAEEMEKEEALCGHSEKLAMALGIISSGHGKIIRVTKNLRVCGDCHEMAKF 840
Query: 590 ISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
+SK+ REI +RD NR+H FK G CSC W
Sbjct: 841 MSKLTRREIVLRDSNRFHQFKDGHCSCRGFW 871
Score = 219 bits (559), Expect = 3e-57
Identities = 131/405 (32%), Positives = 215/405 (52%), Gaps = 10/405 (2%)
Query: 52 SLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQ 111
S+LQ C DSK+L GK++ G + +L +KL +Y L A +FD++ +
Sbjct: 99 SVLQLCADSKSLKDGKEVDNFIRGNGFVIDSNLGSKLSLMYTNCGDLKEASRVFDEVKIE 158
Query: 112 NLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIH 171
WN+L+ A +G +I L+ KM+ G+ D++T V K+ S+L ++ G +H
Sbjct: 159 KALFWNILMNELAKSGDFSGSIGLFKKMMSSGVEMDSYTFSCVSKSFSSLRSVHGGEQLH 218
Query: 172 EYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPD 231
+++KSG+ VG +L+ Y K V A +VFD++ RD + WNS++ Y NG +
Sbjct: 219 GFILKSGFGERNSVGNSLVAFYLKNQRVDSARKVFDEMTERDVISWNSIINGYVSNGLAE 278
Query: 232 ESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALI 291
+ +S+ +M +G+ AT+V+V + AD + GR +H G + F D+ L+
Sbjct: 279 KGLSVFVQMLVSGIEIDLATIVSVFAGCADSRLISLGRAVHSIGVKACFSREDRFCNTLL 338
Query: 292 DMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHI 350
DMY+KCG + A A+F + ++ VVS+ ++I GYA GLA A+ LF++M +E PD
Sbjct: 339 DMYSKCGDLDSAKAVFREMSDRSVVSYTSMIAGYAREGLAGEAVKLFEEMEEEGISPDVY 398
Query: 351 TFVGVLAACSRGRLLDEGRALYN-LMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIR 409
T VL C+R RLLDEG+ ++ + D G V + ++D+ CG + EA +
Sbjct: 399 TVTAVLNCCARYRLLDEGKRVHEWIKENDLGFDIFVSN--ALMDMYAKCGSMQEAELVFS 456
Query: 410 NMSVKPDSGVWGALL----NSCKIHGNVELAELALEKLIELEPDD 450
M VK D W ++ +C + + L L LE+ PD+
Sbjct: 457 EMRVK-DIISWNTIIGGYSKNCYANEALSLFNLLLEEK-RFSPDE 499
Score = 188 bits (477), Expect = 9e-48
Identities = 116/386 (30%), Positives = 199/386 (51%), Gaps = 4/386 (1%)
Query: 48 YGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDK 107
Y ++ + +S ++++ G+QLH G + LV Y + + +AR +FD+
Sbjct: 196 YTFSCVSKSFSSLRSVHGGEQLHGFILKSGFGERNSVGNSLVAFYLKNQRVDSARKVFDE 255
Query: 108 IPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEG 167
+ ++++ WN +I GY NG + + ++ +ML G+ D T+ V C+ I G
Sbjct: 256 MTERDVISWNSIINGYVSNGLAEKGLSVFVQMLVSGIEIDLATIVSVFAGCADSRLISLG 315
Query: 168 RSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQN 227
R++H +K+ + R+ L+DMY+KCG + A VF ++ R V + SM+A YA+
Sbjct: 316 RAVHSIGVKACFSREDRFCNTLLDMYSKCGDLDSAKAVFREMSDRSVVSYTSMIAGYARE 375
Query: 228 GHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVK 287
G E++ L EM G+ P T+ V++ A L G+ +H + + + V
Sbjct: 376 GLAGEAVKLFEEMEEEGISPDVYTVTAVLNCCARYRLLDEGKRVHEWIKENDLGFDIFVS 435
Query: 288 TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDR- 346
AL+DMYAKCGS++ A +F +R K ++SWN II GY+ + A AL LF+ + +E R
Sbjct: 436 NALMDMYAKCGSMQEAELVFSEMRVKDIISWNTIIGGYSKNCYANEALSLFNLLLEEKRF 495
Query: 347 -PDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAY 405
PD T VL AC+ D+GR ++ ++R+ G ++D+ CG L A+
Sbjct: 496 SPDERTVACVLPACASLSAFDKGREIHGYIMRN-GYFSDRHVANSLVDMYAKCGALLLAH 554
Query: 406 DLIRNMSVKPDSGVWGALLNSCKIHG 431
L +++ K D W ++ +HG
Sbjct: 555 MLFDDIASK-DLVSWTVMIAGYGMHG 579
Score = 143 bits (360), Expect = 3e-34
Identities = 89/297 (29%), Positives = 154/297 (50%), Gaps = 4/297 (1%)
Query: 117 NVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIK 176
N +R + +G +NA+ L + + P TL VL+ C+ ++ +G+ + ++
Sbjct: 65 NTQLRRFCESGNLENAVKLLCVSGKWDIDPR--TLCSVLQLCADSKSLKDGKEVDNFIRG 122
Query: 177 SGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISL 236
+G+ D +G+ L MY CG + +A RVFD++ + A+ WN ++ A++G SI L
Sbjct: 123 NGFVIDSNLGSKLSLMYTNCGDLKEASRVFDEVKIEKALFWNILMNELAKSGDFSGSIGL 182
Query: 237 CREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAK 296
++M ++GV T V S + + + G ++HGF + GF + V +L+ Y K
Sbjct: 183 FKKMMSSGVEMDSYTFSCVSKSFSSLRSVHGGEQLHGFILKSGFGERNSVGNSLVAFYLK 242
Query: 297 CGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGV 355
V A +F+ + E+ V+SWN+II GY +GLA L +F +M D T V V
Sbjct: 243 NQRVDSARKVFDEMTERDVISWNSIINGYVSNGLAEKGLSVFVQMLVSGIEIDLATIVSV 302
Query: 356 LAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMS 412
A C+ RL+ GRA++++ V+ + + ++D+ CG LD A + R MS
Sbjct: 303 FAGCADSRLISLGRAVHSIGVKAC-FSREDRFCNTLLDMYSKCGDLDSAKAVFREMS 358
>At3g12770 unknown protein
Length = 719
Score = 450 bits (1158), Expect = e-127
Identities = 230/575 (40%), Positives = 358/575 (62%), Gaps = 4/575 (0%)
Query: 48 YGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFD- 106
+ + LL++C L G+ +HAQ + LG + + L+ LYA L +AR +F+
Sbjct: 120 FTFPHLLKACSGLSHLQMGRFVHAQVFRLGFDADVFVQNGLIALYAKCRRLGSARTVFEG 179
Query: 107 -KIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIG 165
+P++ + W ++ YA NG A+ ++ +M ++PD L VL A + L +
Sbjct: 180 LPLPERTIVSWTAIVSAYAQNGEPMEALEIFSQMRKMDVKPDWVALVSVLNAFTCLQDLK 239
Query: 166 EGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYA 225
+GRSIH V+K G E + + +L MYAKCG V A +FDK+ + +LWN+M++ YA
Sbjct: 240 QGRSIHASVVKMGLEIEPDLLISLNTMYAKCGQVATAKILFDKMKSPNLILWNAMISGYA 299
Query: 226 QNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDK 285
+NG+ E+I + EM VRP ++ + IS+ A V L R ++ + R ++ +
Sbjct: 300 KNGYAREAIDMFHEMINKDVRPDTISITSAISACAQVGSLEQARSMYEYVGRSDYRDDVF 359
Query: 286 VKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED 345
+ +ALIDM+AKCGSV+ A +F+R ++ VV W+A+I GY +HG A A+ L+ M +
Sbjct: 360 ISSALIDMFAKCGSVEGARLVFDRTLDRDVVVWSAMIVGYGLHGRAREAISLYRAMERGG 419
Query: 346 -RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEA 404
P+ +TF+G+L AC+ ++ EG +N M D+ I P QHY C+IDLLG G LD+A
Sbjct: 420 VHPNDVTFLGLLMACNHSGMVREGWWFFNRMA-DHKINPQQQHYACVIDLLGRAGHLDQA 478
Query: 405 YDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQS 464
Y++I+ M V+P VWGALL++CK H +VEL E A ++L ++P ++G+YV L+N+YA +
Sbjct: 479 YEVIKCMPVQPGVTVWGALLSACKKHRHVELGEYAAQQLFSIDPSNTGHYVQLSNLYAAA 538
Query: 465 GKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLM 524
W+ V ++R M +K + K++ CSW+EV+ ++ AF GD SH + I +++ +E +
Sbjct: 539 RLWDRVAEVRVRMKEKGLNKDVGCSWVEVRGRLEAFRVGDKSHPRYEEIERQVEWIESRL 598
Query: 525 HEAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCH 584
E G+ + + HD+ ++E +CSHSER+AIA+GLIST GT L ITKNLR C +CH
Sbjct: 599 KEGGFVANKDASLHDLNDEEAEETLCSHSERIAIAYGLISTPQGTPLRITKNLRACVNCH 658
Query: 585 VAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDH 619
A K ISK+++REI VRD NR+H FK G+CSCGD+
Sbjct: 659 AATKLISKLVDREIVVRDTNRFHHFKDGVCSCGDY 693
Score = 236 bits (601), Expect = 4e-62
Identities = 139/408 (34%), Positives = 231/408 (56%), Gaps = 14/408 (3%)
Query: 50 YTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIP 109
Y SL+ S L KQ+HA+ LG+ ++ L TKL+H + + AR +FD +P
Sbjct: 24 YASLIDSATHKAQL---KQIHARLLVLGLQFSGFLITKLIHASSSFGDITFARQVFDDLP 80
Query: 110 KQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRS 169
+ +F WN +IRGY+ N +A+++Y M + PD+FT P +LKACS LS + GR
Sbjct: 81 RPQIFPWNAIIRGYSRNNHFQDALLMYSNMQLARVSPDSFTFPHLLKACSGLSHLQMGRF 140
Query: 170 IHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVV--RDAVLWNSMLAAYAQN 227
+H V + G++ D+FV LI +YAKC + A VF+ + + R V W ++++AYAQN
Sbjct: 141 VHAQVFRLGFDADVFVQNGLIALYAKCRRLGSARTVFEGLPLPERTIVSWTAIVSAYAQN 200
Query: 228 GHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVK 287
G P E++ + +M V+P LV+V+++ + L GR IH + G + +
Sbjct: 201 GEPMEALEIFSQMRKMDVKPDWVALVSVLNAFTCLQDLKQGRSIHASVVKMGLEIEPDLL 260
Query: 288 TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKM-RKEDR 346
+L MYAKCG V A LF++++ ++ WNA+I+GYA +G A A+D+F +M K+ R
Sbjct: 261 ISLNTMYAKCGQVATAKILFDKMKSPNLILWNAMISGYAKNGYAREAIDMFHEMINKDVR 320
Query: 347 PDHITFVGVLAACSRGRLLDEGRALYNLMVR-DYGITPTVQHYTCMIDLLGHCGQLDEAY 405
PD I+ ++AC++ L++ R++Y + R DY V + +ID+ CG ++ A
Sbjct: 321 PDTISITSAISACAQVGSLEQARSMYEYVGRSDY--RDDVFISSALIDMFAKCGSVEGA- 377
Query: 406 DLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIE---LEPDD 450
L+ + ++ D VW A++ +HG A ++L + +E + P+D
Sbjct: 378 RLVFDRTLDRDVVVWSAMIVGYGLHGRAREA-ISLYRAMERGGVHPND 424
>At1g18485 hypothetical protein
Length = 970
Score = 449 bits (1156), Expect = e-126
Identities = 228/566 (40%), Positives = 344/566 (60%), Gaps = 2/566 (0%)
Query: 57 CIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLW 116
C L K+LH YN+ +A V YA SL A+ +F I + + W
Sbjct: 405 CFHESFLPSLKELHCYSLKQEFVYNELVANAFVASYAKCGSLSYAQRVFHGIRSKTVNSW 464
Query: 117 NVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIK 176
N LI G+A + ++ + +M GL PD+FT+ +L ACS L ++ G+ +H ++I+
Sbjct: 465 NALIGGHAQSNDPRLSLDAHLQMKISGLLPDSFTVCSLLSACSKLKSLRLGKEVHGFIIR 524
Query: 177 SGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISL 236
+ ERDLFV +++ +Y CG + +FD + + V WN+++ Y QNG PD ++ +
Sbjct: 525 NWLERDLFVYLSVLSLYIHCGELCTVQALFDAMEDKSLVSWNTVITGYLQNGFPDRALGV 584
Query: 237 CREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAK 296
R+M G++ +++ V + + + L GRE H + +H + + + +LIDMYAK
Sbjct: 585 FRQMVLYGIQLCGISMMPVFGACSLLPSLRLGREAHAYALKHLLEDDAFIACSLIDMYAK 644
Query: 297 CGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDR-PDHITFVGV 355
GS+ + +F L+EK SWNA+I GY +HGLA A+ LF++M++ PD +TF+GV
Sbjct: 645 NGSITQSSKVFNGLKEKSTASWNAMIMGYGIHGLAKEAIKLFEEMQRTGHNPDDLTFLGV 704
Query: 356 LAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLI-RNMSVK 414
L AC+ L+ EG + M +G+ P ++HY C+ID+LG GQLD+A ++ MS +
Sbjct: 705 LTACNHSGLIHEGLRYLDQMKSSFGLKPNLKHYACVIDMLGRAGQLDKALRVVAEEMSEE 764
Query: 415 PDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLR 474
D G+W +LL+SC+IH N+E+ E KL ELEP+ NYV+L+N+YA GKWE V K+R
Sbjct: 765 ADVGIWKSLLSSCRIHQNLEMGEKVAAKLFELEPEKPENYVLLSNLYAGLGKWEDVRKVR 824
Query: 475 QVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTG 534
Q M + ++K+ CSWIE+ KV++F+ G+ + I + LE + + GY PDT
Sbjct: 825 QRMNEMSLRKDAGCSWIELNRKVFSFVVGERFLDGFEEIKSLWSILEMKISKMGYRPDTM 884
Query: 535 SVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIM 594
SV HD+ E+EK + HSE+LA+ +GLI TS GT + + KNLRIC DCH A K ISK+M
Sbjct: 885 SVQHDLSEEEKIEQLRGHSEKLALTYGLIKTSEGTTIRVYKNLRICVDCHNAAKLISKVM 944
Query: 595 EREITVRDVNRYHSFKHGMCSCGDHW 620
EREI VRD R+H FK+G+CSCGD+W
Sbjct: 945 EREIVVRDNKRFHHFKNGVCSCGDYW 970
Score = 184 bits (466), Expect = 2e-46
Identities = 128/455 (28%), Positives = 219/455 (48%), Gaps = 19/455 (4%)
Query: 53 LLQSCIDSKALNPGKQLHAQFYHLGIAYNQD-LATKLVHLYAVSNSLLNARNLFDKIPKQ 111
LLQ+ K + G+++H N D L T+++ +YA+ S ++R +FD + +
Sbjct: 90 LLQASGKRKDIEMGRKIHQLVSGSTRLRNDDVLCTRIITMYAMCGSPDDSRFVFDALRSK 149
Query: 112 NLFLWNVLIRGYAWNGPHDNAIILYHKMLDY-GLRPDNFTLPFVLKACSALSAIGEGRSI 170
NLF WN +I Y+ N +D + + +M+ L PD+FT P V+KAC+ +S +G G ++
Sbjct: 150 NLFQWNAVISSYSRNELYDEVLETFIEMISTTDLLPDHFTYPCVIKACAGMSDVGIGLAV 209
Query: 171 HEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHP 230
H V+K+G D+FVG AL+ Y G V DA ++FD + R+ V WNSM+ ++ NG
Sbjct: 210 HGLVVKTGLVEDVFVGNALVSFYGTHGFVTDALQLFDIMPERNLVSWNSMIRVFSDNGFS 269
Query: 231 DESISLCRE-MAANG---VRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKV 286
+ES L E M NG P ATLVTV+ A + G+ +HG+ + +
Sbjct: 270 EESFLLLGEMMEENGDGAFMPDVATLVTVLPVCAREREIGLGKGVHGWAVKLRLDKELVL 329
Query: 287 KTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKM---RK 343
AL+DMY+KCG + A +F+ K VVSWN ++ G++ G G D+ +M +
Sbjct: 330 NNALMDMYSKCGCITNAQMIFKMNNNKNVVSWNTMVGGFSAEGDTHGTFDVLRQMLAGGE 389
Query: 344 EDRPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDE 403
+ + D +T + + C L + L+ ++ + + + CG L
Sbjct: 390 DVKADEVTILNAVPVCFHESFLPSLKELHCYSLKQEFVYNELV-ANAFVASYAKCGSLSY 448
Query: 404 AYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQ 463
A + + K + W AL+ H L+L+ ++++ +L + +
Sbjct: 449 AQRVFHGIRSKTVNS-WNALIGG---HAQSNDPRLSLDAHLQMKISG-----LLPDSFTV 499
Query: 464 SGKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVY 498
KL+ + + K + I +W+E VY
Sbjct: 500 CSLLSACSKLKSLRLGKEVHGFIIRNWLERDLFVY 534
Score = 181 bits (460), Expect = 8e-46
Identities = 106/362 (29%), Positives = 187/362 (51%), Gaps = 8/362 (2%)
Query: 47 HYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFD 106
H+ Y ++++C + G +H G+ + + LV Y + +A LFD
Sbjct: 187 HFTYPCVIKACAGMSDVGIGLAVHGLVVKTGLVEDVFVGNALVSFYGTHGFVTDALQLFD 246
Query: 107 KIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKML----DYGLRPDNFTLPFVLKACSALS 162
+P++NL WN +IR ++ NG + + +L +M+ D PD TL VL C+
Sbjct: 247 IMPERNLVSWNSMIRVFSDNGFSEESFLLLGEMMEENGDGAFMPDVATLVTVLPVCARER 306
Query: 163 AIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLA 222
IG G+ +H + +K +++L + AL+DMY+KCGC+ +A +F ++ V WN+M+
Sbjct: 307 EIGLGKGVHGWAVKLRLDKELVLNNALMDMYSKCGCITNAQMIFKMNNNKNVVSWNTMVG 366
Query: 223 AYAQNGHPDESISLCREMAANG--VRPTEATLVTVISSSADVACLPYGREIHGFGWRHGF 280
++ G + + R+M A G V+ E T++ + + LP +E+H + + F
Sbjct: 367 GFSAEGDTHGTFDVLRQMLAGGEDVKADEVTILNAVPVCFHESFLPSLKELHCYSLKQEF 426
Query: 281 QSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDK 340
N+ V A + YAKCGS+ A +F +R K V SWNA+I G+A +LD +
Sbjct: 427 VYNELVANAFVASYAKCGSLSYAQRVFHGIRSKTVNSWNALIGGHAQSNDPRLSLDAHLQ 486
Query: 341 MRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCG 399
M+ PD T +L+ACS+ + L G+ ++ ++R++ + + Y ++ L HCG
Sbjct: 487 MKISGLLPDSFTVCSLLSACSKLKSLRLGKEVHGFIIRNW-LERDLFVYLSVLSLYIHCG 545
Query: 400 QL 401
+L
Sbjct: 546 EL 547
Score = 144 bits (363), Expect = 1e-34
Identities = 114/448 (25%), Positives = 206/448 (45%), Gaps = 23/448 (5%)
Query: 52 SLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQ 111
++L C + + GK +H L + L L+ +Y+ + NA+ +F +
Sbjct: 297 TVLPVCAREREIGLGKGVHGWAVKLRLDKELVLNNALMDMYSKCGCITNAQMIFKMNNNK 356
Query: 112 NLFLWNVLIRGYAWNGPHDNAIILYHKMLDYG--LRPDNFTLPFVLKACSALSAIGEGRS 169
N+ WN ++ G++ G + +ML G ++ D T+ + C S + +
Sbjct: 357 NVVSWNTMVGGFSAEGDTHGTFDVLRQMLAGGEDVKADEVTILNAVPVCFHESFLPSLKE 416
Query: 170 IHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGH 229
+H Y +K + + V A + YAKCG + A RVF I + WN+++ +AQ+
Sbjct: 417 LHCYSLKQEFVYNELVANAFVASYAKCGSLSYAQRVFHGIRSKTVNSWNALIGGHAQSND 476
Query: 230 PDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTA 289
P S+ +M +G+ P T+ +++S+ + + L G+E+HGF R+ + + V +
Sbjct: 477 PRLSLDAHLQMKISGLLPDSFTVCSLLSACSKLKSLRLGKEVHGFIIRNWLERDLFVYLS 536
Query: 290 LIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPD 348
++ +Y CG + ALF+ + +K +VSWN +ITGY +G AL +F +M +
Sbjct: 537 VLSLYIHCGELCTVQALFDAMEDKSLVSWNTVITGYLQNGFPDRALGVFRQMVLYGIQLC 596
Query: 349 HITFVGVLAACSRGRLLDEGR-----ALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDE 403
I+ + V ACS L GR AL +L+ D I +ID+ G + +
Sbjct: 597 GISMMPVFGACSLLPSLRLGREAHAYALKHLLEDDAFIA------CSLIDMYAKNGSITQ 650
Query: 404 AYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELE-----PDDSGNYVILA 458
+ + + K + W A++ IHG LA+ A++ E++ PDD +L
Sbjct: 651 SSKVFNGLKEK-STASWNAMIMGYGIHG---LAKEAIKLFEEMQRTGHNPDDLTFLGVLT 706
Query: 459 NMYAQSGKWEGVEKLRQVMIDKRIKKNI 486
EG+ L Q+ +K N+
Sbjct: 707 ACNHSGLIHEGLRYLDQMKSSFGLKPNL 734
>At2g01510 hypothetical protein
Length = 584
Score = 444 bits (1142), Expect = e-125
Identities = 227/557 (40%), Positives = 340/557 (60%), Gaps = 3/557 (0%)
Query: 67 KQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAWN 126
K++HA G + L T+L+ V + AR +FD++ K +FLWN L +GY N
Sbjct: 28 KKIHAIVLRTGFSEKNSLLTQLLENLVVIGDMCYARQVFDEMHKPRIFLWNTLFKGYVRN 87
Query: 127 GPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVG 186
+++LY KM D G+RPD FT PFV+KA S L G ++H +V+K G+ V
Sbjct: 88 QLPFESLLLYKKMRDLGVRPDEFTYPFVVKAISQLGDFSCGFALHAHVVKYGFGCLGIVA 147
Query: 187 AALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVR 246
L+ MY K G + A +F+ + V+D V WN+ LA Q G+ ++ +M A+ V+
Sbjct: 148 TELVMMYMKFGELSSAEFLFESMQVKDLVAWNAFLAVCVQTGNSAIALEYFNKMCADAVQ 207
Query: 247 PTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALAL 306
T+V+++S+ + L G EI+ + N V+ A +DM+ KCG+ + A L
Sbjct: 208 FDSFTVVSMLSACGQLGSLEIGEEIYDRARKEEIDCNIIVENARLDMHLKCGNTEAARVL 267
Query: 307 FERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLL 365
FE ++++ VVSW+ +I GYAM+G + AL LF M+ E RP+++TF+GVL+ACS L+
Sbjct: 268 FEEMKQRNVVSWSTMIVGYAMNGDSREALTLFTTMQNEGLRPNYVTFLGVLSACSHAGLV 327
Query: 366 DEGRALYNLMVR--DYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGAL 423
+EG+ ++LMV+ D + P +HY CM+DLLG G L+EAY+ I+ M V+PD+G+WGAL
Sbjct: 328 NEGKRYFSLMVQSNDKNLEPRKEHYACMVDLLGRSGLLEEAYEFIKKMPVEPDTGIWGAL 387
Query: 424 LNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIK 483
L +C +H ++ L + + L+E PD +V+L+N+YA +GKW+ V+K+R M K
Sbjct: 388 LGACAVHRDMILGQKVADVLVETAPDIGSYHVLLSNIYAAAGKWDCVDKVRSKMRKLGTK 447
Query: 484 KNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTGSVFHDVEED 543
K A S +E + K++ F GD SH S AIY +L + + + GY PDT SVFHDVE +
Sbjct: 448 KVAAYSSVEFEGKIHFFNRGDKSHPQSKAIYEKLDEILKKIRKMGYVPDTCSVFHDVEME 507
Query: 544 EKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIMEREITVRDV 603
EK + HSE+LAIAFGLI PG + + KNLR C+DCH KF+S + EI +RD
Sbjct: 508 EKECSLSHHSEKLAIAFGLIKGRPGHPIRVMKNLRTCDDCHAFSKFVSSLTSTEIIMRDK 567
Query: 604 NRYHSFKHGMCSCGDHW 620
NR+H F++G+CSC + W
Sbjct: 568 NRFHHFRNGVCSCKEFW 584
Score = 105 bits (263), Expect = 6e-23
Identities = 80/317 (25%), Positives = 145/317 (45%), Gaps = 17/317 (5%)
Query: 66 GKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAW 125
G LHA G +AT+LV +Y L +A LF+ + ++L WN +
Sbjct: 128 GFALHAHVVKYGFGCLGIVATELVMMYMKFGELSSAEFLFESMQVKDLVAWNAFLAVCVQ 187
Query: 126 NGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFV 185
G A+ ++KM ++ D+FT+ +L AC L ++ G I++ K + ++ V
Sbjct: 188 TGNSAIALEYFNKMCADAVQFDSFTVVSMLSACGQLGSLEIGEEIYDRARKEEIDCNIIV 247
Query: 186 GAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGV 245
A +DM+ KCG A +F+++ R+ V W++M+ YA NG E+++L M G+
Sbjct: 248 ENARLDMHLKCGNTEAARVLFEEMKQRNVVSWSTMIVGYAMNGDSREALTLFTTMQNEGL 307
Query: 246 RPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDK-------VKTALIDMYAKCG 298
RP T + V+S+ + + G+ QSNDK ++D+ + G
Sbjct: 308 RPNYVTFLGVLSACSHAGLVNEGKRYFSL----MVQSNDKNLEPRKEHYACMVDLLGRSG 363
Query: 299 SVKVALALFERLR-EKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDRPD----HITFV 353
++ A +++ E W A++ A+H + + D + E PD H+
Sbjct: 364 LLEEAYEFIKKMPVEPDTGIWGALLGACAVHRDMILGQKVADVL-VETAPDIGSYHVLLS 422
Query: 354 GVLAACSRGRLLDEGRA 370
+ AA + +D+ R+
Sbjct: 423 NIYAAAGKWDCVDKVRS 439
>At4g33170 putative protein
Length = 990
Score = 437 bits (1125), Expect = e-123
Identities = 221/575 (38%), Positives = 344/575 (59%), Gaps = 3/575 (0%)
Query: 48 YGYTSLLQSCID-SKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFD 106
Y TS+L++ + L+ KQ+H + + ++T L+ Y+ + + A LF+
Sbjct: 417 YTMTSVLKAASSLPEGLSLSKQVHVHAIKINNVSDSFVSTALIDAYSRNRCMKEAEILFE 476
Query: 107 KIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGE 166
+ +L WN ++ GY + + L+ M G R D+FTL V K C L AI +
Sbjct: 477 R-HNFDLVAWNAMMAGYTQSHDGHKTLKLFALMHKQGERSDDFTLATVFKTCGFLFAINQ 535
Query: 167 GRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQ 226
G+ +H Y IKSG++ DL+V + ++DMY KCG + A FD I V D V W +M++ +
Sbjct: 536 GKQVHAYAIKSGYDLDLWVSSGILDMYVKCGDMSAAQFAFDSIPVPDDVAWTTMISGCIE 595
Query: 227 NGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKV 286
NG + + + +M GV P E T+ T+ +S+ + L GR+IH + ++ V
Sbjct: 596 NGEEERAFHVFSQMRLMGVLPDEFTIATLAKASSCLTALEQGRQIHANALKLNCTNDPFV 655
Query: 287 KTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED- 345
T+L+DMYAKCGS+ A LF+R+ + +WNA++ G A HG L LF +M+
Sbjct: 656 GTSLVDMYAKCGSIDDAYCLFKRIEMMNITAWNAMLVGLAQHGEGKETLQLFKQMKSLGI 715
Query: 346 RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAY 405
+PD +TF+GVL+ACS L+ E M DYGI P ++HY+C+ D LG G + +A
Sbjct: 716 KPDKVTFIGVLSACSHSGLVSEAYKHMRSMHGDYGIKPEIEHYSCLADALGRAGLVKQAE 775
Query: 406 DLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSG 465
+LI +MS++ + ++ LL +C++ G+ E + KL+ELEP DS YV+L+NMYA +
Sbjct: 776 NLIESMSMEASASMYRTLLAACRVQGDTETGKRVATKLLELEPLDSSAYVLLSNMYAAAS 835
Query: 466 KWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMH 525
KW+ ++ R +M ++KK+ SWIEVKNK++ F+ D S+ ++ IY ++K + +
Sbjct: 836 KWDEMKLARTMMKGHKVKKDPGFSWIEVKNKIHIFVVDDRSNRQTELIYRKVKDMIRDIK 895
Query: 526 EAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHV 585
+ GY P+T DVEE+EK + HSE+LA+AFGL+ST P T + + KNLR+C DCH
Sbjct: 896 QEGYVPETDFTLVDVEEEEKERALYYHSEKLAVAFGLLSTPPSTPIRVIKNLRVCGDCHN 955
Query: 586 AIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
A+K+I+K+ REI +RD NR+H FK G+CSCGD+W
Sbjct: 956 AMKYIAKVYNREIVLRDANRFHRFKDGICSCGDYW 990
Score = 176 bits (445), Expect = 5e-44
Identities = 112/377 (29%), Positives = 192/377 (50%), Gaps = 9/377 (2%)
Query: 58 IDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWN 117
+DS AL G+Q+H LG+ ++ L+++Y AR +FD + +++L WN
Sbjct: 328 VDSLAL--GQQVHCMALKLGLDLMLTVSNSLINMYCKLRKFGFARTVFDNMSERDLISWN 385
Query: 118 VLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSAL-SAIGEGRSIHEYVIK 176
+I G A NG A+ L+ ++L GL+PD +T+ VLKA S+L + + +H + IK
Sbjct: 386 SVIAGIAQNGLEVEAVCLFMQLLRCGLKPDQYTMTSVLKAASSLPEGLSLSKQVHVHAIK 445
Query: 177 SGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISL 236
D FV ALID Y++ C+ +A +F++ D V WN+M+A Y Q+ +++ L
Sbjct: 446 INNVSDSFVSTALIDAYSRNRCMKEAEILFERHNF-DLVAWNAMMAGYTQSHDGHKTLKL 504
Query: 237 CREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAK 296
M G R + TL TV + + + G+++H + + G+ + V + ++DMY K
Sbjct: 505 FALMHKQGERSDDFTLATVFKTCGFLFAINQGKQVHAYAIKSGYDLDLWVSSGILDMYVK 564
Query: 297 CGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGV 355
CG + A F+ + V+W +I+G +G A +F +MR PD T +
Sbjct: 565 CGDMSAAQFAFDSIPVPDDVAWTTMISGCIENGEEERAFHVFSQMRLMGVLPDEFTIATL 624
Query: 356 LAACSRGRLLDEGRALY-NLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVK 414
A S L++GR ++ N + + P V T ++D+ CG +D+AY L + + +
Sbjct: 625 AKASSCLTALEQGRQIHANALKLNCTNDPFVG--TSLVDMYAKCGSIDDAYCLFKRIEMM 682
Query: 415 PDSGVWGALLNSCKIHG 431
+ W A+L HG
Sbjct: 683 -NITAWNAMLVGLAQHG 698
Score = 117 bits (292), Expect = 2e-26
Identities = 91/358 (25%), Positives = 170/358 (47%), Gaps = 11/358 (3%)
Query: 123 YAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERD 182
Y +G + + + M++ + D T +L + ++ G+ +H +K G +
Sbjct: 290 YLHSGQYSALLKCFADMVESDVECDQVTFILMLATAVKVDSLALGQQVHCMALKLGLDLM 349
Query: 183 LFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAA 242
L V +LI+MY K A VFD + RD + WNS++A AQNG E++ L ++
Sbjct: 350 LTVSNSLINMYCKLRKFGFARTVFDNMSERDLISWNSVIAGIAQNGLEVEAVCLFMQLLR 409
Query: 243 NGVRPTEATLVTVISSSADV-ACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVK 301
G++P + T+ +V+ +++ + L +++H + S+ V TALID Y++ +K
Sbjct: 410 CGLKPDQYTMTSVLKAASSLPEGLSLSKQVHVHAIKINNVSDSFVSTALIDAYSRNRCMK 469
Query: 302 VALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKE-DRPDHITFVGVLAACS 360
A LFER +V+WNA++ GY L LF M K+ +R D T V C
Sbjct: 470 EAEILFER-HNFDLVAWNAMMAGYTQSHDGHKTLKLFALMHKQGERSDDFTLATVFKTCG 528
Query: 361 RGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVW 420
+++G+ ++ ++ G + + ++D+ CG + A ++ V PD W
Sbjct: 529 FLFAINQGKQVHAYAIKS-GYDLDLWVSSGILDMYVKCGDMSAAQFAFDSIPV-PDDVAW 586
Query: 421 GALLNSCKIHGNVELA--ELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQV 476
+++ C +G E A + +L+ + PD+ +A + S +E+ RQ+
Sbjct: 587 TTMISGCIENGEEERAFHVFSQMRLMGVLPDE----FTIATLAKASSCLTALEQGRQI 640
Score = 107 bits (268), Expect = 2e-23
Identities = 114/465 (24%), Positives = 192/465 (40%), Gaps = 48/465 (10%)
Query: 35 ASVDSFPPQPTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAV 94
AS S + +G+ L++ I S L GK HA+ + L L+ +Y+
Sbjct: 30 ASPSSSSSSSSQWFGF---LRNAITSSDLMLGKCTHARILTFEENPERFLINNLISMYSK 86
Query: 95 SNSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPH-----DNAIILYHKMLDYGLRPDNF 149
SL AR +FDK+P ++L WN ++ YA + A +L+ + +
Sbjct: 87 CGSLTYARRVFDKMPDRDLVSWNSILAAYAQSSECVVENIQQAFLLFRILRQDVVYTSRM 146
Query: 150 TLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKI 209
TL +LK C + S H Y K G + D FV AL+++Y K G V + +F+++
Sbjct: 147 TLSPMLKLCLHSGYVWASESFHGYACKIGLDGDEFVAGALVNIYLKFGKVKEGKVLFEEM 206
Query: 210 VVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGR 269
RD VLWN ML AY + G +E+I L ++G+ P E TL + S D
Sbjct: 207 PYRDVVLWNLMLKAYLEMGFKEEAIDLSSAFHSSGLNPNEITLRLLARISGD-------- 258
Query: 270 EIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHG 329
+ G VK + ++ N ++ Y G
Sbjct: 259 ------------------------DSDAGQVKSFANGNDASSVSEIIFRNKGLSEYLHSG 294
Query: 330 LAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHY 388
L F M + D D +TF+ +LA + L G+ ++ M G+ +
Sbjct: 295 QYSALLKCFADMVESDVECDQVTFILMLATAVKVDSLALGQQVH-CMALKLGLDLMLTVS 353
Query: 389 TCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHG-NVELAELALEKL-IEL 446
+I++ + A + NMS + D W +++ +G VE L ++ L L
Sbjct: 354 NSLINMYCKLRKFGFARTVFDNMSER-DLISWNSVIAGIAQNGLEVEAVCLFMQLLRCGL 412
Query: 447 EPDDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACSWI 491
+PD Y + + + A S EG+ +QV + N++ S++
Sbjct: 413 KPD---QYTMTSVLKAASSLPEGLSLSKQVHVHAIKINNVSDSFV 454
>At3g24000 hypothetical protein
Length = 633
Score = 437 bits (1124), Expect = e-123
Identities = 217/578 (37%), Positives = 346/578 (59%), Gaps = 2/578 (0%)
Query: 44 PTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARN 103
P Y +LL+ C K L G+ +HA ++ + L+++YA SL AR
Sbjct: 57 PADRRFYNTLLKKCTVFKLLIQGRIVHAHILQSIFRHDIVMGNTLLNMYAKCGSLEEARK 116
Query: 104 LFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSA 163
+F+K+P+++ W LI GY+ + +A++ +++ML +G P+ FTL V+KA +A
Sbjct: 117 VFEKMPQRDFVTWTTLISGYSQHDRPCDALLFFNQMLRFGYSPNEFTLSSVIKAAAAERR 176
Query: 164 IGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAA 223
G +H + +K G++ ++ VG+AL+D+Y + G + DA VFD + R+ V WN+++A
Sbjct: 177 GCCGHQLHGFCVKCGFDSNVHVGSALLDLYTRYGLMDDAQLVFDALESRNDVSWNALIAG 236
Query: 224 YAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSN 283
+A+ ++++ L + M +G RP+ + ++ + + L G+ +H + + G +
Sbjct: 237 HARRSGTEKALELFQGMLRDGFRPSHFSYASLFGACSSTGFLEQGKWVHAYMIKSGEKLV 296
Query: 284 DKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRK 343
L+DMYAK GS+ A +F+RL ++ VVSWN+++T YA HG A+ F++MR+
Sbjct: 297 AFAGNTLLDMYAKSGSIHDARKIFDRLAKRDVVSWNSLLTAYAQHGFGKEAVWWFEEMRR 356
Query: 344 ED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLD 402
RP+ I+F+ VL ACS LLDEG Y LM +D GI P HY ++DLLG G L+
Sbjct: 357 VGIRPNEISFLSVLTACSHSGLLDEGWHYYELMKKD-GIVPEAWHYVTVVDLLGRAGDLN 415
Query: 403 EAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYA 462
A I M ++P + +W ALLN+C++H N EL A E + EL+PDD G +VIL N+YA
Sbjct: 416 RALRFIEEMPIEPTAAIWKALLNACRMHKNTELGAYAAEHVFELDPDDPGPHVILYNIYA 475
Query: 463 QSGKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEG 522
G+W ++R+ M + +KK ACSW+E++N ++ F+A D H + I + + +
Sbjct: 476 SGGRWNDAARVRKKMKESGVKKEPACSWVEIENAIHMFVANDERHPQREEIARKWEEVLA 535
Query: 523 LMHEAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICED 582
+ E GY PDT V V++ E+ + HSE++A+AF L++T PG+ + I KN+R+C D
Sbjct: 536 KIKELGYVPDTSHVIVHVDQQEREVNLQYHSEKIALAFALLNTPPGSTIHIKKNIRVCGD 595
Query: 583 CHVAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
CH AIK SK++ REI VRD NR+H FK G CSC D+W
Sbjct: 596 CHTAIKLASKVVGREIIVRDTNRFHHFKDGNCSCKDYW 633
>At4g02750 hypothetical protein
Length = 781
Score = 437 bits (1123), Expect = e-122
Identities = 224/530 (42%), Positives = 328/530 (61%), Gaps = 9/530 (1%)
Query: 92 YAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTL 151
YA S + AR LFD+ P Q++F W ++ GY N + A L+ KM P+ +
Sbjct: 260 YAQSGKIDEARQLFDESPVQDVFTWTAMVSGYIQNRMVEEARELFDKM------PERNEV 313
Query: 152 PFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVV 211
+ A A GE + + + R++ +I YA+CG + +A +FDK+
Sbjct: 314 SW--NAMLAGYVQGERMEMAKELFDVMPCRNVSTWNTMITGYAQCGKISEAKNLFDKMPK 371
Query: 212 RDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREI 271
RD V W +M+A Y+Q+GH E++ L +M G R ++ + +S+ ADV L G+++
Sbjct: 372 RDPVSWAAMIAGYSQSGHSFEALRLFVQMEREGGRLNRSSFSSALSTCADVVALELGKQL 431
Query: 272 HGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLA 331
HG + G+++ V AL+ MY KCGS++ A LF+ + K +VSWN +I GY+ HG
Sbjct: 432 HGRLVKGGYETGCFVGNALLLMYCKCGSIEEANDLFKEMAGKDIVSWNTMIAGYSRHGFG 491
Query: 332 VGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTC 390
AL F+ M++E +PD T V VL+ACS L+D+GR + M +DYG+ P QHY C
Sbjct: 492 EVALRFFESMKREGLKPDDATMVAVLSACSHTGLVDKGRQYFYTMTQDYGVMPNSQHYAC 551
Query: 391 MIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDD 450
M+DLLG G L++A++L++NM +PD+ +WG LL + ++HGN ELAE A +K+ +EP++
Sbjct: 552 MVDLLGRAGLLEDAHNLMKNMPFEPDAAIWGTLLGASRVHGNTELAETAADKIFAMEPEN 611
Query: 451 SGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNS 510
SG YV+L+N+YA SG+W V KLR M DK +KK SWIE++NK + F GD H
Sbjct: 612 SGMYVLLSNLYASSGRWGDVGKLRVRMRDKGVKKVPGYSWIEIQNKTHTFSVGDEFHPEK 671
Query: 511 DAIYAELKRLEGLMHEAGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTR 570
D I+A L+ L+ M +AGY T V HDVEE+EK MV HSERLA+A+G++ S G
Sbjct: 672 DEIFAFLEELDLRMKKAGYVSKTSVVLHDVEEEEKERMVRYHSERLAVAYGIMRVSSGRP 731
Query: 571 LLITKNLRICEDCHVAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
+ + KNLR+CEDCH AIK++++I R I +RD NR+H FK G CSCGD+W
Sbjct: 732 IRVIKNLRVCEDCHNAIKYMARITGRLIILRDNNRFHHFKDGSCSCGDYW 781
Score = 115 bits (288), Expect = 7e-26
Identities = 102/395 (25%), Positives = 181/395 (45%), Gaps = 40/395 (10%)
Query: 92 YAVSNSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTL 151
Y + +L AR LF+ +P++++ WN ++ GYA NG D+A ++ +M P+ +
Sbjct: 136 YVRNRNLGKARELFEIMPERDVCSWNTMLSGYAQNGCVDDARSVFDRM------PEKNDV 189
Query: 152 PFVLKACSALSAIGEGRSIHE--YVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKI 209
+ + LSA + + E + KS L L+ + K +++A + FD +
Sbjct: 190 SWN----ALLSAYVQNSKMEEACMLFKSRENWALVSWNCLLGGFVKKKKIVEARQFFDSM 245
Query: 210 VVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVAC----- 264
VRD V WN+++ YAQ+G DE+ L E V A + I +
Sbjct: 246 NVRDVVSWNTIITGYAQSGKIDEARQLFDESPVQDVFTWTAMVSGYIQNRMVEEARELFD 305
Query: 265 -LPYGREIHGFGWRHGFQSNDKVKTA-----------------LIDMYAKCGSVKVALAL 306
+P E+ G+ ++++ A +I YA+CG + A L
Sbjct: 306 KMPERNEVSWNAMLAGYVQGERMEMAKELFDVMPCRNVSTWNTMITGYAQCGKISEAKNL 365
Query: 307 FERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKE-DRPDHITFVGVLAACSRGRLL 365
F+++ ++ VSW A+I GY+ G + AL LF +M +E R + +F L+ C+ L
Sbjct: 366 FDKMPKRDPVSWAAMIAGYSQSGHSFEALRLFVQMEREGGRLNRSSFSSALSTCADVVAL 425
Query: 366 DEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLN 425
+ G+ L+ +V+ G ++ + CG ++EA DL + M+ K D W ++
Sbjct: 426 ELGKQLHGRLVKG-GYETGCFVGNALLLMYCKCGSIEEANDLFKEMAGK-DIVSWNTMIA 483
Query: 426 SCKIHGNVELAELALEKLIE--LEPDDSGNYVILA 458
HG E+A E + L+PDD+ +L+
Sbjct: 484 GYSRHGFGEVALRFFESMKREGLKPDDATMVAVLS 518
Score = 95.9 bits (237), Expect = 6e-20
Identities = 80/325 (24%), Positives = 141/325 (42%), Gaps = 58/325 (17%)
Query: 101 ARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSA 160
AR LFD++P+++L WNV+I+GY N
Sbjct: 114 ARKLFDEMPERDLVSWNVMIKGYVRN---------------------------------- 139
Query: 161 LSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSM 220
+G+ R + E + ERD+ ++ YA+ GCV DA VFD++ ++ V WN++
Sbjct: 140 -RNLGKARELFEIMP----ERDVCSWNTMLSGYAQNGCVDDARSVFDRMPEKNDVSWNAL 194
Query: 221 LAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGF 280
L+AY QN +E+ L + + L + V + ++
Sbjct: 195 LSAYVQNSKMEEACMLFKSRENWALVSWNCLLGGFVKKKKIVEARQFFDSMN-------- 246
Query: 281 QSNDKVK-TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFD 339
D V +I YA+ G + A LF+ + V +W A+++GY + + A +LFD
Sbjct: 247 -VRDVVSWNTIITGYAQSGKIDEARQLFDESPVQDVFTWTAMVSGYIQNRMVEEARELFD 305
Query: 340 KMRKEDRPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCG 399
KM + + +++ +LA +G ++ + L+++M V + MI CG
Sbjct: 306 KMPER---NEVSWNAMLAGYVQGERMEMAKELFDVMP-----CRNVSTWNTMITGYAQCG 357
Query: 400 QLDEAYDLIRNMSVKPDSGVWGALL 424
++ EA +L M K D W A++
Sbjct: 358 KISEAKNLFDKMP-KRDPVSWAAMI 381
Score = 80.5 bits (197), Expect = 3e-15
Identities = 50/184 (27%), Positives = 91/184 (49%), Gaps = 2/184 (1%)
Query: 50 YTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIP 109
++S L +C D AL GKQLH + G + L+ +Y S+ A +LF ++
Sbjct: 412 FSSALSTCADVVALELGKQLHGRLVKGGYETGCFVGNALLLMYCKCGSIEEANDLFKEMA 471
Query: 110 KQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRS 169
+++ WN +I GY+ +G + A+ + M GL+PD+ T+ VL ACS + +GR
Sbjct: 472 GKDIVSWNTMIAGYSRHGFGEVALRFFESMKREGLKPDDATMVAVLSACSHTGLVDKGRQ 531
Query: 170 IHEYVIKS-GWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVR-DAVLWNSMLAAYAQN 227
+ + G + A ++D+ + G + DA + + DA +W ++L A +
Sbjct: 532 YFYTMTQDYGVMPNSQHYACMVDLLGRAGLLEDAHNLMKNMPFEPDAAIWGTLLGASRVH 591
Query: 228 GHPD 231
G+ +
Sbjct: 592 GNTE 595
Score = 47.8 bits (112), Expect = 2e-05
Identities = 64/301 (21%), Positives = 125/301 (41%), Gaps = 56/301 (18%)
Query: 190 IDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTE 249
I Y + G +A RVF ++ +V +N M++ Y +NG + + L EM E
Sbjct: 71 ISSYMRTGRCNEALRVFKRMPRWSSVSYNGMISGYLRNGEFELARKLFDEMP-------E 123
Query: 250 ATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFER 309
LV+ W +I Y + ++ A LFE
Sbjct: 124 RDLVS---------------------WN-----------VMIKGYVRNRNLGKARELFEI 151
Query: 310 LREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDRPDHITFVGVLAACSRGRLLDEGR 369
+ E+ V SWN +++GYA +G A +FD+M ++ + +++ +L+A + ++E
Sbjct: 152 MPERDVCSWNTMLSGYAQNGCVDDARSVFDRMPEK---NDVSWNALLSAYVQNSKMEEAC 208
Query: 370 ALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKI 429
L+ ++ + + C++ ++ EA +M+V+ D W ++
Sbjct: 209 MLFKSR-ENWALV----SWNCLLGGFVKKKKIVEARQFFDSMNVR-DVVSWNTIITGYAQ 262
Query: 430 HGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACS 489
G ++ A + E D + + + Y Q+ VE+ R+ + DK ++N S
Sbjct: 263 SGKIDEARQLFD---ESPVQDVFTWTAMVSGYIQN---RMVEEARE-LFDKMPERN-EVS 314
Query: 490 W 490
W
Sbjct: 315 W 315
Score = 47.0 bits (110), Expect = 3e-05
Identities = 44/156 (28%), Positives = 72/156 (45%), Gaps = 11/156 (7%)
Query: 282 SNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKM 341
S+ K I Y + G AL +F+R+ VS+N +I+GY +G A LFD+M
Sbjct: 62 SDIKEWNVAISSYMRTGRCNEALRVFKRMPRWSSVSYNGMISGYLRNGEFELARKLFDEM 121
Query: 342 RKEDRPDHITFVGVLAACSRGRLLDEGRALYNLM-VRDYGITPTVQHYTCMIDLLGHCGQ 400
+ D +++ ++ R R L + R L+ +M RD V + M+ G
Sbjct: 122 PERDL---VSWNVMIKGYVRNRNLGKARELFEIMPERD------VCSWNTMLSGYAQNGC 172
Query: 401 LDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELA 436
+D+A + M K D W ALL++ + +E A
Sbjct: 173 VDDARSVFDRMPEKNDVS-WNALLSAYVQNSKMEEA 207
Score = 45.1 bits (105), Expect = 1e-04
Identities = 41/196 (20%), Positives = 92/196 (46%), Gaps = 19/196 (9%)
Query: 290 LIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDRPDH 349
+I Y + G ++A LF+ + E+ +VSWN +I GY + A +LF+ M + D
Sbjct: 101 MISGYLRNGEFELARKLFDEMPERDLVSWNVMIKGYVRNRNLGKARELFEIMPERDVCSW 160
Query: 350 ITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIR 409
T +L+ ++ +D+ R++++ M ++ + ++ +++EA L +
Sbjct: 161 NT---MLSGYAQNGCVDDARSVFDRMPEKNDVS-----WNALLSAYVQNSKMEEACMLFK 212
Query: 410 NMSVKPDSGVWGALLNSCKIHGNVELAEL--ALEKLIELEPDDSGNYVILANMYAQSGKW 467
+ W + +C + G V+ ++ A + + D ++ + YAQSGK
Sbjct: 213 SRE------NWALVSWNCLLGGFVKKKKIVEARQFFDSMNVRDVVSWNTIITGYAQSGK- 265
Query: 468 EGVEKLRQVMIDKRIK 483
+++ RQ+ + ++
Sbjct: 266 --IDEARQLFDESPVQ 279
>At3g08820 unknown protein
Length = 685
Score = 435 bits (1118), Expect = e-122
Identities = 240/623 (38%), Positives = 361/623 (57%), Gaps = 6/623 (0%)
Query: 4 YSIKKTQHTSF--IFNLFPFSQSFYHSLATHQTASVD-SFPPQPTTHYGYTS--LLQSCI 58
YS HT F IF F ++ H+T + S +G+T +L++C
Sbjct: 63 YSYLLFSHTQFPNIFLYNSLINGFVNNHLFHETLDLFLSIRKHGLYLHGFTFPLVLKACT 122
Query: 59 DSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNV 118
+ + G LH+ G ++ T L+ +Y+ S L +A LFD+IP +++ W
Sbjct: 123 RASSRKLGIDLHSLVVKCGFNHDVAAMTSLLSIYSGSGRLNDAHKLFDEIPDRSVVTWTA 182
Query: 119 LIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSG 178
L GY +G H AI L+ KM++ G++PD++ + VL AC + + G I +Y+ +
Sbjct: 183 LFSGYTTSGRHREAIDLFKKMVEMGVKPDSYFIVQVLSACVHVGDLDSGEWIVKYMEEME 242
Query: 179 WERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCR 238
+++ FV L+++YAKCG + A VFD +V +D V W++M+ YA N P E I L
Sbjct: 243 MQKNSFVRTTLVNLYAKCGKMEKARSVFDSMVEKDIVTWSTMIQGYASNSFPKEGIELFL 302
Query: 239 EMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCG 298
+M ++P + ++V +SS A + L G RH F +N + ALIDMYAKCG
Sbjct: 303 QMLQENLKPDQFSIVGFLSSCASLGALDLGEWGISLIDRHEFLTNLFMANALIDMYAKCG 362
Query: 299 SVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLA 357
++ +F+ ++EK +V NA I+G A +G + +F + K PD TF+G+L
Sbjct: 363 AMARGFEVFKEMKEKDIVIMNAAISGLAKNGHVKLSFAVFGQTEKLGISPDGSTFLGLLC 422
Query: 358 ACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDS 417
C L+ +G +N + Y + TV+HY CM+DL G G LD+AY LI +M ++P++
Sbjct: 423 GCVHAGLIQDGLRFFNAISCVYALKRTVEHYGCMVDLWGRAGMLDDAYRLICDMPMRPNA 482
Query: 418 GVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVM 477
VWGALL+ C++ + +LAE L++LI LEP ++GNYV L+N+Y+ G+W+ ++R +M
Sbjct: 483 IVWGALLSGCRLVKDTQLAETVLKELIALEPWNAGNYVQLSNIYSVGGRWDEAAEVRDMM 542
Query: 478 IDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTGSVF 537
K +KK SWIE++ KV+ FLA D SH SD IYA+L+ L M G+ P T VF
Sbjct: 543 NKKGMKKIPGYSWIELEGKVHEFLADDKSHPLSDKIYAKLEDLGNEMRLMGFVPTTEFVF 602
Query: 538 HDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIMERE 597
DVEE+EK ++ HSE+LA+A GLIST G + + KNLR+C DCH +K ISKI RE
Sbjct: 603 FDVEEEEKERVLGYHSEKLAVALGLISTDHGQVIRVVKNLRVCGDCHEVMKLISKITRRE 662
Query: 598 ITVRDVNRYHSFKHGMCSCGDHW 620
I VRD NR+H F +G CSC D+W
Sbjct: 663 IVVRDNNRFHCFTNGSCSCNDYW 685
Score = 58.9 bits (141), Expect = 8e-09
Identities = 65/273 (23%), Positives = 113/273 (40%), Gaps = 38/273 (13%)
Query: 247 PTEATLVTVISSSADVAC-LPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALA 305
P+ + V I + VAC + + ++IH H + + L+ K +
Sbjct: 7 PSATSKVQQIKTLISVACTVNHLKQIHVSLINHHLHHDTFLVNLLLKRTLFFRQTKYSYL 66
Query: 306 LFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDRPDH-ITFVGVLAACSRGRL 364
LF + + +N++I G+ + L LDLF +RK H TF VL AC+R
Sbjct: 67 LFSHTQFPNIFLYNSLINGFVNNHLFHETLDLFLSIRKHGLYLHGFTFPLVLKACTRASS 126
Query: 365 LDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDL----------------- 407
G L++L+V+ G V T ++ + G+L++A+ L
Sbjct: 127 RKLGIDLHSLVVK-CGFNHDVAAMTSLLSIYSGSGRLNDAHKLFDEIPDRSVVTWTALFS 185
Query: 408 -----------------IRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELE-PD 449
+ M VKPDS +L++C G+++ E ++ + E+E
Sbjct: 186 GYTTSGRHREAIDLFKKMVEMGVKPDSYFIVQVLSACVHVGDLDSGEWIVKYMEEMEMQK 245
Query: 450 DSGNYVILANMYAQSGKWEGVEKLRQVMIDKRI 482
+S L N+YA+ GK E + M++K I
Sbjct: 246 NSFVRTTLVNLYAKCGKMEKARSVFDSMVEKDI 278
>At4g37170 putative protein
Length = 691
Score = 434 bits (1116), Expect = e-122
Identities = 223/613 (36%), Positives = 352/613 (57%), Gaps = 37/613 (6%)
Query: 41 PPQPTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLN 100
PP T Y +L+Q C ++AL GK++H G + +L+ +YA SL++
Sbjct: 83 PPAST----YCNLIQVCSQTRALEEGKKVHEHIRTSGFVPGIVIWNRLLRMYAKCGSLVD 138
Query: 101 ARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLD------------------- 141
AR +FD++P ++L WNV++ GYA G + A L+ +M +
Sbjct: 139 ARKVFDEMPNRDLCSWNVMVNGYAEVGLLEEARKLFDEMTEKDSYSWTAMVTGYVKKDQP 198
Query: 142 ------YGL-------RPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAA 188
Y L RP+ FT+ + A +A+ I G+ IH +++++G + D + ++
Sbjct: 199 EEALVLYSLMQRVPNSRPNIFTVSIAVAAAAAVKCIRRGKEIHGHIVRAGLDSDEVLWSS 258
Query: 189 LIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPT 248
L+DMY KCGC+ +A +FDKIV +D V W SM+ Y ++ E SL E+ + RP
Sbjct: 259 LMDMYGKCGCIDEARNIFDKIVEKDVVSWTSMIDRYFKSSRWREGFSLFSELVGSCERPN 318
Query: 249 EATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFE 308
E T V+++ AD+ G+++HG+ R GF ++L+DMY KCG+++ A + +
Sbjct: 319 EYTFAGVLNACADLTTEELGKQVHGYMTRVGFDPYSFASSSLVDMYTKCGNIESAKHVVD 378
Query: 309 RLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDE 367
+ +VSW ++I G A +G AL FD + K +PDH+TFV VL+AC+ L+++
Sbjct: 379 GCPKPDLVSWTSLIGGCAQNGQPDEALKYFDLLLKSGTKPDHVTFVNVLSACTHAGLVEK 438
Query: 368 GRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSC 427
G + + + ++ T HYTC++DLL G+ ++ +I M +KP +W ++L C
Sbjct: 439 GLEFFYSITEKHRLSHTSDHYTCLVDLLARSGRFEQLKSVISEMPMKPSKFLWASVLGGC 498
Query: 428 KIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVMIDKRIKKNIA 487
+GN++LAE A ++L ++EP++ YV +AN+YA +GKWE K+R+ M + + K
Sbjct: 499 STYGNIDLAEEAAQELFKIEPENPVTYVTMANIYAAAGKWEEEGKMRKRMQEIGVTKRPG 558
Query: 488 CSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTGSVFHDVEEDEKTS 547
SW E+K K + F+A D SH + I L+ L M E GY P T V HDVE+++K
Sbjct: 559 SSWTEIKRKRHVFIAADTSHPMYNQIVEFLRELRKKMKEEGYVPATSLVLHDVEDEQKEE 618
Query: 548 MVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIMEREITVRDVNRYH 607
+ HSE+LA+AF ++ST GT + + KNLR C DCH AIKFIS I +R+ITVRD R+H
Sbjct: 619 NLVYHSEKLAVAFAILSTEEGTAIKVFKNLRSCVDCHGAIKFISNITKRKITVRDSTRFH 678
Query: 608 SFKHGMCSCGDHW 620
F++G CSCGD+W
Sbjct: 679 CFENGQCSCGDYW 691
Score = 147 bits (370), Expect = 2e-35
Identities = 95/332 (28%), Positives = 160/332 (47%), Gaps = 35/332 (10%)
Query: 138 KMLDYGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCG 197
++L +P T +++ CS A+ EG+ +HE++ SG+ + + L+ MYAKCG
Sbjct: 75 QLLGRAKKPPASTYCNLIQVCSQTRALEEGKKVHEHIRTSGFVPGIVIWNRLLRMYAKCG 134
Query: 198 CVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAAN-------------- 243
++DA +VFD++ RD WN M+ YA+ G +E+ L EM
Sbjct: 135 SLVDARKVFDEMPNRDLCSWNVMVNGYAEVGLLEEARKLFDEMTEKDSYSWTAMVTGYVK 194
Query: 244 ------------------GVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDK 285
RP T+ ++++A V C+ G+EIHG R G S++
Sbjct: 195 KDQPEEALVLYSLMQRVPNSRPNIFTVSIAVAAAAAVKCIRRGKEIHGHIVRAGLDSDEV 254
Query: 286 VKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKE- 344
+ ++L+DMY KCG + A +F+++ EK VVSW ++I Y LF ++
Sbjct: 255 LWSSLMDMYGKCGCIDEARNIFDKIVEKDVVSWTSMIDRYFKSSRWREGFSLFSELVGSC 314
Query: 345 DRPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEA 404
+RP+ TF GVL AC+ + G+ ++ M R G P + ++D+ CG ++ A
Sbjct: 315 ERPNEYTFAGVLNACADLTTEELGKQVHGYMTR-VGFDPYSFASSSLVDMYTKCGNIESA 373
Query: 405 YDLIRNMSVKPDSGVWGALLNSCKIHGNVELA 436
++ KPD W +L+ C +G + A
Sbjct: 374 KHVVDGCP-KPDLVSWTSLIGGCAQNGQPDEA 404
Score = 71.6 bits (174), Expect = 1e-12
Identities = 54/234 (23%), Positives = 104/234 (44%), Gaps = 13/234 (5%)
Query: 239 EMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCG 298
++ +P +T +I + L G+++H GF + L+ MYAKCG
Sbjct: 75 QLLGRAKKPPASTYCNLIQVCSQTRALEEGKKVHEHIRTSGFVPGIVIWNRLLRMYAKCG 134
Query: 299 SVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDRPDHITFVGVLAA 358
S+ A +F+ + + + SWN ++ GYA GL A LFD+M ++ D ++ ++
Sbjct: 135 SLVDARKVFDEMPNRDLCSWNVMVNGYAEVGLLEEARKLFDEMTEK---DSYSWTAMVTG 191
Query: 359 CSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCG-----QLDEAYDLIRNMSV 413
+ +E LY+LM R P + +T I + + E + I +
Sbjct: 192 YVKKDQPEEALVLYSLMQRVPNSRPNI--FTVSIAVAAAAAVKCIRRGKEIHGHIVRAGL 249
Query: 414 KPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKW 467
D +W +L++ G ++ A +K++E D ++ + + Y +S +W
Sbjct: 250 DSDEVLWSSLMDMYGKCGCIDEARNIFDKIVE---KDVVSWTSMIDRYFKSSRW 300
>At2g03880 putative selenium-binding protein
Length = 630
Score = 434 bits (1115), Expect = e-122
Identities = 225/572 (39%), Positives = 348/572 (60%), Gaps = 6/572 (1%)
Query: 50 YTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIP 109
Y+ L++ CI ++A++ G + Y G L L+++Y N L +A LFD++P
Sbjct: 64 YSELIKCCISNRAVHEGNLICRHLYFNGHRPMMFLVNVLINMYVKFNLLNDAHQLFDQMP 123
Query: 110 KQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGRS 169
++N+ W +I Y+ H A+ L ML +RP+ +T VL++C+ +S + R
Sbjct: 124 QRNVISWTTMISAYSKCKIHQKALELLVLMLRDNVRPNVYTYSSVLRSCNGMSDV---RM 180
Query: 170 IHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGH 229
+H +IK G E D+FV +ALID++AK G DA VFD++V DA++WNS++ +AQN
Sbjct: 181 LHCGIIKEGLESDVFVRSALIDVFAKLGEPEDALSVFDEMVTGDAIVWNSIIGGFAQNSR 240
Query: 230 PDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTA 289
D ++ L + M G +ATL +V+ + +A L G + H + + + + A
Sbjct: 241 SDVALELFKRMKRAGFIAEQATLTSVLRACTGLALLELGMQAHVHIVK--YDQDLILNNA 298
Query: 290 LIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPD 348
L+DMY KCGS++ AL +F +++E+ V++W+ +I+G A +G + AL LF++M+ +P+
Sbjct: 299 LVDMYCKCGSLEDALRVFNQMKERDVITWSTMISGLAQNGYSQEALKLFERMKSSGTKPN 358
Query: 349 HITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLI 408
+IT VGVL ACS LL++G + M + YGI P +HY CMIDLLG G+LD+A L+
Sbjct: 359 YITIVGVLFACSHAGLLEDGWYYFRSMKKLYGIDPVREHYGCMIDLLGKAGKLDDAVKLL 418
Query: 409 RNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWE 468
M +PD+ W LL +C++ N+ LAE A +K+I L+P+D+G Y +L+N+YA S KW+
Sbjct: 419 NEMECEPDAVTWRTLLGACRVQRNMVLAEYAAKKVIALDPEDAGTYTLLSNIYANSQKWD 478
Query: 469 GVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAG 528
VE++R M D+ IKK CSWIEV +++AF+ GD SH + +L +L + G
Sbjct: 479 SVEEIRTRMRDRGIKKEPGCSWIEVNKQIHAFIIGDNSHPQIVEVSKKLNQLIHRLTGIG 538
Query: 529 YAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIK 588
Y P+T V D+E ++ + HSE+LA+AFGL++ + I KNLRIC DCHV K
Sbjct: 539 YVPETNFVLQDLEGEQMEDSLRHHSEKLALAFGLMTLPIEKVIRIRKNLRICGDCHVFCK 598
Query: 589 FISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
SK+ R I +RD RYH F+ G CSCGD+W
Sbjct: 599 LASKLEIRSIVIRDPIRYHHFQDGKCSCGDYW 630
Score = 114 bits (285), Expect = 2e-25
Identities = 81/304 (26%), Positives = 158/304 (51%), Gaps = 10/304 (3%)
Query: 142 YGLRPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMD 201
+GL D+ T ++K C + A+ EG I ++ +G +F+ LI+MY K + D
Sbjct: 55 HGLWADSATYSELIKCCISNRAVHEGNLICRHLYFNGHRPMMFLVNVLINMYVKFNLLND 114
Query: 202 AGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSAD 261
A ++FD++ R+ + W +M++AY++ +++ L M + VRP T +V+ S
Sbjct: 115 AHQLFDQMPQRNVISWTTMISAYSKCKIHQKALELLVLMLRDNVRPNVYTYSSVLRSCNG 174
Query: 262 VACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAI 321
++ + R +H + G +S+ V++ALID++AK G + AL++F+ + + WN+I
Sbjct: 175 MSDV---RMLHCGIIKEGLESDVFVRSALIDVFAKLGEPEDALSVFDEMVTGDAIVWNSI 231
Query: 322 ITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLAACSRGRLLDEG-RALYNLMVRDY 379
I G+A + + AL+LF +M++ + T VL AC+ LL+ G +A +++ D
Sbjct: 232 IGGFAQNSRSDVALELFKRMKRAGFIAEQATLTSVLRACTGLALLELGMQAHVHIVKYDQ 291
Query: 380 GITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELA 439
+ ++D+ CG L++A + M + D W +++ +G + A
Sbjct: 292 DLILN----NALVDMYCKCGSLEDALRVFNQMKER-DVITWSTMISGLAQNGYSQEALKL 346
Query: 440 LEKL 443
E++
Sbjct: 347 FERM 350
>At4g13650 unknown protein
Length = 1024
Score = 433 bits (1113), Expect = e-121
Identities = 214/574 (37%), Positives = 338/574 (58%), Gaps = 1/574 (0%)
Query: 48 YGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDK 107
Y Y S+L++CI L G+Q+H+Q N + + L+ +YA L A ++ +
Sbjct: 451 YTYPSILKTCIRLGDLELGEQIHSQIIKTNFQLNAYVCSVLIDMYAKLGKLDTAWDILIR 510
Query: 108 IPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEG 167
+++ W +I GY D A+ + +MLD G+R D L + AC+ L A+ EG
Sbjct: 511 FAGKDVVSWTTMIAGYTQYNFDDKALTTFRQMLDRGIRSDEVGLTNAVSACAGLQALKEG 570
Query: 168 RSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQN 227
+ IH SG+ DL AL+ +Y++CG + ++ F++ D + WN++++ + Q+
Sbjct: 571 QQIHAQACVSGFSSDLPFQNALVTLYSRCGKIEESYLAFEQTEAGDNIAWNALVSGFQQS 630
Query: 228 GHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVK 287
G+ +E++ + M G+ T + + ++++ A + G+++H + G+ S +V
Sbjct: 631 GNNEEALRVFVRMNREGIDNNNFTFGSAVKAASETANMKQGKQVHAVITKTGYDSETEVC 690
Query: 288 TALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-R 346
ALI MYAKCGS+ A F + K VSWNAII Y+ HG ALD FD+M + R
Sbjct: 691 NALISMYAKCGSISDAEKQFLEVSTKNEVSWNAIINAYSKHGFGSEALDSFDQMIHSNVR 750
Query: 347 PDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYD 406
P+H+T VGVL+ACS L+D+G A + M +YG++P +HY C++D+L G L A +
Sbjct: 751 PNHVTLVGVLSACSHIGLVDKGIAYFESMNSEYGLSPKPEHYVCVVDMLTRAGLLSRAKE 810
Query: 407 LIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGK 466
I+ M +KPD+ VW LL++C +H N+E+ E A L+ELEP+DS YV+L+N+YA S K
Sbjct: 811 FIQEMPIKPDALVWRTLLSACVVHKNMEIGEFAAHHLLELEPEDSATYVLLSNLYAVSKK 870
Query: 467 WEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHE 526
W+ + RQ M +K +KK SWIEVKN +++F GD +H +D I+ + L E
Sbjct: 871 WDARDLTRQKMKEKGVKKEPGQSWIEVKNSIHSFYVGDQNHPLADEIHEYFQDLTKRASE 930
Query: 527 AGYAPDTGSVFHDVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVA 586
GY D S+ ++++ ++K ++ HSE+LAI+FGL+S + + KNLR+C DCH
Sbjct: 931 IGYVQDCFSLLNELQHEQKDPIIFIHSEKLAISFGLLSLPATVPINVMKNLRVCNDCHAW 990
Query: 587 IKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
IKF+SK+ REI VRD R+H F+ G CSC D+W
Sbjct: 991 IKFVSKVSNREIIVRDAYRFHHFEGGACSCKDYW 1024
Score = 183 bits (465), Expect = 2e-46
Identities = 109/396 (27%), Positives = 196/396 (48%), Gaps = 6/396 (1%)
Query: 37 VDSFPPQPTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSN 96
+D P T SL+ +C L G+QLHA LG A N + L++LYA
Sbjct: 342 LDGLEPDSNT---LASLVVACSADGTLFRGQQLHAYTTKLGFASNNKIEGALLNLYAKCA 398
Query: 97 SLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLK 156
+ A + F + +N+ LWNV++ Y N+ ++ +M + P+ +T P +LK
Sbjct: 399 DIETALDYFLETEVENVVLWNVMLVAYGLLDDLRNSFRIFRQMQIEEIVPNQYTYPSILK 458
Query: 157 ACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVL 216
C L + G IH +IK+ ++ + +V + LIDMYAK G + A + + +D V
Sbjct: 459 TCIRLGDLELGEQIHSQIIKTNFQLNAYVCSVLIDMYAKLGKLDTAWDILIRFAGKDVVS 518
Query: 217 WNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGW 276
W +M+A Y Q D++++ R+M G+R E L +S+ A + L G++IH
Sbjct: 519 WTTMIAGYTQYNFDDKALTTFRQMLDRGIRSDEVGLTNAVSACAGLQALKEGQQIHAQAC 578
Query: 277 RHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALD 336
GF S+ + AL+ +Y++CG ++ + FE+ ++WNA+++G+ G AL
Sbjct: 579 VSGFSSDLPFQNALVTLYSRCGKIEESYLAFEQTEAGDNIAWNALVSGFQQSGNNEEALR 638
Query: 337 LFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLL 395
+F +M +E ++ TF + A S + +G+ ++ ++ + G + +I +
Sbjct: 639 VFVRMNREGIDNNNFTFGSAVKAASETANMKQGKQVHAVITKT-GYDSETEVCNALISMY 697
Query: 396 GHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHG 431
CG + +A +S K + W A++N+ HG
Sbjct: 698 AKCGSISDAEKQFLEVSTKNEVS-WNAIINAYSKHG 732
Score = 179 bits (455), Expect = 3e-45
Identities = 122/420 (29%), Positives = 206/420 (49%), Gaps = 4/420 (0%)
Query: 46 THYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLF 105
T Y ++S+L +C ++L G+QLH LG + + + LV LY +L++A ++F
Sbjct: 247 TPYAFSSVLSACKKIESLEIGEQLHGLVLKLGFSSDTYVCNALVSLYFHLGNLISAEHIF 306
Query: 106 DKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIG 165
+ +++ +N LI G + G + A+ L+ +M GL PD+ TL ++ ACSA +
Sbjct: 307 SNMSQRDAVTYNTLINGLSQCGYGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTLF 366
Query: 166 EGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYA 225
G+ +H Y K G+ + + AL+++YAKC + A F + V + VLWN ML AY
Sbjct: 367 RGQQLHAYTTKLGFASNNKIEGALLNLYAKCADIETALDYFLETEVENVVLWNVMLVAYG 426
Query: 226 QNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDK 285
S + R+M + P + T +++ + + L G +IH + FQ N
Sbjct: 427 LLDDLRNSFRIFRQMQIEEIVPNQYTYPSILKTCIRLGDLELGEQIHSQIIKTNFQLNAY 486
Query: 286 VKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED 345
V + LIDMYAK G + A + R K VVSW +I GY + AL F +M
Sbjct: 487 VCSVLIDMYAKLGKLDTAWDILIRFAGKDVVSWTTMIAGYTQYNFDDKALTTFRQMLDRG 546
Query: 346 -RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEA 404
R D + ++AC+ + L EG+ ++ G + + ++ L CG+++E+
Sbjct: 547 IRSDEVGLTNAVSACAGLQALKEGQQIH-AQACVSGFSSDLPFQNALVTLYSRCGKIEES 605
Query: 405 YDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQS 464
Y L + D+ W AL++ + GN E A L + + E D+ N+ + + A S
Sbjct: 606 Y-LAFEQTEAGDNIAWNALVSGFQQSGNNEEA-LRVFVRMNREGIDNNNFTFGSAVKAAS 663
Score = 163 bits (413), Expect = 2e-40
Identities = 96/363 (26%), Positives = 192/363 (52%), Gaps = 7/363 (1%)
Query: 50 YTSLLQSCID-SKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKI 108
++ +L++C S A + +Q+HA+ + G+ + + L+ LY+ + + AR +FD +
Sbjct: 149 FSGVLEACRGGSVAFDVVEQIHARILYQGLRDSTVVCNPLIDLYSRNGFVDLARRVFDGL 208
Query: 109 PKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGEGR 168
++ W +I G + N AI L+ M G+ P + VL AC + ++ G
Sbjct: 209 RLKDHSSWVAMISGLSKNECEAEAIRLFCDMYVLGIMPTPYAFSSVLSACKKIESLEIGE 268
Query: 169 SIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNG 228
+H V+K G+ D +V AL+ +Y G ++ A +F + RDAV +N+++ +Q G
Sbjct: 269 QLHGLVLKLGFSSDTYVCNALVSLYFHLGNLISAEHIFSNMSQRDAVTYNTLINGLSQCG 328
Query: 229 HPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKT 288
+ ++++ L + M +G+ P TL +++ + + L G+++H + + GF SN+K++
Sbjct: 329 YGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTLFRGQQLHAYTTKLGFASNNKIEG 388
Query: 289 ALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RP 347
AL+++YAKC ++ AL F + VV WN ++ Y + + +F +M+ E+ P
Sbjct: 389 ALLNLYAKCADIETALDYFLETEVENVVLWNVMLVAYGLLDDLRNSFRIFRQMQIEEIVP 448
Query: 348 DHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTC--MIDLLGHCGQLDEAY 405
+ T+ +L C R L+ G +++ +++ + Y C +ID+ G+LD A+
Sbjct: 449 NQYTYPSILKTCIRLGDLELGEQIHSQIIK---TNFQLNAYVCSVLIDMYAKLGKLDTAW 505
Query: 406 DLI 408
D++
Sbjct: 506 DIL 508
Score = 160 bits (404), Expect = 3e-39
Identities = 133/504 (26%), Positives = 232/504 (45%), Gaps = 42/504 (8%)
Query: 1 MSFYSIKKTQHTSFIFNLFPFSQSFYHSLATHQTASVDSFPPQPTTHYGYTSLLQSCIDS 60
M F ++ T+ SF S+ S + SV++ +P H LL+ C+ +
Sbjct: 1 MVFLTLCGTRRASFAAISVYISED--ESFQEKRIDSVENRGIRPN-HQTLKWLLEGCLKT 57
Query: 61 K-ALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFDKIPKQNLFLWNVL 119
+L+ G++LH+Q LG+ N L+ KL Y L A +FD++P++ +F WN +
Sbjct: 58 NGSLDEGRKLHSQILKLGLDSNGCLSEKLFDFYLFKGDLYGAFKVFDEMPERTIFTWNKM 117
Query: 120 IRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALS-AIGEGRSIHEYVIKSG 178
I+ A L+ +M+ + P+ T VL+AC S A IH ++ G
Sbjct: 118 IKELASRNLIGEVFGLFVRMVSENVTPNEGTFSGVLEACRGGSVAFDVVEQIHARILYQG 177
Query: 179 WERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCR 238
V LID+Y++ G V A RVFD + ++D W +M++ ++N E+I L
Sbjct: 178 LRDSTVVCNPLIDLYSRNGFVDLARRVFDGLRLKDHSSWVAMISGLSKNECEAEAIRLFC 237
Query: 239 EMAANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCG 298
+M G+ PT +V+S+ + L G ++HG + GF S+ V AL+ +Y G
Sbjct: 238 DMYVLGIMPTPYAFSSVLSACKKIESLEIGEQLHGLVLKLGFSSDTYVCNALVSLYFHLG 297
Query: 299 SVKVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKED-RPDHITFVGVLA 357
++ A +F + ++ V++N +I G + G A++LF +M + PD T ++
Sbjct: 298 NLISAEHIFSNMSQRDAVTYNTLINGLSQCGYGEKAMELFKRMHLDGLEPDSNTLASLVV 357
Query: 358 ACSRGRLLDEGR-------------------ALYNLMVR--------DYGITPTVQH--- 387
ACS L G+ AL NL + DY + V++
Sbjct: 358 ACSADGTLFRGQQLHAYTTKLGFASNNKIEGALLNLYAKCADIETALDYFLETEVENVVL 417
Query: 388 YTCMIDLLGHCGQLDEAYDLIRNMSVK---PDSGVWGALLNSCKIHGNVELAELALEKLI 444
+ M+ G L ++ + R M ++ P+ + ++L +C G++EL E ++I
Sbjct: 418 WNVMLVAYGLLDDLRNSFRIFRQMQIEEIVPNQYTYPSILKTCIRLGDLELGEQIHSQII 477
Query: 445 ELEPDDSGNYV--ILANMYAQSGK 466
+ YV +L +MYA+ GK
Sbjct: 478 KTN-FQLNAYVCSVLIDMYAKLGK 500
Score = 70.5 bits (171), Expect = 3e-12
Identities = 42/179 (23%), Positives = 90/179 (49%), Gaps = 2/179 (1%)
Query: 47 HYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFD 106
++ + S +++ ++ + GKQ+HA G ++ L+ +YA S+ +A F
Sbjct: 652 NFTFGSAVKAASETANMKQGKQVHAVITKTGYDSETEVCNALISMYAKCGSISDAEKQFL 711
Query: 107 KIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGE 166
++ +N WN +I Y+ +G A+ + +M+ +RP++ TL VL ACS + + +
Sbjct: 712 EVSTKNEVSWNAIINAYSKHGFGSEALDSFDQMIHSNVRPNHVTLVGVLSACSHIGLVDK 771
Query: 167 GRSIHEYV-IKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVR-DAVLWNSMLAA 223
G + E + + G ++DM + G + A ++ ++ DA++W ++L+A
Sbjct: 772 GIAYFESMNSEYGLSPKPEHYVCVVDMLTRAGLLSRAKEFIQEMPIKPDALVWRTLLSA 830
>At2g02980 unknown protein
Length = 603
Score = 431 bits (1109), Expect = e-121
Identities = 218/527 (41%), Positives = 336/527 (63%), Gaps = 2/527 (0%)
Query: 96 NSLLNARNLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVL 155
+S+ AR+LF+ + + ++ ++N + RGY+ L+ ++L+ G+ PDN+T P +L
Sbjct: 77 SSMSYARHLFEAMSEPDIVIFNSMARGYSRFTNPLEVFSLFVEILEDGILPDNYTFPSLL 136
Query: 156 KACSALSAIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAV 215
KAC+ A+ EGR +H +K G + +++V LI+MY +C V A VFD+IV V
Sbjct: 137 KACAVAKALEEGRQLHCLSMKLGLDDNVYVCPTLINMYTECEDVDSARCVFDRIVEPCVV 196
Query: 216 LWNSMLAAYAQNGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHGFG 275
+N+M+ YA+ P+E++SL REM ++P E TL++V+SS A + L G+ IH +
Sbjct: 197 CYNAMITGYARRNRPNEALSLFREMQGKYLKPNEITLLSVLSSCALLGSLDLGKWIHKYA 256
Query: 276 WRHGFQSNDKVKTALIDMYAKCGSVKVALALFERLREKRVVSWNAIITGYAMHGLAVGAL 335
+H F KV TALIDM+AKCGS+ A+++FE++R K +W+A+I YA HG A ++
Sbjct: 257 KKHSFCKYVKVNTALIDMFAKCGSLDDAVSIFEKMRYKDTQAWSAMIVAYANHGKAEKSM 316
Query: 336 DLFDKMRKED-RPDHITFVGVLAACSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDL 394
+F++MR E+ +PD ITF+G+L ACS ++EGR ++ MV +GI P+++HY M+DL
Sbjct: 317 LMFERMRSENVQPDEITFLGLLNACSHTGRVEEGRKYFSQMVSKFGIVPSIKHYGSMVDL 376
Query: 395 LGHCGQLDEAYDLIRNMSVKPDSGVWGALLNSCKIHGNVELAELALEKLIELEPDDSGNY 454
L G L++AY+ I + + P +W LL +C H N++LAE E++ EL+ G+Y
Sbjct: 377 LSRAGNLEDAYEFIDKLPISPTPMLWRILLAACSSHNNLDLAEKVSERIFELDDSHGGDY 436
Query: 455 VILANMYAQSGKWEGVEKLRQVMIDKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIY 514
VIL+N+YA++ KWE V+ LR+VM D++ K CS IEV N V+ F +GD S + ++
Sbjct: 437 VILSNLYARNKKWEYVDSLRKVMKDRKAVKVPGCSSIEVNNVVHEFFSGDGVKSATTKLH 496
Query: 515 AELKRLEGLMHEAGYAPDTGSVFH-DVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLI 573
L + + +GY PDT V H ++ + EK + HSE+LAI FGL++T PGT + +
Sbjct: 497 RALDEMVKELKLSGYVPDTSMVVHANMNDQEKEITLRYHSEKLAITFGLLNTPPGTTIRV 556
Query: 574 TKNLRICEDCHVAIKFISKIMEREITVRDVNRYHSFKHGMCSCGDHW 620
KNLR+C DCH A K IS I R++ +RDV R+H F+ G CSCGD W
Sbjct: 557 VKNLRVCRDCHNAAKLISLIFGRKVVLRDVQRFHHFEDGKCSCGDFW 603
Score = 143 bits (361), Expect = 2e-34
Identities = 84/297 (28%), Positives = 161/297 (53%), Gaps = 6/297 (2%)
Query: 47 HYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNARNLFD 106
+Y + SLL++C +KAL G+QLH LG+ N + L+++Y + +AR +FD
Sbjct: 129 NYTFPSLLKACAVAKALEEGRQLHCLSMKLGLDDNVYVCPTLINMYTECEDVDSARCVFD 188
Query: 107 KIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALSAIGE 166
+I + + +N +I GYA + A+ L+ +M L+P+ TL VL +C+ L ++
Sbjct: 189 RIVEPCVVCYNAMITGYARRNRPNEALSLFREMQGKYLKPNEITLLSVLSSCALLGSLDL 248
Query: 167 GRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQ 226
G+ IH+Y K + + + V ALIDM+AKCG + DA +F+K+ +D W++M+ AYA
Sbjct: 249 GKWIHKYAKKHSFCKYVKVNTALIDMFAKCGSLDDAVSIFEKMRYKDTQAWSAMIVAYAN 308
Query: 227 NGHPDESISLCREMAANGVRPTEATLVTVISSSADVACLPYGREIHG-FGWRHGFQSNDK 285
+G ++S+ + M + V+P E T + ++++ + + GR+ + G + K
Sbjct: 309 HGKAEKSMLMFERMRSENVQPDEITFLGLLNACSHTGRVEEGRKYFSQMVSKFGIVPSIK 368
Query: 286 VKTALIDMYAKCGSVKVALALFERLR-EKRVVSWNAIITGYAMHGLAVGALDLFDKM 341
+++D+ ++ G+++ A ++L + W ++ + H LDL +K+
Sbjct: 369 HYGSMVDLLSRAGNLEDAYEFIDKLPISPTPMLWRILLAACSSH----NNLDLAEKV 421
>At4g21070 putative protein (fragment)
Length = 1495
Score = 430 bits (1106), Expect = e-120
Identities = 222/547 (40%), Positives = 342/547 (61%), Gaps = 8/547 (1%)
Query: 67 KQLHAQFYHLGIAYNQDLATKLVHLYAVS----NSLLNARNLFDKIPKQ-NLFLWNVLIR 121
+Q+HA G++ + K + Y VS + A +F KI K N+F+WN LIR
Sbjct: 34 RQIHAFSIRHGVSISDAELGKHLIFYLVSLPSPPPMSYAHKVFSKIEKPINVFIWNTLIR 93
Query: 122 GYAWNGPHDNAIILYHKMLDYGL-RPDNFTLPFVLKACSALSAIGEGRSIHEYVIKSGWE 180
GYA G +A LY +M GL PD T PF++KA + ++ + G +IH VI+SG+
Sbjct: 94 GYAEIGNSISAFSLYREMRVSGLVEPDTHTYPFLIKAVTTMADVRLGETIHSVVIRSGFG 153
Query: 181 RDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLAAYAQNGHPDESISLCREM 240
++V +L+ +YA CG V A +VFDK+ +D V WNS++ +A+NG P+E+++L EM
Sbjct: 154 SLIYVQNSLLHLYANCGDVASAYKVFDKMPEKDLVAWNSVINGFAENGKPEEALALYTEM 213
Query: 241 AANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQSNDKVKTALIDMYAKCGSV 300
+ G++P T+V+++S+ A + L G+ +H + + G N L+D+YA+CG V
Sbjct: 214 NSKGIKPDGFTIVSLLSACAKIGALTLGKRVHVYMIKVGLTRNLHSSNVLLDLYARCGRV 273
Query: 301 KVALALFERLREKRVVSWNAIITGYAMHGLAVGALDLFDKMRKEDR--PDHITFVGVLAA 358
+ A LF+ + +K VSW ++I G A++G A++LF M + P ITFVG+L A
Sbjct: 274 EEAKTLFDEMVDKNSVSWTSLIVGLAVNGFGKEAIELFKYMESTEGLLPCEITFVGILYA 333
Query: 359 CSRGRLLDEGRALYNLMVRDYGITPTVQHYTCMIDLLGHCGQLDEAYDLIRNMSVKPDSG 418
CS ++ EG + M +Y I P ++H+ CM+DLL GQ+ +AY+ I++M ++P+
Sbjct: 334 CSHCGMVKEGFEYFRRMREEYKIEPRIEHFGCMVDLLARAGQVKKAYEYIKSMPMQPNVV 393
Query: 419 VWGALLNSCKIHGNVELAELALEKLIELEPDDSGNYVILANMYAQSGKWEGVEKLRQVMI 478
+W LL +C +HG+ +LAE A ++++LEP+ SG+YV+L+NMYA +W V+K+R+ M+
Sbjct: 394 IWRTLLGACTVHGDSDLAEFARIQILQLEPNHSGDYVLLSNMYASEQRWSDVQKIRKQML 453
Query: 479 DKRIKKNIACSWIEVKNKVYAFLAGDVSHSNSDAIYAELKRLEGLMHEAGYAPDTGSVFH 538
+KK S +EV N+V+ FL GD SH SDAIYA+LK + G + GY P +V+
Sbjct: 454 RDGVKKVPGHSLVEVGNRVHEFLMGDKSHPQSDAIYAKLKEMTGRLRSEGYVPQISNVYV 513
Query: 539 DVEEDEKTSMVCSHSERLAIAFGLISTSPGTRLLITKNLRICEDCHVAIKFISKIMEREI 598
DVEE+EK + V HSE++AIAF LIST + + + KNLR+C DCH+AIK +SK+ REI
Sbjct: 514 DVEEEEKENAVVYHSEKIAIAFMLISTPERSPITVVKNLRVCADCHLAIKLVSKVYNREI 573
Query: 599 TVRDVNR 605
VRD +R
Sbjct: 574 VVRDRSR 580
Score = 154 bits (389), Expect = 1e-37
Identities = 90/295 (30%), Positives = 155/295 (52%), Gaps = 14/295 (4%)
Query: 43 QPTTHYGYTSLLQSCIDSKALNPGKQLHAQFYHLGIAYNQDLATKLVHLYAVSNSLLNAR 102
+P TH Y L+++ + G+ +H+ G + L+HLYA + +A
Sbjct: 118 EPDTHT-YPFLIKAVTTMADVRLGETIHSVVIRSGFGSLIYVQNSLLHLYANCGDVASAY 176
Query: 103 NLFDKIPKQNLFLWNVLIRGYAWNGPHDNAIILYHKMLDYGLRPDNFTLPFVLKACSALS 162
+FDK+P+++L WN +I G+A NG + A+ LY +M G++PD FT+ +L AC+ +
Sbjct: 177 KVFDKMPEKDLVAWNSVINGFAENGKPEEALALYTEMNSKGIKPDGFTIVSLLSACAKIG 236
Query: 163 AIGEGRSIHEYVIKSGWERDLFVGAALIDMYAKCGCVMDAGRVFDKIVVRDAVLWNSMLA 222
A+ G+ +H Y+IK G R+L L+D+YA+CG V +A +FD++V +++V W S++
Sbjct: 237 ALTLGKRVHVYMIKVGLTRNLHSSNVLLDLYARCGRVEEAKTLFDEMVDKNSVSWTSLIV 296
Query: 223 AYAQNGHPDESISLCREM-AANGVRPTEATLVTVISSSADVACLPYGREIHGFGWRHGFQ 281
A NG E+I L + M + G+ P E T V ++ AC G GF + +
Sbjct: 297 GLAVNGFGKEAIELFKYMESTEGLLPCEITFVGIL-----YACSHCGMVKEGFEYFRRMR 351
Query: 282 SNDKVKT------ALIDMYAKCGSVKVALALFERL-REKRVVSWNAIITGYAMHG 329
K++ ++D+ A+ G VK A + + + VV W ++ +HG
Sbjct: 352 EEYKIEPRIEHFGCMVDLLARAGQVKKAYEYIKSMPMQPNVVIWRTLLGACTVHG 406
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,222,398
Number of Sequences: 26719
Number of extensions: 602517
Number of successful extensions: 7296
Number of sequences better than 10.0: 443
Number of HSP's better than 10.0 without gapping: 380
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 1441
Number of HSP's gapped (non-prelim): 1867
length of query: 620
length of database: 11,318,596
effective HSP length: 105
effective length of query: 515
effective length of database: 8,513,101
effective search space: 4384247015
effective search space used: 4384247015
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149489.3


