

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.11 - phase: 0
(526 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
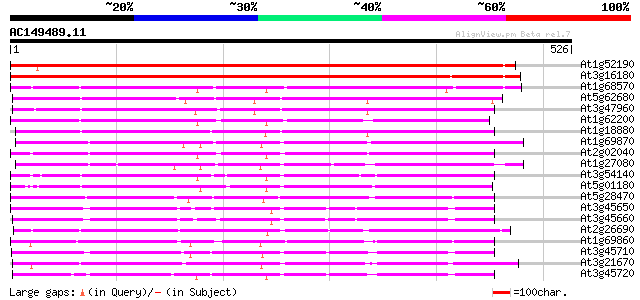
Score E
Sequences producing significant alignments: (bits) Value
At1g52190 peptide transporter like protein 555 e-158
At3g16180 putative transport protein 552 e-157
At1g68570 peptide transporter like 314 7e-86
At5g62680 peptide transporter 312 3e-85
At3g47960 putative peptide transporter 308 5e-84
At1g62200 unknown protein (At1g62200) 302 3e-82
At1g18880 unknown protein 298 4e-81
At1g69870 unknown protein 297 1e-80
At2g02040 histidine transport protein (PTR2-B) 290 1e-78
At1g27080 nitrite transporter, putative 288 5e-78
At3g54140 peptide transport - like protein 282 3e-76
At5g01180 oligopeptide transporter - like protein 280 1e-75
At5g28470 peptide transporter - like protein 272 3e-73
At3g45650 unknown protein 268 5e-72
At3g45660 putative protein 268 7e-72
At2g26690 putative nitrate transporter 263 2e-70
At1g69860 putative peptide transporter 257 1e-68
At3g45710 putative transporter protein 253 2e-67
At3g21670 nitrate transporter 251 9e-67
At3g45720 putative transporter protein 248 4e-66
>At1g52190 peptide transporter like protein
Length = 607
Score = 555 bits (1431), Expect = e-158
Identities = 277/480 (57%), Positives = 358/480 (73%), Gaps = 7/480 (1%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEG--CESATPGQLAMLFSALILIAIGNGGIS-CSLAFG 57
M LLWLTAM+P +P C+ + G C S+T QLA+L+SA LI+IG+GGI CSLAFG
Sbjct: 107 MVLLWLTAMLPQVKPSPCDPTAAGSHCGSSTASQLALLYSAFALISIGSGGIRPCSLAFG 166
Query: 58 ADQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLI 117
ADQ++ K+NP N RVLE FF WYYA + +AV+IA TGIVYIQ+HLGWK+GFGVPA+LMLI
Sbjct: 167 ADQLDNKENPKNERVLESFFGWYYASSAVAVLIAFTGIVYIQEHLGWKIGFGVPAVLMLI 226
Query: 118 STVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPK-TSPEFYHQQKDSELVV 176
+ +LF LASPLYV SLFTG AQ VAAYK RKL LP S + Y+ KDSE+
Sbjct: 227 AALLFILASPLYVTRGVTKSLFTGLAQAIVAAYKKRKLSLPDHHDSFDCYYHMKDSEIKA 286
Query: 177 PTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSI 236
P+ KLRFLNKAC+I + E++I SDG A+N W LCT D+VEELKA+IKVIP+WST I MSI
Sbjct: 287 PSQKLRFLNKACLISNREEEIGSDGFALNPWRLCTTDKVEELKALIKVIPIWSTGIMMSI 346
Query: 237 NIG-GSFGLLQAKSLDRHIIS-SSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIR 294
N SF LLQA S+DR + S+F+VPAGSF + I+A+ +W+I+YDR +IPLASKIR
Sbjct: 347 NTSQSSFQLLQATSMDRRLSRHGSSFQVPAGSFGMFTIIALALWVILYDRAVIPLASKIR 406
Query: 295 GKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQ 354
G+P +S K RMG+GLF +FL + +A+ E+ RRK+AI +GY N+++ V+ +SAMWL PQ
Sbjct: 407 GRPFRLSVKLRMGLGLFMSFLAMAISAMVESFRRKKAISQGYANNSNAVVDISAMWLVPQ 466
Query: 355 LCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSG 414
L G+AE IGQ EF+Y EFPKSMSS+AASL GL M V +L++S+VL+ + T
Sbjct: 467 YVLHGLAEALTAIGQTEFFYTEFPKSMSSIAASLFGLGMAVASLLASVVLNAVNELTSRN 526
Query: 415 GNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEE 474
G E WVSDNINKGH++ YYWV+ ++ +N++YY++CSW+YGP VD+V N + NG + EE
Sbjct: 527 GKESWVSDNINKGHYNYYYWVLAIMSFINVIYYVICSWSYGPLVDQVRN-GRVNGVREEE 585
>At3g16180 putative transport protein
Length = 591
Score = 552 bits (1422), Expect = e-157
Identities = 271/483 (56%), Positives = 357/483 (73%), Gaps = 6/483 (1%)
Query: 1 MSLLWLTAMIPSARPPACNHPS-EGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGA 58
M +LWLTAM+P +P C + C SAT QLA+L++A LI+IG+GGI CSLAFGA
Sbjct: 109 MVVLWLTAMLPQVKPSPCVATAGTNCSSATSSQLALLYTAFALISIGSGGIRPCSLAFGA 168
Query: 59 DQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIS 118
DQ++ K+NP N RVLE FF WYYA +++AV+IA T IVYIQDHLGWK+GFG+PAILML++
Sbjct: 169 DQLDNKENPKNERVLESFFGWYYASSSVAVLIAFTVIVYIQDHLGWKIGFGIPAILMLLA 228
Query: 119 TVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPK-TSPEFYHQQKDSELVVP 177
LF ASPLYVK SLFTG AQV AAY R L LP S + Y++ KDSEL P
Sbjct: 229 GFLFVFASPLYVKRDVSKSLFTGLAQVVAAAYVKRNLTLPDHHDSRDCYYRLKDSELKAP 288
Query: 178 TDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSIN 237
+DKLRFLNKAC I + ++D+ SDG A+N+W LCT DQVE+LKA++KVIP+WST I MSIN
Sbjct: 289 SDKLRFLNKACAISNRDEDLGSDGLALNQWRLCTTDQVEKLKALVKVIPVWSTGIMMSIN 348
Query: 238 IG-GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGK 296
+ SF LLQAKS+DR + S+S F++PAGSF + I+A++ W+++YDR ++PLASKIRG+
Sbjct: 349 VSQNSFQLLQAKSMDRRLSSNSTFQIPAGSFGMFTIIALISWVVLYDRAILPLASKIRGR 408
Query: 297 PVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLC 356
PV ++ K RMG+GLF +FL + +A E RRK AI +G ND + + +SAMWL PQ
Sbjct: 409 PVRVNVKIRMGLGLFISFLAMAVSATVEHYRRKTAISQGLANDANSTVSISAMWLVPQYV 468
Query: 357 LAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGN 416
L G+AE IGQ EF+Y EFPKSMSS+AASL GL M V N+++S++L+ +++++ GN
Sbjct: 469 LHGLAEALTGIGQTEFFYTEFPKSMSSIAASLFGLGMAVANILASVILNAVKNSSKQ-GN 527
Query: 417 EGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEEST 476
W+ DNINKGH+D YYWV+ ++ +N++YY+VCSW+YGPTVD+V N K NG + EE
Sbjct: 528 VSWIEDNINKGHYDYYYWVLAILSFVNVIYYVVCSWSYGPTVDQVRN-DKVNGMRKEEEE 586
Query: 477 EFK 479
K
Sbjct: 587 VIK 589
>At1g68570 peptide transporter like
Length = 596
Score = 314 bits (805), Expect = 7e-86
Identities = 186/491 (37%), Positives = 290/491 (58%), Gaps = 20/491 (4%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGAD 59
M+LL ++A+IP+ RPP C E C A QL++L+ AL+L A+G+GGI C +AFGAD
Sbjct: 109 MTLLTISAIIPTLRPPPCKG-EEVCVVADTAQLSILYVALLLGALGSGGIRPCVVAFGAD 167
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + D PN +F+WYY AV++A+T +V+IQD++GW +G G+P + M +S
Sbjct: 168 QFDESD-PNQTTKTWNYFNWYYFCMGAAVLLAVTVLVWIQDNVGWGLGLGIPTVAMFLSV 226
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSEL----- 174
+ F LY + S FT QV VAA++ RKL + S +++ + D+ +
Sbjct: 227 IAFVGGFQLYRHLVPAGSPFTRLIQVGVAAFRKRKLRMVSDPSLLYFNDEIDAPISLGGK 286
Query: 175 VVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM 234
+ T + FL+KA ++ E+D G N W L TV +VEELK++I++ P+ ++ I +
Sbjct: 287 LTHTKHMSFLDKAAIVT--EEDNLKPGQIPNHWRLSTVHRVEELKSVIRMGPIGASGILL 344
Query: 235 SINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASK 292
G+F L QAK+++RH+ +S F++PAGS SV VA+L II YDRV + +A K
Sbjct: 345 ITAYAQQGTFSLQQAKTMNRHLTNS--FQIPAGSMSVFTTVAMLTTIIFYDRVFVKVARK 402
Query: 293 IRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLA 352
G I+ RMGIG + + + A E R+ AI+ G L+ H ++ +S +WL
Sbjct: 403 FTGLERGITFLHRMGIGFVISIIATLVAGFVEVKRKSVAIEHGLLDKPHTIVPISFLWLI 462
Query: 353 PQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIE--ST 410
PQ L G+AE F IG EF+Y + P+SM S A +L +A+ +GN VS+L+++++ S
Sbjct: 463 PQYGLHGVAEAFMSIGHLEFFYDQAPESMRSTATALFWMAISIGNYVSTLLVTLVHKFSA 522
Query: 411 TPSGGNEGWVSD-NINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENG 469
P G N W+ D N+N+G + +YW+I + A+NL+YYL C+ Y +V + SKE+
Sbjct: 523 KPDGSN--WLPDNNLNRGRLEYFYWLITVLQAVNLVYYLWCAKIYTYKPVQVHH-SKEDS 579
Query: 470 SKVEESTEFKH 480
S V+E + +
Sbjct: 580 SPVKEELQLSN 590
>At5g62680 peptide transporter
Length = 616
Score = 312 bits (799), Expect = 3e-85
Identities = 176/474 (37%), Positives = 273/474 (57%), Gaps = 19/474 (4%)
Query: 3 LLWLTAMIPSARPPACNHPSEG-CESATPGQLAMLFSALILIAIGNGGIS-CSLAFGADQ 60
++ LTA +P P AC ++ C + GQ+A L L + +G GGI C+LAFGADQ
Sbjct: 131 VILLTAAVPQLHPAACGTAADSICNGPSGGQIAFLLMGLGFLVVGAGGIRPCNLAFGADQ 190
Query: 61 VNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTV 120
N K + +R ++ FF+WY+ T A I++LT +VY+Q ++ W +G +PA+LM ++ +
Sbjct: 191 FNPKSE-SGKRGIDSFFNWYFFTFTFAQILSLTLVVYVQSNVSWTIGLTIPAVLMFLACL 249
Query: 121 LFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSP-----EFYHQQKDSELV 175
+FF LYVKIK S G AQV A K R L P P +Y + + +
Sbjct: 250 IFFAGDKLYVKIKASGSPLAGIAQVIAVAIKKRGLK--PAKQPWLNLYNYYPPKYANSKL 307
Query: 176 VPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLW--STAIT 233
TD+ RFL+KA ++ E + DG + W LCT+ QVEE+K I++V+P+W S+
Sbjct: 308 KYTDQFRFLDKAAILTP-EDKLQPDGKPADPWKLCTMQQVEEVKCIVRVLPIWFASSIYY 366
Query: 234 MSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKI 293
++I ++ + QA DR + S F +PA ++ V L+ + ++I++YDRVL+P +I
Sbjct: 367 LTITQQMTYPVFQALQSDRRL-GSGGFVIPAATYVVFLMTGMTVFIVVYDRVLVPTMRRI 425
Query: 294 RGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKE---GYLNDTHGVLKMSAMW 350
G I+ +R+G G+FF L+ A E RR A+ + G + MSAMW
Sbjct: 426 TGLDTGITLLQRIGTGIFFATASLVVAGFVEERRRTFALTKPTLGMAPRKGEISSMSAMW 485
Query: 351 LAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIEST 410
L PQL LAG+AE F IGQ EFYYK+FP++M S A S+ + GV + + S +++ + T
Sbjct: 486 LIPQLSLAGVAEAFAAIGQMEFYYKQFPENMRSFAGSIFYVGGGVSSYLGSFLIATVHRT 545
Query: 411 TPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCS--WAYGPTVDEVS 462
T + W+++++NKG D +Y++I GI A+N Y+LV S + Y + DEV+
Sbjct: 546 TQNSSGGNWLAEDLNKGRLDLFYFMIAGILAVNFAYFLVMSRWYRYKGSDDEVT 599
>At3g47960 putative peptide transporter
Length = 636
Score = 308 bits (789), Expect = 5e-84
Identities = 173/461 (37%), Positives = 267/461 (57%), Gaps = 13/461 (2%)
Query: 3 LLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGADQV 61
++ LTA IPS P AC + CE + GQ+ L L + +G GGI C+LAFGADQ
Sbjct: 149 VILLTAAIPSLHPVACGNKIS-CEGPSVGQILFLLMGLGFLVVGAGGIRPCNLAFGADQF 207
Query: 62 NRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVL 121
N K + ++ + FF+WY+ T A II+LT +VYIQ ++ W +G +P LM ++ V+
Sbjct: 208 NPKSE-SGKKGINSFFNWYFFTFTFAQIISLTAVVYIQSNVSWTIGLIIPVALMFLACVI 266
Query: 122 FFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDS---ELVVPT 178
FF LYVK+K S G A+V AA K R L + Y+ + + T
Sbjct: 267 FFAGDRLYVKVKASGSPLAGIARVIAAAIKKRGLKPVKQPWVNLYNHIPSNYANTTLKYT 326
Query: 179 DKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLW--STAITMSI 236
D+ RFL+KA ++ E+ + SDG+A + W LCT+ QVEE+K I++VIP+W ST ++I
Sbjct: 327 DQFRFLDKAAIMTPEEK-LNSDGTASDPWKLCTLQQVEEVKCIVRVIPIWFASTIYYLAI 385
Query: 237 NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGK 296
I ++ + QA DR + S F +PA ++ V L+ + ++II YDRVL+P ++ G
Sbjct: 386 TIQMTYPVFQALQSDRRL-GSGGFRIPAATYVVFLMTGMTVFIIFYDRVLVPSLRRVTGL 444
Query: 297 PVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKE---GYLNDTHGVLKMSAMWLAP 353
IS +R+G G F + L+ + E RR A+ + G T + MSA+WL P
Sbjct: 445 ETGISLLQRIGAGFTFAIMSLLVSGFIEERRRNFALTKPTLGMAPRTGEISSMSALWLIP 504
Query: 354 QLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPS 413
QL LAGIAE F IGQ EFYYK+FP++M S A S+ + GV + ++S ++S + TT
Sbjct: 505 QLTLAGIAEAFAAIGQMEFYYKQFPENMKSFAGSIFYVGAGVSSYLASFLISTVHRTTAH 564
Query: 414 GGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
+ W+++++NK D +Y+++ G+ +N+ Y+L+ + Y
Sbjct: 565 SPSGNWLAEDLNKAKLDYFYFMLTGLMVVNMAYFLLMARWY 605
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 302 bits (773), Expect = 3e-82
Identities = 180/460 (39%), Positives = 268/460 (58%), Gaps = 26/460 (5%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEG-CESATPGQLAMLFSALILIAIGNGGIS-CSLAFGA 58
M+LL L+A +P +P AC + C AT Q A+ F+ L LIA+G GGI C +FGA
Sbjct: 138 MALLTLSASLPVLKPAACAGVAAALCSPATTVQYAVFFTGLYLIALGTGGIKPCVSSFGA 197
Query: 59 DQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIS 118
DQ + D P R FF+W+Y I I+ T +V++Q+++GW +GF +P + M +S
Sbjct: 198 DQFDDTD-PRERVRKASFFNWFYFSINIGSFISSTLLVWVQENVGWGLGFLIPTVFMGVS 256
Query: 119 TVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSEL---- 174
FF+ +PLY K S T QV VAAY+ KL LP S + ++K+S +
Sbjct: 257 IASFFIGTPLYRFQKPGGSPITRVCQVLVAAYRKLKLNLPEDISFLYETREKNSMIAGSR 316
Query: 175 -VVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAIT 233
+ TD +FL+KA VI ++E + G+ N W LCTV QVEE+K +I++ P+W++ I
Sbjct: 317 KIQHTDGYKFLDKAAVISEYE---SKSGAFSNPWKLCTVTQVEEVKTLIRMFPIWASGIV 373
Query: 234 MSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLAS 291
S+ + + Q +S++R I S FE+P SF V + +LI I IYDR L+P
Sbjct: 374 YSVLYSQISTLFVQQGRSMNRIIRS---FEIPPASFGVFDTLIVLISIPIYDRFLVPFVR 430
Query: 292 KIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWL 351
+ G P ++ +RMGIGLF + L + AAI ETVR + A + MS W
Sbjct: 431 RFTGIPKGLTDLQRMGIGLFLSVLSIAAAAIVETVRLQLA---------QDFVAMSIFWQ 481
Query: 352 APQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTT 411
PQ L GIAE+F IG+ EF+Y E P +M SV ++L+ L VG+ +SSL+L+++ T
Sbjct: 482 IPQYILMGIAEVFFFIGRVEFFYDESPDAMRSVCSALALLNTAVGSYLSSLILTLVAYFT 541
Query: 412 PSGGNEGWVSDNINKGHFDKYYWVIVGINALNL-LYYLVC 450
GG +GWV D++NKGH D ++W++V + +N+ +Y L+C
Sbjct: 542 ALGGKDGWVPDDLNKGHLDYFFWLLVSLGLVNIPVYALIC 581
>At1g18880 unknown protein
Length = 587
Score = 298 bits (764), Expect = 4e-81
Identities = 166/457 (36%), Positives = 263/457 (57%), Gaps = 11/457 (2%)
Query: 6 LTAMIPSARPPAC-NHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGADQVNR 63
LTA+I P C C + GQ+ L A++L+ IG GGI C+L FGADQ +
Sbjct: 110 LTAVIHPLHPAQCAKEIGSVCNGPSIGQIMFLAGAMVLLVIGAGGIRPCNLPFGADQFDP 169
Query: 64 KDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFF 123
K +R +E FF+WY+ T A +++LT IVY+Q ++ W +G +PAILML+ ++FF
Sbjct: 170 KTK-EGKRGIESFFNWYFFTFTFAQMVSLTLIVYVQSNVSWSIGLAIPAILMLLGCIIFF 228
Query: 124 LASPLYVKIKQKTSLFTGFAQVSVAAYKNRKL-PLPPKTSPEFYHQQKDSELVVPTDKLR 182
S LYVK+K S +V V A K R+L P+ P + + + T++ R
Sbjct: 229 AGSKLYVKVKASGSPIHSITRVIVVAIKKRRLKPVGPNELYNYIASDFKNSKLGHTEQFR 288
Query: 183 FLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINI--GG 240
FL+K+ I+ + + DGS ++ W LC++ QVEE+K +I+V+P+W +A +
Sbjct: 289 FLDKSA-IQTQDDKLNKDGSPVDAWKLCSMQQVEEVKCVIRVLPVWLSAALFYLAYIQQT 347
Query: 241 SFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVII 300
++ + Q+ DR + +F++PAGS++V L++ + I+I IYDRVL+P K G+ I
Sbjct: 348 TYTIFQSLQSDRRL-GPGSFQIPAGSYTVFLMLGMTIFIPIYDRVLVPFLRKYTGRDGGI 406
Query: 301 SPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKE---GYLNDTHGVLKMSAMWLAPQLCL 357
+ +R+G GLF ++ +AI E RRK A+ + G + MS MWL PQL L
Sbjct: 407 TQLQRVGAGLFLCITSMMVSAIVEQYRRKVALTKPTLGLAPRKGAISSMSGMWLIPQLVL 466
Query: 358 AGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNE 417
GIA+ +GQ EFYYK+FP++M S A SL +G+ + +S+ +LS + TT
Sbjct: 467 MGIADALAGVGQMEFYYKQFPENMRSFAGSLYYCGIGLASYLSTFLLSAVHDTTEGFSGG 526
Query: 418 GWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
W+ +++NKG + +Y+++ G+ LNL Y+L+ S Y
Sbjct: 527 SWLPEDLNKGRLEYFYFLVAGMMTLNLAYFLLVSHWY 563
>At1g69870 unknown protein
Length = 620
Score = 297 bits (760), Expect = 1e-80
Identities = 170/486 (34%), Positives = 271/486 (54%), Gaps = 18/486 (3%)
Query: 6 LTAMIPSARPPACNHPSE-GCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGADQVNR 63
LTA P P +CN C Q+ +L L +++G+GGI CS+ FG DQ ++
Sbjct: 142 LTASFPQLHPASCNSQDPLSCGGPNKLQIGVLLLGLCFLSVGSGGIRPCSIPFGVDQFDQ 201
Query: 64 KDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFF 123
+ + V FF+WYY T+ +II T +VYIQD + W +GF +P LM ++ V+FF
Sbjct: 202 RTEEGVKGVAS-FFNWYYMTFTVVLIITQTVVVYIQDQVSWIIGFSIPTGLMALAVVMFF 260
Query: 124 LASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKT--SPEFYHQQKDSELVVP---T 178
YV +K + S+F+G AQV VAA K RKL LP + + +Y S ++ +
Sbjct: 261 AGMKRYVYVKPEGSIFSGIAQVIVAARKKRKLKLPAEDDGTVTYYDPAIKSSVLSKLHRS 320
Query: 179 DKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SI 236
++ R L+KA V+ E D+ +G ++W LC+V +VEE+K +I+++P+WS I ++
Sbjct: 321 NQFRCLDKAAVVI--EGDLTPEGPPADKWRLCSVQEVEEVKCLIRIVPIWSAGIISLAAM 378
Query: 237 NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGK 296
G+F + QA +DR++ FE+PAGS SVI ++ I I++ YDRV +P +I G
Sbjct: 379 TTQGTFTVSQALKMDRNL--GPKFEIPAGSLSVISLLTIGIFLPFYDRVFVPFMRRITGH 436
Query: 297 PVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLC 356
I+ +R+G G+ F +I A I E +RR +I G D G+ MS WL+PQL
Sbjct: 437 KSGITLLQRIGTGIVFAIFSMIVAGIVERMRRIRSINAG---DPTGMTPMSVFWLSPQLI 493
Query: 357 LAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGN 416
L G+ E FN+IGQ EF+ +FP+ M S+A SL L+ + +SS +++++ +
Sbjct: 494 LMGLCEAFNIIGQIEFFNSQFPEHMRSIANSLFSLSFAGSSYLSSFLVTVVHKFSGGHDR 553
Query: 417 EGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD-EVSNVSKENGSKVEES 475
W++ N+N G D +Y++I + +NL+Y+ C+ Y V + + ++ S E
Sbjct: 554 PDWLNKNLNAGKLDYFYYLIAVLGVVNLVYFWYCARGYRYKVGLPIEDFEEDKSSDDVEM 613
Query: 476 TEFKHM 481
T K M
Sbjct: 614 TSKKSM 619
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 290 bits (742), Expect = 1e-78
Identities = 162/462 (35%), Positives = 262/462 (56%), Gaps = 18/462 (3%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGAD 59
MS L L+A +P+ +P C + C SATP Q AM F L LIA+G GGI C +FGAD
Sbjct: 127 MSALTLSASVPALKPAECI--GDFCPSATPAQYAMFFGGLYLIALGTGGIKPCVSSFGAD 184
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + D+ R FF+W+Y I +++ + +V+IQ++ GW +GFG+P + M ++
Sbjct: 185 QFDDTDSRERVRKAS-FFNWFYFSINIGALVSSSLLVWIQENRGWGLGFGIPTVFMGLAI 243
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSEL----- 174
FF +PLY K S T +QV VA+++ + +P + + Q K+S +
Sbjct: 244 ASFFFGTPLYRFQKPGGSPITRISQVVVASFRKSSVKVPEDATLLYETQDKNSAIAGSRK 303
Query: 175 VVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM 234
+ TD ++L+KA VI + E + G N W LCTV QVEELK +I++ P+W++ I
Sbjct: 304 IEHTDDCQYLDKAAVISEEE---SKSGDYSNSWRLCTVTQVEELKILIRMFPIWASGIIF 360
Query: 235 SINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASK 292
S + + Q ++++ I +F++P + +++IW+ +YDR ++PLA K
Sbjct: 361 SAVYAQMSTMFVQQGRAMNCKI---GSFQLPPAALGTFDTASVIIWVPLYDRFIVPLARK 417
Query: 293 IRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLA 352
G + +RMGIGLF + L + AAI E +R A G L ++ + +S +W
Sbjct: 418 FTGVDKGFTEIQRMGIGLFVSVLCMAAAAIVEIIRLHMANDLG-LVESGAPVPISVLWQI 476
Query: 353 PQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTP 412
PQ + G AE+F IGQ EF+Y + P +M S+ ++L+ L +GN +SSL+L+++ T
Sbjct: 477 PQYFILGAAEVFYFIGQLEFFYDQSPDAMRSLCSALALLTNALGNYLSSLILTLVTYFTT 536
Query: 413 SGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
G EGW+SDN+N GH D ++W++ G++ +N+ Y + Y
Sbjct: 537 RNGQEGWISDNLNSGHLDYFFWLLAGLSLVNMAVYFFSAARY 578
>At1g27080 nitrite transporter, putative
Length = 534
Score = 288 bits (737), Expect = 5e-78
Identities = 169/485 (34%), Positives = 274/485 (55%), Gaps = 36/485 (7%)
Query: 6 LTAMIPSARPPACNHPS-EGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGADQVNR 63
LTA +P PP CN+P + C+ QL +LF L ++IG+GGI CS+ FG DQ ++
Sbjct: 63 LTACLPQLHPPPCNNPHPDECDDPNKLQLGILFLGLGFLSIGSGGIRPCSIPFGVDQFDQ 122
Query: 64 KDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFF 123
+ + V FF+WYY T+ +I + T +VY+Q + W +GF +P LM + VLFF
Sbjct: 123 RTEQGLKGVAS-FFNWYYLTLTMVLIFSHTVVVYLQT-VSWVIGFSIPTSLMACAVVLFF 180
Query: 124 LASPLYVKIKQKTSLFTGFAQVSVAAYKNR--KLPLPPKTSPEFYHQQKDSELVVP---T 178
+ YV +K + S+F+G A+V VAA K R K+ L + E+Y ++ T
Sbjct: 181 VGMRFYVYVKPEGSVFSGIARVIVAARKKRDLKISLVDDGTEEYYEPPVKPGVLSKLPLT 240
Query: 179 DKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAIT--MSI 236
D+ +FL+KA VI D D+ S+G N+W LC++ +VEE+K +I+V+P+WS I +++
Sbjct: 241 DQFKFLDKAAVILDG--DLTSEGVPANKWRLCSIQEVEEVKCLIRVVPVWSAGIISIVAM 298
Query: 237 NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGK 296
+F + QA +DRH+ +FE+PA S +VI + I IW+ IY+ +L+P ++R
Sbjct: 299 TTQATFMVFQATKMDRHM--GPHFEIPAASITVISYITIGIWVPIYEHLLVPFLWRMRKF 356
Query: 297 PVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLC 356
V + +RMGIG+ F L + TA E VRR A + + +MS WLA L
Sbjct: 357 RVTLL--QRMGIGIVFAILSMFTAGFVEGVRRTRATE---------MTQMSVFWLALPLI 405
Query: 357 LAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGN 416
L G+ E FN IG EF+ +FP+ M S+A SL L+ N +SSL+++ + + + +
Sbjct: 406 LMGLCESFNFIGLIEFFNSQFPEHMRSIANSLFPLSFAAANYLSSLLVTTVHKVSGTKDH 465
Query: 417 EGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEEST 476
W++ ++++G D +Y++I + +NL+Y+ C+ Y + GS++E+
Sbjct: 466 PDWLNKDLDRGKLDYFYYLIAVLGVVNLVYFWYCAHRY----------QYKAGSQIEDFN 515
Query: 477 EFKHM 481
E K +
Sbjct: 516 EEKSL 520
>At3g54140 peptide transport - like protein
Length = 570
Score = 282 bits (722), Expect = 3e-76
Identities = 166/462 (35%), Positives = 259/462 (55%), Gaps = 19/462 (4%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGAD 59
M+LL L+A +P +P CN ++ C + Q A+ F AL +IA+G GGI C +FGAD
Sbjct: 110 MTLLTLSASVPGLKPGNCN--ADTCHPNS-SQTAVFFVALYMIALGTGGIKPCVSSFGAD 166
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + D N + FF+W+Y + +IA T +V+IQ ++GW GFGVP + M+I+
Sbjct: 167 QFDENDE-NEKIKKSSFFNWFYFSINVGALIAATVLVWIQMNVGWGWGFGVPTVAMVIAV 225
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSEL----- 174
FF S Y + S T QV VAA++ + +P S F +S +
Sbjct: 226 CFFFFGSRFYRLQRPGGSPLTRIFQVIVAAFRKISVKVPEDKSLLFETADDESNIKGSRK 285
Query: 175 VVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM 234
+V TD L+F +KA V + + D DG +N W LC+V QVEELK+II ++P+W+T I
Sbjct: 286 LVHTDNLKFFDKAAV--ESQSDSIKDGE-VNPWRLCSVTQVEELKSIITLLPVWATGIVF 342
Query: 235 SINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASK 292
+ + +LQ ++D+H+ NFE+P+ S S+ V++L W +YD+ +IPLA K
Sbjct: 343 ATVYSQMSTMFVLQGNTMDQHM--GKNFEIPSASLSLFDTVSVLFWTPVYDQFIIPLARK 400
Query: 293 IRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLA 352
+ +RMGIGL + +ITA + E VR + +K D + MS W
Sbjct: 401 FTRNERGFTQLQRMGIGLVVSIFAMITAGVLEVVRL-DYVKTHNAYDQKQI-HMSIFWQI 458
Query: 353 PQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTP 412
PQ L G AE+F IGQ EF+Y + P +M S+ ++LS + +GN +S+++++++ T
Sbjct: 459 PQYLLIGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTTVALGNYLSTVLVTVVMKITK 518
Query: 413 SGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
G GW+ DN+N+GH D +++++ ++ LN L YL S Y
Sbjct: 519 KNGKPGWIPDNLNRGHLDYFFYLLATLSFLNFLVYLWISKRY 560
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 280 bits (717), Expect = 1e-75
Identities = 162/461 (35%), Positives = 256/461 (55%), Gaps = 21/461 (4%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGAD 59
M+LL ++A +P P C+ E C AT GQ A+ F AL LIA+G GGI C +FGAD
Sbjct: 111 MTLLTISASVPGLTP-TCS--GETCH-ATAGQTAITFIALYLIALGTGGIKPCVSSFGAD 166
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + D + FF+W+Y + +IA + +V+IQ ++GW G GVP + M I+
Sbjct: 167 QFDDTDE-KEKESKSSFFNWFYFVINVGAMIASSVLVWIQMNVGWGWGLGVPTVAMAIAV 225
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVP-- 177
V FF S Y K S T QV VA+ + K+ +P S + +Q +S ++
Sbjct: 226 VFFFAGSNFYRLQKPGGSPLTRMLQVIVASCRKSKVKIPEDESLLYENQDAESSIIGSRK 285
Query: 178 ---TDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM 234
T L F +KA V + + A+ S+ W LCTV QVEELKA+I+++P+W+T I
Sbjct: 286 LEHTKILTFFDKAAVETESDNKGAAKSSS---WKLCTVTQVEELKALIRLLPIWATGIVF 342
Query: 235 SINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASK 292
+ G+ +LQ +LD+H+ NF++P+ S S+ +++L W +YD++++P A K
Sbjct: 343 ASVYSQMGTVFVLQGNTLDQHM--GPNFKIPSASLSLFDTLSVLFWAPVYDKLIVPFARK 400
Query: 293 IRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLA 352
G + +R+GIGL + +++A I E R N+ + M+ W
Sbjct: 401 YTGHERGFTQLQRIGIGLVISIFSMVSAGILEVARLNYVQTHNLYNEE--TIPMTIFWQV 458
Query: 353 PQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTP 412
PQ L G AE+F IGQ EF+Y + P +M S+ ++LS A+ GN +S+ +++++ T
Sbjct: 459 PQYFLVGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTAIAFGNYLSTFLVTLVTKVTR 518
Query: 413 SGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYL-VCSW 452
SGG GW++ N+N GH D ++W++ G++ LN L YL + W
Sbjct: 519 SGGRPGWIAKNLNNGHLDYFFWLLAGLSFLNFLVYLWIAKW 559
>At5g28470 peptide transporter - like protein
Length = 561
Score = 272 bits (696), Expect = 3e-73
Identities = 156/462 (33%), Positives = 256/462 (54%), Gaps = 19/462 (4%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGAD 59
M + LTA +PS RP AC PS QL +LFS L L+AIG GG+ C++AFGAD
Sbjct: 103 MGIFALTAALPSLRPDACIDPSNCSNQPAKWQLGVLFSGLGLLAIGAGGVRPCNIAFGAD 162
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + LE FF+W+Y T+A++IALTG+VYIQ ++ W +GF +P + +S
Sbjct: 163 QFDTSTKKGKAH-LETFFNWWYFSFTVALVIALTGVVYIQTNISWVIGFVIPTACLALSI 221
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFY----HQQKDSELV 175
F + Y+ K + S+F +V AA K RK + P + FY + + LV
Sbjct: 222 TTFVIGQHTYICAKAEGSVFADIVKVVTAACKKRK--VKPGSDITFYIGPSNDGSPTTLV 279
Query: 176 VPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMS 235
+LRF +KA ++ + ++ DG+A +W LC+V QV+ LK + ++P+W T I
Sbjct: 280 RDKHRLRFFDKASIV-TNPNELNEDGNAKYKWRLCSVQQVKNLKCVTAILPVWVTGIACF 338
Query: 236 I--NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKI 293
I + +G+LQA +D+ NF+VPAG +++ ++ + IWI +Y+ V+IP+ +I
Sbjct: 339 ILTDQQNIYGILQAMQMDK-TFGPHNFQVPAGWMNLVSMITLAIWISLYECVIIPIVKQI 397
Query: 294 RGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAP 353
G+ ++ K R+ IG+ + +I A E RR A+K G V +S + L P
Sbjct: 398 TGRKKRLTLKHRIEIGIVMGIICMIVAGFQEKKRRASALKNGSF-----VSPVSIVMLLP 452
Query: 354 QLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPS 413
Q LAG+ E F+ + EF P+ M +VA ++ L+ + + + +L++++I++ T
Sbjct: 453 QFALAGLTEAFSAVALMEFLTVRMPEHMRAVAGAIFFLSSSIASYICTLLINVIDAVTRK 512
Query: 414 GGNEGWVSD-NINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
G + W+ D ++NK + Y+++I GI NLLY+ + + Y
Sbjct: 513 EG-KSWLGDKDLNKNRLENYFFIIAGIQVANLLYFRLFASRY 553
>At3g45650 unknown protein
Length = 558
Score = 268 bits (685), Expect = 5e-72
Identities = 161/457 (35%), Positives = 251/457 (54%), Gaps = 27/457 (5%)
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAF-GAD 59
++LL LTA + + RP C S C+S + QL +L++A+ L +IG GG +LA GA+
Sbjct: 108 VALLTLTASLDTLRPRPCETASILCQSPSKTQLGVLYTAITLASIGTGGTRFTLATAGAN 167
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + + + FF+W++ T +A I+ T IVY +D++ W +GFG+ S
Sbjct: 168 QYEKTKDQGS------FFNWFFFTTYLAGAISATAIVYTEDNISWTLGFGLSVAANFFSF 221
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPTD 179
++F Y K S FT V AA + RK + T+ + YH + + +PT
Sbjct: 222 LVFVSGKRFYKHDKPLGSPFTSLLCVIFAALRKRKAVV--STNEKDYHNES---ITMPTK 276
Query: 180 KLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG 239
RF N+A + E ++ DG+ N W LC+V QVE+ KA+I++IPL I +S I
Sbjct: 277 SFRFFNRAAL--KQEDEVKPDGTIRNPWRLCSVQQVEDFKAVIRIIPLALATIFLSTPIA 334
Query: 240 GSFGL--LQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKP 297
L LQ +DR + +F++PAGS VI +++ ++II+ DRVL P K+ GK
Sbjct: 335 MQLSLTVLQGLVMDRRL--GPSFKIPAGSLQVITLLSTCLFIIVNDRVLYPFYQKLTGKH 392
Query: 298 VIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCL 357
++P +R+GIG FN L + AI E +R + +++G+ + V MS +WL P L +
Sbjct: 393 --LTPLQRVGIGHAFNILSMAVTAIVE-AKRLKIVQKGHFLGSSSVADMSVLWLFPPLVI 449
Query: 358 AGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNE 417
GI E F+ G Y+EFP+SM S A S++ + +G+ S+ ++ +I+ TT
Sbjct: 450 VGIGEAFHFPGNVALCYQEFPESMRSTATSITSVVIGICFYTSTALIDLIQRTT------ 503
Query: 418 GWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
W+ D+IN G D YW++V LNL Y+LVCSW Y
Sbjct: 504 AWLPDDINHGRVDNVYWILVIGGVLNLGYFLVCSWLY 540
>At3g45660 putative protein
Length = 557
Score = 268 bits (684), Expect = 7e-72
Identities = 160/455 (35%), Positives = 250/455 (54%), Gaps = 27/455 (5%)
Query: 3 LLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAF-GADQV 61
LL LT + S RP AC S+ C++ T QL +L++A+ L +G GG+ +LA GA+Q
Sbjct: 109 LLALTTLFDSLRPQACETASKLCQAPTNIQLGVLYTAITLGCVGAGGLRFTLATAGANQY 168
Query: 62 NRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVL 121
+ + + FF+W++ +A I+ T IVY ++++ W GFG+ L+ ++
Sbjct: 169 EKTKDQGS------FFNWFFFTWYLAASISATAIVYAEENISWSFGFGLCVAANLLGLIV 222
Query: 122 FFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPTDKL 181
F Y K S FT +V AA + RK + T+ + YH + PT
Sbjct: 223 FISGKKFYKHDKPLGSPFTSLLRVIFAAIRKRKAVV--STNEKDYHSESKK---TPTKSF 277
Query: 182 RFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGS 241
RF N+A + +D E + SDG+ N+W LC+V QVE+ KA+I++IPL + +S I
Sbjct: 278 RFFNRAALKQDDE--VNSDGTIHNQWRLCSVQQVEDFKAVIRIIPLVLAILFLSTPIAMQ 335
Query: 242 FGL--LQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVI 299
GL LQ +DR + +F++PAGS VI +++ ++II+ DR L P K+ GK
Sbjct: 336 LGLTVLQGLVMDRRL--GPHFKIPAGSLQVITLLSTCLFIIVNDRFLYPFYQKLTGK--F 391
Query: 300 ISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAG 359
+P +R+GIG FN L + AI E +R + +++G+ + V MS +WL P L + G
Sbjct: 392 PTPIQRVGIGHVFNILSMAVTAIVE-AKRLKIVQKGHFLGSSSVADMSVLWLFPPLVIVG 450
Query: 360 IAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGW 419
I E F+ G Y+EFP+SM S A S++ + +G+ S+ ++ +I+ TT W
Sbjct: 451 IGEAFHFPGNVALCYQEFPESMRSTATSITSVLIGICFYTSTALIDLIQKTT------AW 504
Query: 420 VSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
+ D+IN G D YW++V LNL Y+LVCSW Y
Sbjct: 505 LPDDINHGRVDNVYWILVIGGVLNLGYFLVCSWFY 539
>At2g26690 putative nitrate transporter
Length = 586
Score = 263 bits (672), Expect = 2e-70
Identities = 153/470 (32%), Positives = 262/470 (55%), Gaps = 17/470 (3%)
Query: 4 LWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-FGADQVN 62
L + +P RPP C+H E C AT Q+ +L+ +L LIA+G GG+ S++ FG+DQ +
Sbjct: 122 LAVATKLPELRPPTCHH-GEACIPATAFQMTILYVSLYLIALGTGGLKSSISGFGSDQFD 180
Query: 63 RKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLF 122
KD P + + FF+ ++ F ++ ++A+T +VY+QD +G +G+ + M I+ V+F
Sbjct: 181 DKD-PKEKAHMAFFFNRFFFFISMGTLLAVTVLVYMQDEVGRSWAYGICTVSMAIAIVIF 239
Query: 123 FLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPTDKLR 182
+ Y K + S QV AA++ RK+ LP ++ Y + + TD+
Sbjct: 240 LCGTKRYRYKKSQGSPVVQIFQVIAAAFRKRKMELP-QSIVYLYEDNPEGIRIEHTDQFH 298
Query: 183 FLNKACVIKDHEQDIASDGSAI-NRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG- 240
L+KA ++ + + + DG AI N W L +V +VEE+K +++++P+W+T I
Sbjct: 299 LLDKAAIVAEGDFEQTLDGVAIPNPWKLSSVTKVEEVKMMVRLLPIWATTIIFWTTYAQM 358
Query: 241 -SFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVI 299
+F + QA ++ R+I +F++PAGS +V + AILI + +YDR ++P K +GKP
Sbjct: 359 ITFSVEQASTMRRNI---GSFKIPAGSLTVFFVAAILITLAVYDRAIMPFWKKWKGKPGF 415
Query: 300 ISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAG 359
S +R+ IGL + + AA+ E R A + + L +S L PQ L G
Sbjct: 416 -SSLQRIAIGLVLSTAGMAAAALVEQKRLSVA-----KSSSQKTLPISVFLLVPQFFLVG 469
Query: 360 IAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGW 419
E F GQ +F+ + PK M +++ L + +G VSS ++SI++ T + + GW
Sbjct: 470 AGEAFIYTGQLDFFITQSPKGMKTMSTGLFLTTLSLGFFVSSFLVSIVKRVTSTSTDVGW 529
Query: 420 VSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENG 469
++DNIN G D +YW++V ++ +N + Y++C+ + PT + +V KENG
Sbjct: 530 LADNINHGRLDYFYWLLVILSGINFVVYIICALWFKPTKGK-DSVEKENG 578
>At1g69860 putative peptide transporter
Length = 555
Score = 257 bits (657), Expect = 1e-68
Identities = 154/464 (33%), Positives = 261/464 (56%), Gaps = 31/464 (6%)
Query: 1 MSLLWLTAMIPSARPPAC--NHPSEGCESATPGQLAMLFSALILIAIGNGGI-SCSLAFG 57
M +L T+++P+ RPP C + + C + QL +L S L L+++G GGI SCS+ F
Sbjct: 104 MLVLTFTSLVPNLRPPPCTADQITGQCIPYSYSQLYVLLSGLFLLSVGTGGIRSCSVPFS 163
Query: 58 ADQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLI 117
DQ + R FFSWYY TI ++++T ++Y+Q+++ W +GF +P +L
Sbjct: 164 LDQFD-DSTEEGREGSRSFFSWYYTTHTIVQLVSMTLVLYVQNNISWGIGFAIPTVLNFF 222
Query: 118 STVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQ-----QKDS 172
+ +L F+ + YV +K + S+F+G +V VAAYK RK TS YHQ S
Sbjct: 223 ALLLLFVGTRYYVFVKPEGSVFSGVFKVLVAAYKKRKARF---TSGIDYHQPLLETDLQS 279
Query: 173 ELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAI 232
+V TD+ RFLNKA ++ + +D + W CTV Q+E++K+II +IP+++++I
Sbjct: 280 NKLVLTDQFRFLNKAVIVMN------NDEAGNEEWRTCTVRQIEDIKSIISIIPIFASSI 333
Query: 233 T--MSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLA 290
+++N +F + QA +D +S + +P S +VI ++ I IW+ Y+ VL+
Sbjct: 334 IGFLAMNQQQTFTVSQALKMDLQFPGTS-YLIPPASITVISLLNIGIWLPFYETVLVRHI 392
Query: 291 SKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMW 350
I + IS +++GIG F+ ++ + I E RR ++ +GV KMS W
Sbjct: 393 ENITKQNGGISLLQKVGIGNIFSISTMLISGIVERKRRDLSL--------NGV-KMSVFW 443
Query: 351 LAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIEST 410
L PQ L G ++F ++G EF+ K+ P +M S+ SL L + + + +SS ++SI+ S
Sbjct: 444 LTPQQVLMGFYQVFTIVGLTEFFNKQVPINMRSIGNSLLYLGLSLASYLSSAMVSIVHSV 503
Query: 411 TPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
T GG + W++D+I+K D +Y+ I ++ LN +++ C+ Y
Sbjct: 504 TARGG-QSWLTDDIDKSKLDCFYYFIAALSTLNFIFFFWCARRY 546
>At3g45710 putative transporter protein
Length = 556
Score = 253 bits (645), Expect = 2e-67
Identities = 158/460 (34%), Positives = 244/460 (52%), Gaps = 33/460 (7%)
Query: 2 SLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQV 61
SLL L + P C S C+S + QL +L+ AL L+ IG+ G +LA
Sbjct: 104 SLLTLITSLNYLMPRPCETGSILCQSPSKLQLGILYVALALVIIGSAGTRFTLAAAGANQ 163
Query: 62 NRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVL 121
+K R FF+W++ I I T IVY QD+ WK+GFG+ A+ LIS ++
Sbjct: 164 YKKPKEQGR-----FFNWFFLALYIGAITGTTAIVYTQDNASWKLGFGLCAVANLISFIV 218
Query: 122 FFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQ-----QKDSELVV 176
F Y K S +T +V VAA RK + K E YHQ + + +
Sbjct: 219 FIAGVRFYKHDKPLGSPYTSLIRVLVAATMKRKAVISSKD--EDYHQYGLGKEAKTYTTM 276
Query: 177 PTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMS- 235
P+ RFLN+A +K+ E S S+ N W LC+V +VE+ KAI++++PLW+ + +S
Sbjct: 277 PSKSFRFLNRAA-LKNKEDLNTSGDSSNNMWRLCSVQEVEDFKAILRLVPLWAAVMFLST 335
Query: 236 -INIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIR 294
+ + S +LQA +DR + S +FEV AGS VI++V ++I++ + ++ P+ K+
Sbjct: 336 PVAVQMSMTVLQALVMDRKL--SPHFEVSAGSLQVIVLVFGCVFIMLNNWIIYPMYQKLI 393
Query: 295 GKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQ 354
GKP ++P +++GIG F L + +A+ E R K G+ MS +WL P
Sbjct: 394 GKP--LTPLQQVGIGHVFTILSMAISAVVEAKRLKTVENGGH--------PMSVLWLVPA 443
Query: 355 LCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSG 414
L + GI E F+ +Y EFP+S+ + A SL+ + +G+ +S+ V+ +I+ TT
Sbjct: 444 LVMVGIGEAFHFPANVAVFYGEFPESLKNTATSLTSVVIGISFYLSTAVIDVIQRTT--- 500
Query: 415 GNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
W+ ++IN G D YWV+V LNL Y+LVCSW Y
Sbjct: 501 ---SWLPNDINHGRVDNVYWVVVIGGVLNLGYFLVCSWFY 537
>At3g21670 nitrate transporter
Length = 590
Score = 251 bits (640), Expect = 9e-67
Identities = 156/481 (32%), Positives = 261/481 (53%), Gaps = 25/481 (5%)
Query: 3 LLWLTAMIPSARPPACN---HPSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-FGA 58
LL + I S RPP C+ C A QLA+L+ AL IA+G GGI +++ FG+
Sbjct: 117 LLTVATTISSMRPPICDDFRRLHHQCIEANGHQLALLYVALYTIALGGGGIKSNVSGFGS 176
Query: 59 DQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIS 118
DQ + D P + + FF+ +Y ++ + A+ +VY+QD++G G+G+ A M+++
Sbjct: 177 DQFDTSD-PKEEKQMIFFFNRFYFSISVGSLFAVIALVYVQDNVGRGWGYGISAATMVVA 235
Query: 119 TVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPT 178
++ + Y K K S FT +V A+K RK P S D+ V T
Sbjct: 236 AIVLLCGTKRYRFKKPKGSPFTTIWRVGFLAWKKRKESYPAHPS---LLNGYDNTTVPHT 292
Query: 179 DKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SI 236
+ L+ L+KA + K+ + D + W + TV QVEE+K ++K++P+W+T I
Sbjct: 293 EMLKCLDKAAISKNESSPSSKDFEEKDPWIVSTVTQVEEVKLVMKLVPIWATNILFWTIY 352
Query: 237 NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGK 296
+ +F + QA +DR + +F VPAGS+S LI+ IL++ + +RV +PL ++ K
Sbjct: 353 SQMTTFTVEQATFMDRKL---GSFTVPAGSYSAFLILTILLFTSLNERVFVPLTRRLTKK 409
Query: 297 PVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLC 356
P I+ +R+G+GL F+ + AA+ E RR+ A+ ND K+SA WL PQ
Sbjct: 410 PQGITSLQRIGVGLVFSMAAMAVAAVIENARREAAVN----NDK----KISAFWLVPQYF 461
Query: 357 LAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGN 416
L G E F +GQ EF+ +E P+ M S++ L + +G VSSL++S+++ T +
Sbjct: 462 LVGAGEAFAYVGQLEFFIREAPERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVT----D 517
Query: 417 EGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEEST 476
+ W+ N+NK + +YW++V + ALN L ++V + + D ++ V ++ S +E T
Sbjct: 518 KSWLRSNLNKARLNYFYWLLVVLGALNFLIFIVFAMKHQYKADVITVVVTDDDSVEKEVT 577
Query: 477 E 477
+
Sbjct: 578 K 578
>At3g45720 putative transporter protein
Length = 555
Score = 248 bits (634), Expect = 4e-66
Identities = 151/458 (32%), Positives = 244/458 (52%), Gaps = 33/458 (7%)
Query: 3 LLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQVN 62
LL L + + RP C S C+S + L +L++AL L+ G G +LA N
Sbjct: 107 LLTLISSFENLRPRPCETGSILCQSPSKLHLGVLYAALALVTAGTSGTRVALASAG--AN 164
Query: 63 RKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLF 122
+ D P ++ FF+WY+ II+ T IVY Q++ W++GFG+ A LIS ++F
Sbjct: 165 QYDKPRDKGS---FFNWYFLTVNTGAIISATAIVYTQENASWRLGFGLCAAANLISFIVF 221
Query: 123 FLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSE----LVVPT 178
Y K S FT +V VAA K+ + + E YH++ + E + +P+
Sbjct: 222 ISGKRFYKHDKPMGSPFTSLIRVLVAAIL--KIKVVTSSKEEDYHREVEKESKTCIGMPS 279
Query: 179 DKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINI 238
RFLN+A +K + DG N W LC+V++VE+ K++++V+PLW + + +I
Sbjct: 280 KSFRFLNRAA-LKSEKDLNQEDGLCHNPWRLCSVEEVEDFKSVLRVLPLWLAILFVGTSI 338
Query: 239 G--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGK 296
G S +LQA DR + S F+VPAGS VI++++ +++++ + + P+ KI K
Sbjct: 339 GVQASMTVLQALVTDRGL--DSKFKVPAGSLQVIVLISSCVFLVLNNWTIYPIYQKITHK 396
Query: 297 PVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLC 356
++P +++GIG FN L + +AI E R K E MS +WL P L
Sbjct: 397 Q--LTPLQQVGIGQVFNILSMAISAIVEAKRLKTVENEH---------PMSVLWLLPPLV 445
Query: 357 LAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGN 416
+ GI + F+ + +Y EFP+SM + A S++ +A G+ +S+ ++++I+ TT
Sbjct: 446 IVGIGDAFHYMANVAVFYGEFPESMRNTATSVTSVAFGISFYLSTALINLIQRTT----- 500
Query: 417 EGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
W+ D+IN G D YWV+V LNL Y+ VCSW +
Sbjct: 501 -AWLPDDINHGRVDNVYWVLVIGGVLNLGYFFVCSWYF 537
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,966,515
Number of Sequences: 26719
Number of extensions: 508597
Number of successful extensions: 1927
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1589
Number of HSP's gapped (non-prelim): 78
length of query: 526
length of database: 11,318,596
effective HSP length: 104
effective length of query: 422
effective length of database: 8,539,820
effective search space: 3603804040
effective search space used: 3603804040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149489.11


