

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.2 - phase: 0
(365 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
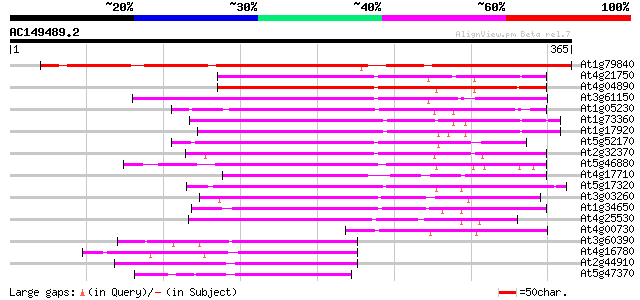
Score E
Sequences producing significant alignments: (bits) Value
At1g79840 homeobox protein (GLABRA2) 348 3e-96
At4g21750 L1 specific homeobox gene ATML1/ovule-specific homeobo... 189 2e-48
At4g04890 putative homeotic protein 187 8e-48
At3g61150 homeobox protein 184 7e-47
At1g05230 homeobox like protein 182 2e-46
At1g73360 putative homeobox protein 179 2e-45
At1g17920 unknown protein 172 2e-43
At5g52170 homeodomain transcription factor-like 170 1e-42
At2g32370 putative homeodomain transcription factor 165 3e-41
At5g46880 homeobox protein 162 3e-40
At4g17710 GLABRA2 like protein 144 8e-35
At5g17320 homeobox protein 142 4e-34
At3g03260 unknown protein 122 4e-28
At1g34650 hypothetical protein 117 1e-26
At4g25530 homeodomain-containing transcription factor FWA 101 6e-22
At4g00730 homeodomain protein AHDP 77 1e-14
At3g60390 homeobox-leucine zipper protein HAT3 74 2e-13
At4g16780 DNA-binding homeotic protein Athb-2 72 4e-13
At2g44910 homeodomain transcription factor (ATHB-4) 71 8e-13
At5g47370 homeobox-leucine zipper protein-like 69 4e-12
>At1g79840 homeobox protein (GLABRA2)
Length = 747
Score = 348 bits (893), Expect = 3e-96
Identities = 192/348 (55%), Positives = 241/348 (69%), Gaps = 36/348 (10%)
Query: 21 LSMCADMSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEE 80
+SM DMS+ P KDFF SPALSLSLAGIFR+ + E + V++ +
Sbjct: 1 MSMAVDMSSKQP-----TKDFFSSPALSLSLAGIFRNASSGSTNPEEDFLGRRVVDDED- 54
Query: 81 GSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKR 140
R E+SSE SGP +S+S ++ EG++ +D+E E+++G G K+KR
Sbjct: 55 -------RTVEMSSENSGPTRSRSEEDLEGEDHDDEEEEEEDGAAGNKG-----TNKRKR 102
Query: 141 KKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER 200
KKYHRHT++QIR MEALFKE+PHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER
Sbjct: 103 KKYHRHTTDQIRHMEALFKETPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER 162
Query: 201 HENSLLKSEIEKLREKNKTLRETINKA--CCPNCGVPTTNRDGTMATEEQQLRIENAKLK 258
HENSLLK+E+EKLRE+NK +RE+ +KA CPNCG L +EN+KLK
Sbjct: 163 HENSLLKAELEKLREENKAMRESFSKANSSCPNCG-----------GGPDDLHLENSKLK 211
Query: 259 AEVERLRAALGKYASGTMSP-SCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAMEE 317
AE+++LRAALG+ T P S S DQE+ SLDFYTG+F L++SRI ++ N+A E
Sbjct: 212 AELDKLRAALGR----TPYPLQASCSDDQEHRLGSLDFYTGVFALEKSRIAEISNRATLE 267
Query: 318 LIKMATMGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRSIEA 365
L KMAT GEPMWLRS+ETGREILNYDEY+KEF + +++IEA
Sbjct: 268 LQKMATSGEPMWLRSVETGREILNYDEYLKEFPQAQASSFPGRKTIEA 315
>At4g21750 L1 specific homeobox gene ATML1/ovule-specific homeobox
protein A20
Length = 762
Score = 189 bits (480), Expect = 2e-48
Identities = 104/248 (41%), Positives = 147/248 (58%), Gaps = 39/248 (15%)
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ K+K+YHRHT QI+ +E+ FKE PHPD+KQR++LS++L L P QVKFWFQN+RTQ+K
Sbjct: 59 RPNKKKRYHRHTQRQIQELESFFKECPHPDDKQRKELSRELSLEPLQVKFWFQNKRTQMK 118
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENA 255
A ERHEN +LKSE +KLR +N ++ ++ A CPNCG P G M+ +EQ LRIENA
Sbjct: 119 AQHERHENQILKSENDKLRAENNRYKDALSNATCPNCGGPAA--IGEMSFDEQHLRIENA 176
Query: 256 KLKAEVERLRAALGKY------ASGTMSPSCSTSHDQENIKSSLDFYTGIFC-------- 301
+L+ E++R+ A KY A+ + P S+SH + SLD G F
Sbjct: 177 RLREEIDRISAIAAKYVGKPLMANSSSFPQLSSSHHIPS--RSLDLEVGNFGNNNNSHTG 234
Query: 302 --------------------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILN 341
D+ I+++ AMEEL++MA G+P+W+ S + EILN
Sbjct: 235 FVGEMFGSSDILRSVSIPSEADKPMIVELAVAAMEELVRMAQTGDPLWVSS-DNSVEILN 293
Query: 342 YDEYMKEF 349
+EY + F
Sbjct: 294 EEEYFRTF 301
>At4g04890 putative homeotic protein
Length = 743
Score = 187 bits (475), Expect = 8e-48
Identities = 100/237 (42%), Positives = 143/237 (60%), Gaps = 26/237 (10%)
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ K+K+YHRHT QI+ +E+ FKE PHPD+KQR++LS+ L L P QVKFWFQN+RTQ+K
Sbjct: 59 RPNKKKRYHRHTQRQIQELESFFKECPHPDDKQRKELSRDLNLEPLQVKFWFQNKRTQMK 118
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENA 255
A ERHEN +LKS+ +KLR +N +E ++ A CPNCG P G M+ +EQ LRIENA
Sbjct: 119 AQSERHENQILKSDNDKLRAENNRYKEALSNATCPNCGGPAA--IGEMSFDEQHLRIENA 176
Query: 256 KLKAEVERLRAALGKYASGTM----------SPSCSTSHDQENIKSSLDFYTGIFC---- 301
+L+ E++R+ A KY + +PS S + N + F ++
Sbjct: 177 RLREEIDRISAIAAKYVGKPLGSSFAPLAIHAPSRSLDLEVGNFGNQTGFVGEMYGTGDI 236
Query: 302 ---------LDESRIMDVVNQAMEELIKMATMGEPMWLRSLETGREILNYDEYMKEF 349
D+ I+++ AMEEL++MA G+P+WL S + EILN +EY + F
Sbjct: 237 LRSVSIPSETDKPIIVELAVAAMEELVRMAQTGDPLWL-STDNSVEILNEEEYFRTF 292
>At3g61150 homeobox protein
Length = 808
Score = 184 bits (467), Expect = 7e-47
Identities = 115/318 (36%), Positives = 167/318 (52%), Gaps = 57/318 (17%)
Query: 81 GSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKR 140
G ++G + E+S + S G+++E D+ GD + KK+
Sbjct: 52 GLSLGLQTNGEMSRNGEIMESNVSRKSSRGEDVESRSESDNAEAVSGDDLDTSDRPLKKK 111
Query: 141 KKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER 200
K+YHRHT +QI+ +E++FKE HPDEKQR LS++L L PRQVKFWFQNRRTQ+K ER
Sbjct: 112 KRYHRHTPKQIQDLESVFKECAHPDEKQRLDLSRRLNLDPRQVKFWFQNRRTQMKTQIER 171
Query: 201 HENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAE 260
HEN+LL+ E +KLR +N ++RE + C NCG P G ++ EEQ LRIEN++LK E
Sbjct: 172 HENALLRQENDKLRAENMSVREAMRNPMCGNCGGPAV--IGEISMEEQHLRIENSRLKDE 229
Query: 261 VERLRAALGKY------------------------------------------------A 272
++R+ A GK+ +
Sbjct: 230 LDRVCALTGKFLGRSNGSHHIPDSALVLGVGVGSGGCNVGGGFTLSSPLLPQASPRFEIS 289
Query: 273 SGTMSPSCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAMEELIKMATMGEPMWLRS 332
+GT S +T + Q+ + S DF SR +D+ AM+EL+KMA EP+W+RS
Sbjct: 290 NGTGSGLVATVNRQQPVSVS-DFD------QRSRYLDLALAAMDELVKMAQTREPLWVRS 342
Query: 333 LETGREILNYDEYMKEFA 350
++G E+LN +EY F+
Sbjct: 343 SDSGFEVLNQEEYDTSFS 360
>At1g05230 homeobox like protein
Length = 721
Score = 182 bits (462), Expect = 2e-46
Identities = 112/263 (42%), Positives = 149/263 (56%), Gaps = 31/263 (11%)
Query: 106 DEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPD 165
DE++ + E+ EG D D ++ NKKK+ YHRHT QI+ MEA FKE PHPD
Sbjct: 35 DEFDSPNTKSGS-ENQEGGSGNDQDPLHPNKKKR---YHRHTQLQIQEMEAFFKECPHPD 90
Query: 166 EKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETIN 225
+KQR+QLS++L L P QVKFWFQN+RTQ+K ERHENS L++E EKLR N RE +
Sbjct: 91 DKQRKQLSRELNLEPLQVKFWFQNKRTQMKNHHERHENSHLRAENEKLRNDNLRYREALA 150
Query: 226 KACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGT------MSPS 279
A CPNCG PT G M+ +E QLR+ENA+L+ E++R+ A KY MSP
Sbjct: 151 NASCPNCGGPTAI--GEMSFDEHQLRLENARLREEIDRISAIAAKYVGKPVSNYPLMSPP 208
Query: 280 CSTSHDQE-------------NIKSSLDFYTGIFCLDESRIMDVVNQAMEELIKMATMGE 326
E N L T D+ I+D+ AMEEL++M + E
Sbjct: 209 PLPPRPLELAMGNIGGEAYGNNPNDLLKSITAPTESDKPVIIDLSVAAMEELMRMVQVDE 268
Query: 327 PMWLRSLETGREILNYDEYMKEF 349
P+W +SL +L+ +EY + F
Sbjct: 269 PLW-KSL-----VLDEEEYARTF 285
>At1g73360 putative homeobox protein
Length = 722
Score = 179 bits (455), Expect = 2e-45
Identities = 109/278 (39%), Positives = 149/278 (53%), Gaps = 42/278 (15%)
Query: 118 GEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLG 177
G GDG G +K+K+YHRHT++QI+ +E+ FKE PHPDEKQR QLS++LG
Sbjct: 11 GSGSGGDGGGSHHHDGSETDRKKKRYHRHTAQQIQRLESSFKECPHPDEKQRNQLSRELG 70
Query: 178 LAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTT 237
LAPRQ+KFWFQNRRTQ+KA ER +NS LK+E +K+R +N +RE + A CPNCG P
Sbjct: 71 LAPRQIKFWFQNRRTQLKAQHERADNSALKAENDKIRCENIAIREALKHAICPNCGGPPV 130
Query: 238 NRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQE---------- 287
+ D +EQ+LRIENA L+ E+ER+ KY +S ST H
Sbjct: 131 SEDPYF--DEQKLRIENAHLREELERMSTIASKYMGRPIS-QLSTLHPMHISPLDLSMTS 187
Query: 288 -------NIKSSLDF--------------------YTGIFCLDESRIMDVVNQAMEELIK 320
SLDF I +D+ + + AMEEL++
Sbjct: 188 LTGCGPFGHGPSLDFDLLPGSSMAVGPNNNLQSQPNLAISDMDKPIMTGIALTAMEELLR 247
Query: 321 MATMGEPMWLRSLETGREILNYDEYMKEFADENSDHGR 358
+ EP+W R+ + R+ILN Y F +S+ G+
Sbjct: 248 LLQTNEPLWTRT-DGCRDILNLGSYENVF-PRSSNRGK 283
>At1g17920 unknown protein
Length = 687
Score = 172 bits (437), Expect = 2e-43
Identities = 99/262 (37%), Positives = 147/262 (55%), Gaps = 29/262 (11%)
Query: 123 GDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQ 182
GD + K KK+K++HRHT QI+ +E+ F E HPDEKQR QLS++LGLAPRQ
Sbjct: 5 GDSQNHDSSETEKKNKKKKRFHRHTPHQIQRLESTFNECQHPDEKQRNQLSRELGLAPRQ 64
Query: 183 VKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGT 242
+KFWFQNRRTQ KA ER +N LK E +K+R +N +RE I A CP+CG N D
Sbjct: 65 IKFWFQNRRTQKKAQHERADNCALKEENDKIRCENIAIREAIKHAICPSCGDSPVNEDSY 124
Query: 243 MATEEQQLRIENAKLKAEVERLRAALGKYASGTMS---PSCSTSH----DQENIKSSLDF 295
+EQ+LRIENA+L+ E+ER+ + K+ +S P + H + + SLDF
Sbjct: 125 F--DEQKLRIENAQLRDELERVSSIAAKFLGRPISHLPPLLNPMHVSPLELFHTGPSLDF 182
Query: 296 -------------------YTGIFCLDESRIMDVVNQAMEELIKMATMGEPMWLRSLETG 336
+ +D+S + ++ AMEEL+++ EP+W+++ +
Sbjct: 183 DLLPGSCSSMSVPSLPSQPNLVLSEMDKSLMTNIAVTAMEELLRLLQTNEPLWIKT-DGC 241
Query: 337 REILNYDEYMKEFADENSDHGR 358
R++LN + Y F ++ G+
Sbjct: 242 RDVLNLENYENMFTRSSTSGGK 263
>At5g52170 homeodomain transcription factor-like
Length = 682
Score = 170 bits (431), Expect = 1e-42
Identities = 100/235 (42%), Positives = 138/235 (58%), Gaps = 14/235 (5%)
Query: 106 DEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPD 165
DE+E L DD D GD D KKK++ KYHRHTS QI+ +E+ FKE PHP+
Sbjct: 26 DEFESRSLSDDSF--DAMSGDEDKQEQRPKKKKRKTKYHRHTSYQIQELESFFKECPHPN 83
Query: 166 EKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETIN 225
EKQR +L K+L L +Q+KFWFQNRRTQ+K ERHEN +LK E EKLR +N L+E++
Sbjct: 84 EKQRLELGKKLTLESKQIKFWFQNRRTQMKTQLERHENVILKQENEKLRLENSFLKESMR 143
Query: 226 KACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMS---PSCST 282
+ C +CG G ++ E+ QLRIENAKLK E++R+ A ++ G++S PS
Sbjct: 144 GSLCIDCGGAVI--PGEVSFEQHQLRIENAKLKEELDRICALANRFIGGSISLEQPSNGG 201
Query: 283 SHDQE-NIKSSLDFYTGIFCLDESRIMDVVNQAMEELIKMATMGEPMWLRSLETG 336
Q I + T + MD+ +AM+EL+K+A + +W E G
Sbjct: 202 IGSQHLPIGHCVSGGTSLM------FMDLAMEAMDELLKLAELETSLWSSKSEKG 250
>At2g32370 putative homeodomain transcription factor
Length = 721
Score = 165 bits (418), Expect = 3e-41
Identities = 105/259 (40%), Positives = 139/259 (53%), Gaps = 28/259 (10%)
Query: 115 DDEGEDDEGDGD------GDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQ 168
+D G+ D G+ G+G G N+ + K+KKY+RHT QI MEA F+E PHPD+KQ
Sbjct: 38 NDSGDQDFDSGNTSSGNHGEGLGNNQAPRHKKKKYNRHTQLQISEMEAFFRECPHPDDKQ 97
Query: 169 RQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKAC 228
R LS QLGL P Q+KFWFQN+RTQ K QER ENS L++ LR +N+ LRE I++A
Sbjct: 98 RYDLSAQLGLDPVQIKFWFQNKRTQNKNQQERFENSELRNLNNHLRSENQRLREAIHQAL 157
Query: 229 CPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGT-----MSPSCSTS 283
CP CG T G M EE LRI NA+L E+++L K + T P S
Sbjct: 158 CPKCGGQTA--IGEMTFEEHHLRILNARLTEEIKQLSVTAEKISRLTGIPVRSHPRVSPP 215
Query: 284 HDQENIKSSLDFYTGIFCLDESR-------------IMDVVNQAMEELIKMATMGEPMWL 330
+ N + + + + SR IM++ AMEEL+ MA + EP+W+
Sbjct: 216 NPPPNFEFGMGSKGNVG--NHSRETTGPADANTKPIIMELAFGAMEELLVMAQVAEPLWM 273
Query: 331 RSLETGREILNYDEYMKEF 349
LN DEY K F
Sbjct: 274 GGFNGTSLALNLDEYEKTF 292
>At5g46880 homeobox protein
Length = 783
Score = 162 bits (410), Expect = 3e-40
Identities = 110/325 (33%), Positives = 161/325 (48%), Gaps = 74/325 (22%)
Query: 75 VEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNK 134
VEE E + GG G ++ D G+E + +E DDE +
Sbjct: 67 VEEMMENGSAGGS---------FGSGSEQAEDPKFGNESDVNELHDDE-----------Q 106
Query: 135 NKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQI 194
K+K+YHRHT+ QI+ MEALFKE+PHPD+KQR++LS +LGL PRQVKFWFQNRRTQ+
Sbjct: 107 PPPAKKKRYHRHTNRQIQEMEALFKENPHPDDKQRKRLSAELGLKPRQVKFWFQNRRTQM 166
Query: 195 KAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIEN 254
KA Q+R+EN +L++E + L+ +N L+ + CP+CG PT D ++ IEN
Sbjct: 167 KAQQDRNENVMLRAENDNLKSENCHLQAELRCLSCPSCGGPTVLGD----IPFNEIHIEN 222
Query: 255 AKLKAEVERLRAALGKYASGTM----------SPSCSTSHDQENIKSSLDFYTGIF---- 300
+L+ E++RL +Y M +PS H Q +++ + Y G F
Sbjct: 223 CRLREELDRLCCIASRYTGRPMQSMPPSQPLINPSPMLPHHQPSLELDMSVYAGNFPEQS 282
Query: 301 ------------------------------CLDESRI--MDVVNQAMEELIKMATMGEPM 328
DE ++ M+ ++EL KM EP+
Sbjct: 283 CTDMMMLPPQDTACFFPDQTANNNNNNNMLLADEEKVIAMEFAVSCVQELTKMCDTEEPL 342
Query: 329 WL--RSLETGREI--LNYDEYMKEF 349
W+ +S + G EI LN +EYM+ F
Sbjct: 343 WIKKKSDKIGGEILCLNEEEYMRLF 367
>At4g17710 GLABRA2 like protein
Length = 661
Score = 144 bits (363), Expect = 8e-35
Identities = 86/213 (40%), Positives = 123/213 (57%), Gaps = 23/213 (10%)
Query: 139 KRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQ 198
K+K+YHRHT+ QI+ MEALFKE+ HPD K R +LSK+LGL+P QVKFWFQN+RTQIKA Q
Sbjct: 88 KKKRYHRHTASQIQQMEALFKENAHPDTKTRLRLSKKLGLSPIQVKFWFQNKRTQIKAQQ 147
Query: 199 ERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLK 258
R +N+ LK+E E L+ +++ ++ C CG LR+ENA+L+
Sbjct: 148 SRSDNAKLKAENETLKTESQNIQSNFQCLFCSTCG--------------HNLRLENARLR 193
Query: 259 AEVERLRAALGKYASGTMSPSCSTSHDQENIKSSLDFYTGIFCLDESRI-MDVVNQAMEE 317
E++RLR+ + +PS S E K++ D + +E I M++ E
Sbjct: 194 QELDRLRSIV-----SMRNPSPSQEITPETNKNNND--NMLIAEEEKAIDMELAVSCARE 246
Query: 318 LIKMATMGEPMW-LRSLETGREILNYDEYMKEF 349
L KM + EP+W + L+ LN +EY K F
Sbjct: 247 LAKMCDINEPLWNKKRLDNESVCLNEEEYKKMF 279
>At5g17320 homeobox protein
Length = 718
Score = 142 bits (357), Expect = 4e-34
Identities = 88/293 (30%), Positives = 146/293 (49%), Gaps = 51/293 (17%)
Query: 116 DEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQ 175
D+ DE + D D N N + ++K YHRHT+EQI +E FKE PHPDE QR+ L ++
Sbjct: 6 DDNSSDERENDVDA---NTNNRHEKKGYHRHTNEQIHRLETYFKECPHPDEFQRRLLGEE 62
Query: 176 LGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVP 235
L L P+Q+KFWFQN+RTQ K+ E+ +N+ L++E K+R +N+++ + +N CP CG
Sbjct: 63 LNLKPKQIKFWFQNKRTQAKSHNEKADNAALRAENIKIRRENESMEDALNNVVCPPCGGR 122
Query: 236 TTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTM------------SPSCSTS 283
R+ + Q+LR +NA LK E ER+ L +Y +M S STS
Sbjct: 123 GPGREDQL-RHLQKLRAQNAYLKDEYERVSNYLKQYGGHSMHNVEATPYLHGPSNHASTS 181
Query: 284 HDQENIKSS----------------------------------LDFYTGIFCLDESRIMD 309
++ + + + + + L++ +++
Sbjct: 182 KNRPALYGTSSNRLPEPSSIFRGPYTRGNMNTTAPPQPRKPLEMQNFQPLSQLEKIAMLE 241
Query: 310 VVNQAMEELIKMATMGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRS 362
+A+ E++ + M + MW +S R +++ Y K F N+ +GRP+ S
Sbjct: 242 AAEKAVSEVLSLIQMDDTMWKKSSIDDRLVIDPGLYEKYFTKTNT-NGRPESS 293
>At3g03260 unknown protein
Length = 699
Score = 122 bits (305), Expect = 4e-28
Identities = 78/254 (30%), Positives = 122/254 (47%), Gaps = 38/254 (14%)
Query: 124 DGDGDGDGVNKN------KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLG 177
D +G G N+ K+ ++ HRHT +QI+ +EA FKE PHPDE+QR QL ++L
Sbjct: 2 DNNGGGSSGNEQYTSGDAKQNGKRTCHRHTPQQIQRLEAYFKECPHPDERQRNQLCRELK 61
Query: 178 LAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTT 237
L P Q+KFWFQN+RTQ K ++R N LL+ E E L+ N+ + + + CP CG P
Sbjct: 62 LEPDQIKFWFQNKRTQSKTQEDRSTNVLLRGENETLQSDNEAMLDALKSVLCPACGGPPF 121
Query: 238 NRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSSLDFYT 297
R+ Q+LR ENA+LK +R+ + ++ P+ T D SLD +
Sbjct: 122 GRE-ERGHNLQKLRFENARLKDHRDRISNFVDQH-----KPNEPTVEDSLAYVPSLDRIS 175
Query: 298 --------------------------GIFCLDESRIMDVVNQAMEELIKMATMGEPMWLR 331
+ D S + ++ A+EEL ++ E W++
Sbjct: 176 YGINGGNMYEPSSSYGPPNFQIIQPRPLAETDMSLLSEIAASAVEELKRLFLAEEQFWVK 235
Query: 332 SLETGREILNYDEY 345
S +++ + Y
Sbjct: 236 SCIDETYVIDTESY 249
>At1g34650 hypothetical protein
Length = 708
Score = 117 bits (293), Expect = 1e-26
Identities = 76/275 (27%), Positives = 126/275 (45%), Gaps = 53/275 (19%)
Query: 119 EDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGL 178
+ D D +G++ N ++ H++ Q++ +EA F E PHPD+ QR+QL +L L
Sbjct: 2 DSSHNDSSSDEEGIDSNNRR------HHSNHQVQRLEAFFHECPHPDDSQRRQLGNELNL 55
Query: 179 APRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTN 238
+Q+KFWFQNRRTQ + E+ +N L+ E K+R N+ + + + CP CG P
Sbjct: 56 KHKQIKFWFQNRRTQARIHNEKADNIALRVENMKIRCVNEAMEKALETVLCPPCGGPHGK 115
Query: 239 RDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSC-------------STSHD 285
+ Q+LR +N LK E ERL + L K+ G PS STS++
Sbjct: 116 EE--QLCNLQKLRTKNVILKTEYERLSSYLTKH-GGYSIPSVDALPDLHGPSTYGSTSNN 172
Query: 286 QENIKSS-------------------------------LDFYTGIFCLDESRIMDVVNQA 314
+ S L + + L+++R+ ++ A
Sbjct: 173 RPASYGSSSNHLPQQSSLLRRPFTRELINTTPLPKPVLLQHFQQLSQLEKNRMFEIAKNA 232
Query: 315 MEELIKMATMGEPMWLRSLETGREILNYDEYMKEF 349
+ E++ + M MW++S GR I++ Y + F
Sbjct: 233 VAEVMSLIQMEHSMWIKSTIDGRAIIDPGNYKRYF 267
>At4g25530 homeodomain-containing transcription factor FWA
Length = 686
Score = 101 bits (252), Expect = 6e-22
Identities = 69/224 (30%), Positives = 108/224 (47%), Gaps = 15/224 (6%)
Query: 117 EGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL 176
E E DE D D GVN + ++ HR T+ Q + +E + E+PHP E+QR +L ++L
Sbjct: 18 EAEGDEIDMINDMSGVNDQDGGRMRRTHRRTAYQTQELENFYMENPHPTEEQRYELGQRL 77
Query: 177 GLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPT 236
+ QVK WFQN+R K + EN L+ E ++L LR + ++ C CG T
Sbjct: 78 NMGVNQVKNWFQNKRNLEKINNDHLENVTLREEHDRLLATQDQLRSAMLRSLCNICGKAT 137
Query: 237 TNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSS---- 292
G E Q+L ENA L+ E+++ + +Y S STS + S+
Sbjct: 138 --NCGDTEYEVQKLMAENANLEREIDQFNS---RYLSHPKQRMVSTSEQAPSSSSNPGIN 192
Query: 293 ----LDFYTGIFCLDE--SRIMDVVNQAMEELIKMATMGEPMWL 330
LDF G ++ S +++ A+ ELI + + P W+
Sbjct: 193 ATPVLDFSGGTRTSEKETSIFLNLAITALRELITLGEVDCPFWM 236
>At4g00730 homeodomain protein AHDP
Length = 590
Score = 77.0 bits (188), Expect = 1e-14
Identities = 48/154 (31%), Positives = 81/154 (52%), Gaps = 24/154 (15%)
Query: 219 TLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYA------ 272
++RE + C NCG P G ++ EE LRIENA+LK E++R+ GK+
Sbjct: 2 SIREAMRNPICTNCGGPAML--GDVSLEEHHLRIENARLKDELDRVCNLTGKFLGHHHNH 59
Query: 273 --SGTMSPSCSTSHDQENIKSSLDFYTGIFCL--------------DESRIMDVVNQAME 316
+ ++ + T+++ + DF G CL +S ++++ AM+
Sbjct: 60 HYNSSLELAVGTNNNGGHFAFPPDFGGGGGCLPPQQQQSTVINGIDQKSVLLELALTAMD 119
Query: 317 ELIKMATMGEPMWLRSLETGREILNYDEYMKEFA 350
EL+K+A EP+W++SL+ R+ LN DEYM+ F+
Sbjct: 120 ELVKLAQSEEPLWVKSLDGERDELNQDEYMRTFS 153
>At3g60390 homeobox-leucine zipper protein HAT3
Length = 315
Score = 73.6 bits (179), Expect = 2e-13
Identities = 53/163 (32%), Positives = 79/163 (47%), Gaps = 9/163 (5%)
Query: 71 SNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSI----DEYEGDELEDDEGEDDE---G 123
S + V+ +EG+ + +SS SG + + G +ED+E E G
Sbjct: 86 STVVVDVEDEGAGVSSPN-STVSSVMSGKKSERELMAAAGAVGGGRVEDNEIERASCSLG 144
Query: 124 DGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQV 183
G D DG RKK R + EQ V+E FKE + KQ+ L+KQL L RQV
Sbjct: 145 GGSDDEDGSGNGDDSSRKKL-RLSKEQALVLEETFKEHSTLNPKQKMALAKQLNLRTRQV 203
Query: 184 KFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
+ WFQNRR + K Q + LK E L ++N+ L++ +++
Sbjct: 204 EVWFQNRRARTKLKQTEVDCEYLKRCCENLTDENRRLQKEVSE 246
>At4g16780 DNA-binding homeotic protein Athb-2
Length = 284
Score = 72.4 bits (176), Expect = 4e-13
Identities = 58/188 (30%), Positives = 86/188 (44%), Gaps = 17/188 (9%)
Query: 48 SLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVE------EISSEYSGPAK 101
S S G+FR E S+ N + + E + I G V E E +G +
Sbjct: 34 SSSSFGLFRRSSWN--ESFTSSVPNSDSSQKETRTFIRGIDVNRPPSTAEYGDEDAGVSS 91
Query: 102 SKS-IDEYEGDELEDDEGEDDEGDG--DGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALF 158
S + G E +E D +G D DG N KK + K +Q ++E F
Sbjct: 92 PNSTVSSSTGKRSEREEDTDPQGSRGISDDEDGDNSRKKLRLSK------DQSAILEETF 145
Query: 159 KESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNK 218
K+ + KQ+Q L+KQLGL RQV+ WFQNRR + K Q + L+ E L E+N+
Sbjct: 146 KDHSTLNPKQKQALAKQLGLRARQVEVWFQNRRARTKLKQTEVDCEFLRRCCENLTEENR 205
Query: 219 TLRETINK 226
L++ + +
Sbjct: 206 RLQKEVTE 213
>At2g44910 homeodomain transcription factor (ATHB-4)
Length = 318
Score = 71.2 bits (173), Expect = 8e-13
Identities = 47/158 (29%), Positives = 77/158 (47%), Gaps = 5/158 (3%)
Query: 69 SISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGD 128
S++ +++EE + V +S A ++ DE E + G G D D
Sbjct: 95 SVAVVDLEEEAAVVSSPNSAVSSLSGNKRDLAVARGGDENEAERASCSRGGGSGGSDDED 154
Query: 129 GDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQ 188
G + ++KK R + +Q V+E FKE + KQ+ L+KQL L RQV+ WFQ
Sbjct: 155 GGNGDGSRKKLRL-----SKDQALVLEETFKEHSTLNPKQKLALAKQLNLRARQVEVWFQ 209
Query: 189 NRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
NRR + K Q + LK + L E+N+ L++ +++
Sbjct: 210 NRRARTKLKQTEVDCEYLKRCCDNLTEENRRLQKEVSE 247
>At5g47370 homeobox-leucine zipper protein-like
Length = 283
Score = 68.9 bits (167), Expect = 4e-12
Identities = 48/141 (34%), Positives = 70/141 (49%), Gaps = 11/141 (7%)
Query: 82 STIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRK 141
STI G+R E +G + DE+ D G G D + DG ++KK R
Sbjct: 81 STISGKRSEREGISGTGVGSGD-----DHDEITPDRGYS-RGTSDEEEDGGETSRKKLRL 134
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERH 201
+ +Q +E FKE + KQ+ L+K+L L RQV+ WFQNRR + K Q
Sbjct: 135 -----SKDQSAFLEETFKEHNTLNPKQKLALAKKLNLTARQVEVWFQNRRARTKLKQTEV 189
Query: 202 ENSLLKSEIEKLREKNKTLRE 222
+ LK +EKL E+N+ L++
Sbjct: 190 DCEYLKRCVEKLTEENRRLQK 210
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,651,646
Number of Sequences: 26719
Number of extensions: 414518
Number of successful extensions: 4757
Number of sequences better than 10.0: 469
Number of HSP's better than 10.0 without gapping: 238
Number of HSP's successfully gapped in prelim test: 238
Number of HSP's that attempted gapping in prelim test: 2965
Number of HSP's gapped (non-prelim): 1106
length of query: 365
length of database: 11,318,596
effective HSP length: 101
effective length of query: 264
effective length of database: 8,619,977
effective search space: 2275673928
effective search space used: 2275673928
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149489.2


