

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.10 - phase: 0
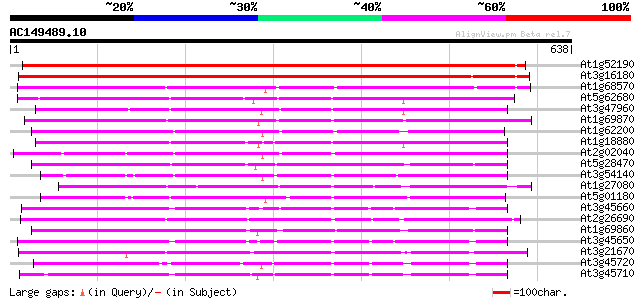
(638 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g52190 peptide transporter like protein 664 0.0
At3g16180 putative transport protein 643 0.0
At1g68570 peptide transporter like 383 e-106
At5g62680 peptide transporter 369 e-102
At3g47960 putative peptide transporter 364 e-101
At1g69870 unknown protein 360 1e-99
At1g62200 unknown protein (At1g62200) 349 2e-96
At1g18880 unknown protein 345 3e-95
At2g02040 histidine transport protein (PTR2-B) 337 2e-92
At5g28470 peptide transporter - like protein 329 3e-90
At3g54140 peptide transport - like protein 323 2e-88
At1g27080 nitrite transporter, putative 318 4e-87
At5g01180 oligopeptide transporter - like protein 317 2e-86
At3g45660 putative protein 314 8e-86
At2g26690 putative nitrate transporter 311 9e-85
At1g69860 putative peptide transporter 310 2e-84
At3g45650 unknown protein 307 1e-83
At3g21670 nitrate transporter 293 3e-79
At3g45720 putative transporter protein 288 5e-78
At3g45710 putative transporter protein 288 6e-78
>At1g52190 peptide transporter like protein
Length = 607
Score = 664 bits (1713), Expect = 0.0
Identities = 332/578 (57%), Positives = 431/578 (74%), Gaps = 7/578 (1%)
Query: 15 EEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQIL 74
E + + + +++KGG++TMPFII NEA K+AS GLLPNMI+YL+ YR + T +L
Sbjct: 9 EAKQIQTNEGKKTKGGIITMPFIIANEAFEKVASYGLLPNMIMYLIRDYRFGVAKGTNVL 68
Query: 75 LLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPS 134
+ SAASNFTP++GAF++DSYLGRFL + I S SFLGM LLWLTAM+P +PS C+ +
Sbjct: 69 FMWSAASNFTPLLGAFLSDSYLGRFLTISIASLSSFLGMVLLWLTAMLPQVKPSPCDPTA 128
Query: 135 EG--CESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSW 191
G C S+T QLA+L+SA LI+IG+GGI CSLAFGADQ++ K+NP N RVLE FF W
Sbjct: 129 AGSHCGSSTASQLALLYSAFALISIGSGGIRPCSLAFGADQLDNKENPKNERVLESFFGW 188
Query: 192 YYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLL 251
YYA +AV+IA TGIVYIQ+HLGW+IGFGVPA LMLI+ +LF LASPLYV SL
Sbjct: 189 YYASSAVAVLIAFTGIVYIQEHLGWKIGFGVPAVLMLIAALLFILASPLYVTRGVTKSLF 248
Query: 252 TGFAQVSVAAYKNRKLSLPPK-TSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIA 310
TG AQ VAAYK RKLSLP S + Y+H KDS++ P+ KLRFLNKAC+I + E++I
Sbjct: 249 TGLAQAIVAAYKKRKLSLPDHHDSFDCYYHMKDSEIKAPSQKLRFLNKACLISNREEEIG 308
Query: 311 SDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG-GSFGLLQAKSLDRHIIS- 368
SDG A+N W LCT D+VEELKA+IKVIP+WST I MSIN SF LLQA S+DR +
Sbjct: 309 SDGFALNPWRLCTTDKVEELKALIKVIPIWSTGIMMSINTSQSSFQLLQATSMDRRLSRH 368
Query: 369 SSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLH 428
S+F+VPAGSF + I+A+ +W+I+YDR +IPLASKIRG+P +S K RMG+GLF +FL
Sbjct: 369 GSSFQVPAGSFGMFTIIALALWVILYDRAVIPLASKIRGRPFRLSVKLRMGLGLFMSFLA 428
Query: 429 LITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKE 488
+ +A+ E+ RRK+AI +GY N+++ V+ +SAMWL PQ L G+AE IGQ EF+Y E
Sbjct: 429 MAISAMVESFRRKKAISQGYANNSNAVVDISAMWLVPQYVLHGLAEALTAIGQTEFFYTE 488
Query: 489 FPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVI 548
FPKSMSS+AASL GL M V +L++S+VL+ + T G E WVSDNINKGH++ YYWV+
Sbjct: 489 FPKSMSSIAASLFGLGMAVASLLASVVLNAVNELTSRNGKESWVSDNINKGHYNYYYWVL 548
Query: 549 VGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEE 586
++ +N++YY++CSW+YGP VD+V N + NG + EE
Sbjct: 549 AIMSFINVIYYVICSWSYGPLVDQVRN-GRVNGVREEE 585
>At3g16180 putative transport protein
Length = 591
Score = 643 bits (1659), Expect = 0.0
Identities = 324/586 (55%), Positives = 425/586 (72%), Gaps = 7/586 (1%)
Query: 11 DAADEEEMVSQQ-KPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGI 69
D + +E + Q +R+KGGL+TMPFII NE K+AS GLL NMILYLM YRL L
Sbjct: 6 DQTESKETLQQPITRRRTKGGLLTMPFIIANEGFEKVASYGLLQNMILYLMSDYRLGLVK 65
Query: 70 STQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSA 129
+L + AA+NF P++GAF++DSYLGRFL + I S S LGM +LWLTAM+P +PS
Sbjct: 66 GQTVLFMWVAATNFMPLVGAFLSDSYLGRFLTIVIASLSSLLGMVVLWLTAMLPQVKPSP 125
Query: 130 CNHPS-EGCESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEI 187
C + C SAT QLA+L++A LI+IG+GGI CSLAFGADQ++ K+NP N RVLE
Sbjct: 126 CVATAGTNCSSATSSQLALLYTAFALISIGSGGIRPCSLAFGADQLDNKENPKNERVLES 185
Query: 188 FFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQR 247
FF WYYA ++AV+IA T IVYIQDHLGW+IGFG+PA LML++ LF ASPLYVK
Sbjct: 186 FFGWYYASSSVAVLIAFTVIVYIQDHLGWKIGFGIPAILMLLAGFLFVFASPLYVKRDVS 245
Query: 248 TSLLTGFAQVSVAAYKNRKLSLPPK-TSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHE 306
SL TG AQV AAY R L+LP S + Y+ KDS+L P+DKLRFLNKAC I + +
Sbjct: 246 KSLFTGLAQVVAAAYVKRNLTLPDHHDSRDCYYRLKDSELKAPSDKLRFLNKACAISNRD 305
Query: 307 QDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG-GSFGLLQAKSLDRH 365
+D+ SDG A+N+W LCT DQVE+LKA++KVIP+WST I MSIN+ SF LLQAKS+DR
Sbjct: 306 EDLGSDGLALNQWRLCTTDQVEKLKALVKVIPVWSTGIMMSINVSQNSFQLLQAKSMDRR 365
Query: 366 IISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFN 425
+ S+S F++PAGSF + I+A++ W+++YDR ++PLASKIRG+PV ++ K RMG+GLF +
Sbjct: 366 LSSNSTFQIPAGSFGMFTIIALISWVVLYDRAILPLASKIRGRPVRVNVKIRMGLGLFIS 425
Query: 426 FLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFY 485
FL + +A E RRK AI +G ND + + +SAMWL PQ L G+AE IGQ EF+
Sbjct: 426 FLAMAVSATVEHYRRKTAISQGLANDANSTVSISAMWLVPQYVLHGLAEALTGIGQTEFF 485
Query: 486 YKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYY 545
Y EFPKSMSS+AASL GL M V N+++S++L+ +++++ GN W+ DNINKGH+D YY
Sbjct: 486 YTEFPKSMSSIAASLFGLGMAVANILASVILNAVKNSSKQ-GNVSWIEDNINKGHYDYYY 544
Query: 546 WVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEESTEFK 591
WV+ ++ +N++YY+VCSW+YGPTVD+V N K NG + EE K
Sbjct: 545 WVLAILSFVNVIYYVVCSWSYGPTVDQVRN-DKVNGMRKEEEEVIK 589
>At1g68570 peptide transporter like
Length = 596
Score = 383 bits (984), Expect = e-106
Identities = 229/595 (38%), Positives = 344/595 (57%), Gaps = 20/595 (3%)
Query: 9 SVDAADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLG 68
S + EEE +P R KGGL+TMPFI NE K+A VG NMI YL L L
Sbjct: 5 SKNKISEEEKQLHGRPNRPKGGLITMPFIFANEICEKLAVVGFHANMISYLTTQLHLPLT 64
Query: 69 ISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPS 128
+ L + S+ TP++GAFIADS+ GRF + S I +GM+LL ++A+IP RP
Sbjct: 65 KAANTLTNFAGTSSLTPLLGAFIADSFAGRFWTITFASIIYQIGMTLLTISAIIPTLRPP 124
Query: 129 ACNHPSEGCESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEI 187
C E C A QL++L+ AL+L A+G+GGI C +AFGADQ + D PN
Sbjct: 125 PCKG-EEVCVVADTAQLSILYVALLLGALGSGGIRPCVVAFGADQFDESD-PNQTTKTWN 182
Query: 188 FFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQR 247
+F+WYY + AV++A+T +V+IQD++GW +G G+P M +S + F LY +
Sbjct: 183 YFNWYYFCMGAAVLLAVTVLVWIQDNVGWGLGLGIPTVAMFLSVIAFVGGFQLYRHLVPA 242
Query: 248 TSLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIP-----TDKLRFLNKACVI 302
S T QV VAA++ RKL + S +++ + D+ + + T + FL+KA ++
Sbjct: 243 GSPFTRLIQVGVAAFRKRKLRMVSDPSLLYFNDEIDAPISLGGKLTHTKHMSFLDKAAIV 302
Query: 303 KDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG--GSFGLLQAK 360
E+D G N W L TV +VEELK++I++ P+ ++ I + G+F L QAK
Sbjct: 303 T--EEDNLKPGQIPNHWRLSTVHRVEELKSVIRMGPIGASGILLITAYAQQGTFSLQQAK 360
Query: 361 SLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGI 420
+++RH+ +S F++PAGS SV VA+L II YDRV + +A K G I+ RMGI
Sbjct: 361 TMNRHLTNS--FQIPAGSMSVFTTVAMLTTIIFYDRVFVKVARKFTGLERGITFLHRMGI 418
Query: 421 GLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIG 480
G + + + A E R+ AI+ G L+ H ++ +S +WL PQ L G+AE F IG
Sbjct: 419 GFVISIIATLVAGFVEVKRKSVAIEHGLLDKPHTIVPISFLWLIPQYGLHGVAEAFMSIG 478
Query: 481 QNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIE--STTPSGGNEGWVSD-NIN 537
EF+Y + P+SM S A +L +A+ +GN VS+L+++++ S P G N W+ D N+N
Sbjct: 479 HLEFFYDQAPESMRSTATALFWMAISIGNYVSTLLVTLVHKFSAKPDGSN--WLPDNNLN 536
Query: 538 KGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEESTEFKH 592
+G + +YW+I + A+NL+YYL C+ Y +V + SKE+ S V+E + +
Sbjct: 537 RGRLEYFYWLITVLQAVNLVYYLWCAKIYTYKPVQVHH-SKEDSSPVKEELQLSN 590
>At5g62680 peptide transporter
Length = 616
Score = 369 bits (947), Expect = e-102
Identities = 217/581 (37%), Positives = 325/581 (55%), Gaps = 21/581 (3%)
Query: 9 SVDAADE-EEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHL 67
SV A D EE V QK +G V MPFIIGNE K+ +G L N+++YL + L
Sbjct: 25 SVTAVDSVEEDVQNQKKVVYRGWKV-MPFIIGNETFEKLGIIGTLSNLLVYLTAVFNLKS 83
Query: 68 GISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARP 127
+ I+ S NF + AF+ D+Y GR+ + + FLG ++ LTA +P P
Sbjct: 84 ITAATIINAFSGTINFGTFVAAFLCDTYFGRYKTLSVAVIACFLGSFVILLTAAVPQLHP 143
Query: 128 SACNHPSEG-CESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVL 185
+AC ++ C + GQ+A L L + +G GGI C+LAFGADQ N K R +
Sbjct: 144 AACGTAADSICNGPSGGQIAFLLMGLGFLVVGAGGIRPCNLAFGADQFNPKSESGK-RGI 202
Query: 186 EIFFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKIT 245
+ FF+WY+ T A I++LT +VY+Q ++ W IG +PA LM ++ ++FF LYVKI
Sbjct: 203 DSFFNWYFFTFTFAQILSLTLVVYVQSNVSWTIGLTIPAVLMFLACLIFFAGDKLYVKIK 262
Query: 246 QRTSLLTGFAQVSVAAYKNRKLSLPPKTSP-----EFYHHKKDSDLVIPTDKLRFLNKAC 300
S L G AQV A K R L P P +Y K + + TD+ RFL+KA
Sbjct: 263 ASGSPLAGIAQVIAVAIKKR--GLKPAKQPWLNLYNYYPPKYANSKLKYTDQFRFLDKAA 320
Query: 301 VIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLW--STAITMSINIGGSFGLLQ 358
++ E + DG + W LCT+ QVEE+K I++V+P+W S+ ++I ++ + Q
Sbjct: 321 ILTP-EDKLQPDGKPADPWKLCTMQQVEEVKCIVRVLPIWFASSIYYLTITQQMTYPVFQ 379
Query: 359 AKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRM 418
A DR + S F +PA ++ V L+ + ++I++YDRVL+P +I G I+ +R+
Sbjct: 380 ALQSDRRL-GSGGFVIPAATYVVFLMTGMTVFIVVYDRVLVPTMRRITGLDTGITLLQRI 438
Query: 419 GIGLFFNFLHLITAAIFETVRRKEAIKE---GYLNDTHGVLKMSAMWLAPQLCLAGIAEM 475
G G+FF L+ A E RR A+ + G + MSAMWL PQL LAG+AE
Sbjct: 439 GTGIFFATASLVVAGFVEERRRTFALTKPTLGMAPRKGEISSMSAMWLIPQLSLAGVAEA 498
Query: 476 FNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDN 535
F IGQ EFYYK+FP++M S A S+ + GV + + S +++ + TT + W++++
Sbjct: 499 FAAIGQMEFYYKQFPENMRSFAGSIFYVGGGVSSYLGSFLIATVHRTTQNSSGGNWLAED 558
Query: 536 INKGHFDKYYWVIVGINALNLLYYLVCS--WAYGPTVDEVS 574
+NKG D +Y++I GI A+N Y+LV S + Y + DEV+
Sbjct: 559 LNKGRLDLFYFMIAGILAVNFAYFLVMSRWYRYKGSDDEVT 599
>At3g47960 putative peptide transporter
Length = 636
Score = 364 bits (935), Expect = e-101
Identities = 205/546 (37%), Positives = 310/546 (56%), Gaps = 13/546 (2%)
Query: 30 GLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPVIGA 89
G MPFIIGNE K+ +G L N+++YL + L + I+ S NF I A
Sbjct: 64 GWKVMPFIIGNETFEKLGIIGTLSNLLVYLTSVFNLKSYTAATIINAFSGTINFGTFIAA 123
Query: 90 FIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQLAMLF 149
F+ D+Y GR+ + + FLG ++ LTA IP+ P AC + CE + GQ+ L
Sbjct: 124 FLCDTYFGRYKTLSVAVIACFLGSFVILLTAAIPSLHPVACGNKIS-CEGPSVGQILFLL 182
Query: 150 SALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGIV 208
L + +G GGI C+LAFGADQ N K + + FF+WY+ T A II+LT +V
Sbjct: 183 MGLGFLVVGAGGIRPCNLAFGADQFNPKSESGK-KGINSFFNWYFFTFTFAQIISLTAVV 241
Query: 209 YIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKLS 268
YIQ ++ W IG +P ALM ++ V+FF LYVK+ S L G A+V AA K R L
Sbjct: 242 YIQSNVSWTIGLIIPVALMFLACVIFFAGDRLYVKVKASGSPLAGIARVIAAAIKKRGLK 301
Query: 269 LPPKTSPEFYHHKKDS---DLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVD 325
+ Y+H + + TD+ RFL+KA ++ E+ + SDG+A + W LCT+
Sbjct: 302 PVKQPWVNLYNHIPSNYANTTLKYTDQFRFLDKAAIMTPEEK-LNSDGTASDPWKLCTLQ 360
Query: 326 QVEELKAIIKVIPLW--STAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVIL 383
QVEE+K I++VIP+W ST ++I I ++ + QA DR + S F +PA ++ V L
Sbjct: 361 QVEEVKCIVRVIPIWFASTIYYLAITIQMTYPVFQALQSDRRL-GSGGFRIPAATYVVFL 419
Query: 384 IVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEA 443
+ + ++II YDRVL+P ++ G IS +R+G G F + L+ + E RR A
Sbjct: 420 MTGMTVFIIFYDRVLVPSLRRVTGLETGISLLQRIGAGFTFAIMSLLVSGFIEERRRNFA 479
Query: 444 IKE---GYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASL 500
+ + G T + MSA+WL PQL LAGIAE F IGQ EFYYK+FP++M S A S+
Sbjct: 480 LTKPTLGMAPRTGEISSMSALWLIPQLTLAGIAEAFAAIGQMEFYYKQFPENMKSFAGSI 539
Query: 501 SGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYL 560
+ GV + ++S ++S + TT + W+++++NK D +Y+++ G+ +N+ Y+L
Sbjct: 540 FYVGAGVSSYLASFLISTVHRTTAHSPSGNWLAEDLNKAKLDYFYFMLTGLMVVNMAYFL 599
Query: 561 VCSWAY 566
+ + Y
Sbjct: 600 LMARWY 605
>At1g69870 unknown protein
Length = 620
Score = 360 bits (924), Expect = 1e-99
Identities = 201/586 (34%), Positives = 329/586 (55%), Gaps = 18/586 (3%)
Query: 18 MVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLS 77
++ +K ++ GG + FI+GNE L ++ S+GLL N ++YL + L + ++ +
Sbjct: 42 ILDAEKVEKKPGGWRAVSFILGNETLERLGSIGLLANFMVYLTKVFHLEQVDAANVINIW 101
Query: 78 SAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSE-G 136
S +N TP++GA+I+D+Y+GRF + S + LG+ + LTA P P++CN
Sbjct: 102 SGFTNLTPLVGAYISDTYVGRFKTIAFASFATLLGLITITLTASFPQLHPASCNSQDPLS 161
Query: 137 CESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSWYYAF 195
C Q+ +L L +++G+GGI CS+ FG DQ +++ + + FF+WYY
Sbjct: 162 CGGPNKLQIGVLLLGLCFLSVGSGGIRPCSIPFGVDQFDQRTE-EGVKGVASFFNWYYMT 220
Query: 196 ITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFA 255
T+ +II T +VYIQD + W IGF +P LM ++ V+FF YV + S+ +G A
Sbjct: 221 FTVVLIITQTVVVYIQDQVSWIIGFSIPTGLMALAVVMFFAGMKRYVYVKPEGSIFSGIA 280
Query: 256 QVSVAAYKNRKLSLPPKTSPEFYHHK---KDSDL--VIPTDKLRFLNKACVIKDHEQDIA 310
QV VAA K RKL LP + ++ K S L + +++ R L+KA V+ E D+
Sbjct: 281 QVIVAARKKRKLKLPAEDDGTVTYYDPAIKSSVLSKLHRSNQFRCLDKAAVVI--EGDLT 338
Query: 311 SDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SINIGGSFGLLQAKSLDRHIIS 368
+G ++W LC+V +VEE+K +I+++P+WS I ++ G+F + QA +DR++
Sbjct: 339 PEGPPADKWRLCSVQEVEEVKCLIRIVPIWSAGIISLAAMTTQGTFTVSQALKMDRNL-- 396
Query: 369 SSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLH 428
FE+PAGS SVI ++ I I++ YDRV +P +I G I+ +R+G G+ F
Sbjct: 397 GPKFEIPAGSLSVISLLTIGIFLPFYDRVFVPFMRRITGHKSGITLLQRIGTGIVFAIFS 456
Query: 429 LITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKE 488
+I A I E +RR +I G D G+ MS WL+PQL L G+ E FN+IGQ EF+ +
Sbjct: 457 MIVAGIVERMRRIRSINAG---DPTGMTPMSVFWLSPQLILMGLCEAFNIIGQIEFFNSQ 513
Query: 489 FPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVI 548
FP+ M S+A SL L+ + +SS +++++ + W++ N+N G D +Y++I
Sbjct: 514 FPEHMRSIANSLFSLSFAGSSYLSSFLVTVVHKFSGGHDRPDWLNKNLNAGKLDYFYYLI 573
Query: 549 VGINALNLLYYLVCSWAYGPTVD-EVSNVSKENGSKVEESTEFKHM 593
+ +NL+Y+ C+ Y V + + ++ S E T K M
Sbjct: 574 AVLGVVNLVYFWYCARGYRYKVGLPIEDFEEDKSSDDVEMTSKKSM 619
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 349 bits (896), Expect = 2e-96
Identities = 207/548 (37%), Positives = 312/548 (56%), Gaps = 26/548 (4%)
Query: 25 QRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFT 84
++ G PFI+GNE ++A G+ N+I Y + +++ T
Sbjct: 50 KKKTGNWKACPFILGNECCERLAYYGIAKNLITYYTSELHESNVSAASDVMIWQGTCYIT 109
Query: 85 PVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEG-CESATPG 143
P+IGA IADSY GR+ + S+I F+GM+LL L+A +P +P+AC + C AT
Sbjct: 110 PLIGAVIADSYWGRYWTIASFSAIYFIGMALLTLSASLPVLKPAACAGVAAALCSPATTV 169
Query: 144 QLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVII 202
Q A+ F+ L LIA+G GGI C +FGADQ + D R FF+W+Y I I I
Sbjct: 170 QYAVFFTGLYLIALGTGGIKPCVSSFGADQFDDTDPRERVRKAS-FFNWFYFSINIGSFI 228
Query: 203 ALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAY 262
+ T +V++Q+++GW +GF +P M +S FF+ +PLY S +T QV VAAY
Sbjct: 229 SSTLLVWVQENVGWGLGFLIPTVFMGVSIASFFIGTPLYRFQKPGGSPITRVCQVLVAAY 288
Query: 263 KNRKLSLPPKTSPEFYHHKKDSDL-----VIPTDKLRFLNKACVIKDHEQDIASDGSAIN 317
+ KL+LP S + +K+S + + TD +FL+KA VI ++E + G+ N
Sbjct: 289 RKLKLNLPEDISFLYETREKNSMIAGSRKIQHTDGYKFLDKAAVISEYE---SKSGAFSN 345
Query: 318 RWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVP 375
W LCTV QVEE+K +I++ P+W++ I S+ + + Q +S++R I S FE+P
Sbjct: 346 PWKLCTVTQVEEVKTLIRMFPIWASGIVYSVLYSQISTLFVQQGRSMNRIIRS---FEIP 402
Query: 376 AGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIF 435
SF V + +LI I IYDR L+P + G P ++ +RMGIGLF + L + AAI
Sbjct: 403 PASFGVFDTLIVLISIPIYDRFLVPFVRRFTGIPKGLTDLQRMGIGLFLSVLSIAAAAIV 462
Query: 436 ETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSS 495
ETVR + A + MS W PQ L GIAE+F IG+ EF+Y E P +M S
Sbjct: 463 ETVRLQLA---------QDFVAMSIFWQIPQYILMGIAEVFFFIGRVEFFYDESPDAMRS 513
Query: 496 VAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALN 555
V ++L+ L VG+ +SSL+L+++ T GG +GWV D++NKGH D ++W++V + +N
Sbjct: 514 VCSALALLNTAVGSYLSSLILTLVAYFTALGGKDGWVPDDLNKGHLDYFFWLLVSLGLVN 573
Query: 556 L-LYYLVC 562
+ +Y L+C
Sbjct: 574 IPVYALIC 581
>At1g18880 unknown protein
Length = 587
Score = 345 bits (886), Expect = 3e-95
Identities = 197/548 (35%), Positives = 310/548 (55%), Gaps = 17/548 (3%)
Query: 30 GLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPVIGA 89
G MPFIIGNE K+ VG N+++YL + + + +++ + SNF ++ A
Sbjct: 22 GWKVMPFIIGNETFEKLGIVGSSSNLVIYLTTVFNMKSITAAKVVNIYGGTSNFGTIVAA 81
Query: 90 FIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSAC-NHPSEGCESATPGQLAML 148
F+ DSY GR+ + FLG + LTA+I P+ C C + GQ+ L
Sbjct: 82 FLCDSYFGRYKTLSFAMIACFLGSVAMDLTAVIHPLHPAQCAKEIGSVCNGPSIGQIMFL 141
Query: 149 FSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGI 207
A++L+ IG GGI C+L FGADQ + K R +E FF+WY+ T A +++LT I
Sbjct: 142 AGAMVLLVIGAGGIRPCNLPFGADQFDPKTKEGK-RGIESFFNWYFFTFTFAQMVSLTLI 200
Query: 208 VYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKL 267
VY+Q ++ W IG +PA LML+ ++FF S LYVK+ S + +V V A K R+L
Sbjct: 201 VYVQSNVSWSIGLAIPAILMLLGCIIFFAGSKLYVKVKASGSPIHSITRVIVVAIKKRRL 260
Query: 268 SLPPKTSPEFYHHK----KDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCT 323
P E Y++ K+S L T++ RFL+K+ I+ + + DGS ++ W LC+
Sbjct: 261 K--PVGPNELYNYIASDFKNSKLG-HTEQFRFLDKSA-IQTQDDKLNKDGSPVDAWKLCS 316
Query: 324 VDQVEELKAIIKVIPLWSTAITMSINI--GGSFGLLQAKSLDRHIISSSNFEVPAGSFSV 381
+ QVEE+K +I+V+P+W +A + ++ + Q+ DR + +F++PAGS++V
Sbjct: 317 MQQVEEVKCVIRVLPVWLSAALFYLAYIQQTTYTIFQSLQSDRRL-GPGSFQIPAGSYTV 375
Query: 382 ILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRK 441
L++ + I+I IYDRVL+P K G+ I+ +R+G GLF ++ +AI E RRK
Sbjct: 376 FLMLGMTIFIPIYDRVLVPFLRKYTGRDGGITQLQRVGAGLFLCITSMMVSAIVEQYRRK 435
Query: 442 EAIKE---GYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAA 498
A+ + G + MS MWL PQL L GIA+ +GQ EFYYK+FP++M S A
Sbjct: 436 VALTKPTLGLAPRKGAISSMSGMWLIPQLVLMGIADALAGVGQMEFYYKQFPENMRSFAG 495
Query: 499 SLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLY 558
SL +G+ + +S+ +LS + TT W+ +++NKG + +Y+++ G+ LNL Y
Sbjct: 496 SLYYCGIGLASYLSTFLLSAVHDTTEGFSGGSWLPEDLNKGRLEYFYFLVAGMMTLNLAY 555
Query: 559 YLVCSWAY 566
+L+ S Y
Sbjct: 556 FLLVSHWY 563
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 337 bits (863), Expect = 2e-92
Identities = 198/573 (34%), Positives = 318/573 (54%), Gaps = 23/573 (4%)
Query: 5 LQLCSVDAADEEEMVSQQKPQRSK-GGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSY 63
LQ + A D + P + K G PFI+GNE ++A G+ N+I YL +
Sbjct: 18 LQEVKLYAEDGSVDFNGNPPLKEKTGNWKACPFILGNECCERLAYYGIAGNLITYL--TT 75
Query: 64 RLHLGISTQILLLSS--AASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAM 121
+LH G + +++ TP+IGA +AD+Y GR+ + S I F+GMS L L+A
Sbjct: 76 KLHQGNVSAATNVTTWQGTCYLTPLIGAVLADAYWGRYWTIACFSGIYFIGMSALTLSAS 135
Query: 122 IPAARPSACNHPSEGCESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPN 180
+PA +P+ C + C SATP Q AM F L LIA+G GGI C +FGADQ + D+
Sbjct: 136 VPALKPAECI--GDFCPSATPAQYAMFFGGLYLIALGTGGIKPCVSSFGADQFDDTDSRE 193
Query: 181 NYRVLEIFFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPL 240
R FF+W+Y I I +++ + +V+IQ++ GW +GFG+P M ++ FF +PL
Sbjct: 194 RVRKAS-FFNWFYFSINIGALVSSSLLVWIQENRGWGLGFGIPTVFMGLAIASFFFGTPL 252
Query: 241 YVKITQRTSLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDL-----VIPTDKLRF 295
Y S +T +QV VA+++ + +P + + K+S + + TD ++
Sbjct: 253 YRFQKPGGSPITRISQVVVASFRKSSVKVPEDATLLYETQDKNSAIAGSRKIEHTDDCQY 312
Query: 296 LNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG--GS 353
L+KA VI + E + G N W LCTV QVEELK +I++ P+W++ I S +
Sbjct: 313 LDKAAVISEEE---SKSGDYSNSWRLCTVTQVEELKILIRMFPIWASGIIFSAVYAQMST 369
Query: 354 FGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIIS 413
+ Q ++++ I +F++P + +++IW+ +YDR ++PLA K G +
Sbjct: 370 MFVQQGRAMNCKI---GSFQLPPAALGTFDTASVIIWVPLYDRFIVPLARKFTGVDKGFT 426
Query: 414 PKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIA 473
+RMGIGLF + L + AAI E +R A G L ++ + +S +W PQ + G A
Sbjct: 427 EIQRMGIGLFVSVLCMAAAAIVEIIRLHMANDLG-LVESGAPVPISVLWQIPQYFILGAA 485
Query: 474 EMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVS 533
E+F IGQ EF+Y + P +M S+ ++L+ L +GN +SSL+L+++ T G EGW+S
Sbjct: 486 EVFYFIGQLEFFYDQSPDAMRSLCSALALLTNALGNYLSSLILTLVTYFTTRNGQEGWIS 545
Query: 534 DNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 566
DN+N GH D ++W++ G++ +N+ Y + Y
Sbjct: 546 DNLNSGHLDYFFWLLAGLSLVNMAVYFFSAARY 578
>At5g28470 peptide transporter - like protein
Length = 561
Score = 329 bits (843), Expect = 3e-90
Identities = 187/550 (34%), Positives = 307/550 (55%), Gaps = 19/550 (3%)
Query: 25 QRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFT 84
++ KGG + +II NE+ K+AS+ L+ N+ +YLM Y L ++ + + N
Sbjct: 15 KKEKGGWRAIKYIIANESFEKLASMSLIGNLSVYLMTKYNLGGVFLVNVINIWFGSCNIL 74
Query: 85 PVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQ 144
+ GAF++D+YLGRF + +GS SF+GM + LTA +P+ RP AC PS Q
Sbjct: 75 TLAGAFVSDAYLGRFWTLLLGSIASFIGMGIFALTAALPSLRPDACIDPSNCSNQPAKWQ 134
Query: 145 LAMLFSALILIAIGNGGI-TCSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIA 203
L +LFS L L+AIG GG+ C++AFGADQ + LE FF+W+Y T+A++IA
Sbjct: 135 LGVLFSGLGLLAIGAGGVRPCNIAFGADQFDTSTKKGKAH-LETFFNWWYFSFTVALVIA 193
Query: 204 LTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYK 263
LTG+VYIQ ++ W IGF +P A + +S F + Y+ S+ +V AA K
Sbjct: 194 LTGVVYIQTNISWVIGFVIPTACLALSITTFVIGQHTYICAKAEGSVFADIVKVVTAACK 253
Query: 264 NRKLSLPPKTSPEFY----HHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRW 319
RK+ P + FY + + LV +LRF +KA ++ + ++ DG+A +W
Sbjct: 254 KRKVK--PGSDITFYIGPSNDGSPTTLVRDKHRLRFFDKASIV-TNPNELNEDGNAKYKW 310
Query: 320 SLCTVDQVEELKAIIKVIPLWSTAITMSI--NIGGSFGLLQAKSLDRHIISSSNFEVPAG 377
LC+V QV+ LK + ++P+W T I I + +G+LQA +D+ NF+VPAG
Sbjct: 311 RLCSVQQVKNLKCVTAILPVWVTGIACFILTDQQNIYGILQAMQMDK-TFGPHNFQVPAG 369
Query: 378 SFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFET 437
+++ ++ + IWI +Y+ V+IP+ +I G+ ++ K R+ IG+ + +I A E
Sbjct: 370 WMNLVSMITLAIWISLYECVIIPIVKQITGRKKRLTLKHRIEIGIVMGIICMIVAGFQEK 429
Query: 438 VRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVA 497
RR A+K G V +S + L PQ LAG+ E F+ + EF P+ M +VA
Sbjct: 430 KRRASALKNGSF-----VSPVSIVMLLPQFALAGLTEAFSAVALMEFLTVRMPEHMRAVA 484
Query: 498 ASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSD-NINKGHFDKYYWVIVGINALNL 556
++ L+ + + + +L++++I++ T G + W+ D ++NK + Y+++I GI NL
Sbjct: 485 GAIFFLSSSIASYICTLLINVIDAVTRKEG-KSWLGDKDLNKNRLENYFFIIAGIQVANL 543
Query: 557 LYYLVCSWAY 566
LY+ + + Y
Sbjct: 544 LYFRLFASRY 553
>At3g54140 peptide transport - like protein
Length = 570
Score = 323 bits (828), Expect = 2e-88
Identities = 196/541 (36%), Positives = 303/541 (55%), Gaps = 23/541 (4%)
Query: 36 FIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLS--SAASNFTPVIGAFIAD 93
FI+GNE ++A G+ N++ YL RL+ G +T ++ S TP+IGAFIAD
Sbjct: 33 FILGNECCERLAYYGMGTNLVNYLES--RLNQGNATAANNVTNWSGTCYITPLIGAFIAD 90
Query: 94 SYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQLAMLFSALI 153
+YLGR+ + I GM+LL L+A +P +P CN ++ C + Q A+ F AL
Sbjct: 91 AYLGRYWTIATFVFIYVSGMTLLTLSASVPGLKPGNCN--ADTCHPNS-SQTAVFFVALY 147
Query: 154 LIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGIVYIQD 212
+IA+G GGI C +FGADQ + D + FF+W+Y I + +IA T +V+IQ
Sbjct: 148 MIALGTGGIKPCVSSFGADQFDENDENEKIKKSS-FFNWFYFSINVGALIAATVLVWIQM 206
Query: 213 HLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKLSLPPK 272
++GW GFGVP M+I+ FF S Y S LT QV VAA++ + +P
Sbjct: 207 NVGWGWGFGVPTVAMVIAVCFFFFGSRFYRLQRPGGSPLTRIFQVIVAAFRKISVKVPED 266
Query: 273 TSPEFYHHKKDSDL-----VIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQV 327
S F +S++ ++ TD L+F +KA V + + D DG +N W LC+V QV
Sbjct: 267 KSLLFETADDESNIKGSRKLVHTDNLKFFDKAAV--ESQSDSIKDGE-VNPWRLCSVTQV 323
Query: 328 EELKAIIKVIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIV 385
EELK+II ++P+W+T I + + +LQ ++D+H+ NFE+P+ S S+ V
Sbjct: 324 EELKSIITLLPVWATGIVFATVYSQMSTMFVLQGNTMDQHM--GKNFEIPSASLSLFDTV 381
Query: 386 AILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIK 445
++L W +YD+ +IPLA K + +RMGIGL + +ITA + E VR + +K
Sbjct: 382 SVLFWTPVYDQFIIPLARKFTRNERGFTQLQRMGIGLVVSIFAMITAGVLEVVRL-DYVK 440
Query: 446 EGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAM 505
D + MS W PQ L G AE+F IGQ EF+Y + P +M S+ ++LS +
Sbjct: 441 THNAYDQKQI-HMSIFWQIPQYLLIGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTTV 499
Query: 506 GVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWA 565
+GN +S+++++++ T G GW+ DN+N+GH D +++++ ++ LN L YL S
Sbjct: 500 ALGNYLSTVLVTVVMKITKKNGKPGWIPDNLNRGHLDYFFYLLATLSFLNFLVYLWISKR 559
Query: 566 Y 566
Y
Sbjct: 560 Y 560
>At1g27080 nitrite transporter, putative
Length = 534
Score = 318 bits (816), Expect = 4e-87
Identities = 187/547 (34%), Positives = 308/547 (56%), Gaps = 36/547 (6%)
Query: 56 ILYLMGSYRLHLGISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSL 115
+LYL + + + + L +NF P++GA I+D+Y+GRF + S S LG+
Sbjct: 1 MLYLRNVFHMEPVEAFNVYYLWMGLTNFAPLLGALISDAYIGRFKTIAYASLFSILGLMT 60
Query: 116 LWLTAMIPAARPSACNHPS-EGCESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQV 173
+ LTA +P P CN+P + C+ QL +LF L ++IG+GGI CS+ FG DQ
Sbjct: 61 VTLTACLPQLHPPPCNNPHPDECDDPNKLQLGILFLGLGFLSIGSGGIRPCSIPFGVDQF 120
Query: 174 NRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVL 233
+++ + + FF+WYY +T+ +I + T +VY+Q + W IGF +P +LM + VL
Sbjct: 121 DQRTE-QGLKGVASFFNWYYLTLTMVLIFSHTVVVYLQT-VSWVIGFSIPTSLMACAVVL 178
Query: 234 FFLASPLYVKITQRTSLLTGFAQVSVAAYKNR--KLSLPPKTSPEFYHHKKDSDLV--IP 289
FF+ YV + S+ +G A+V VAA K R K+SL + E+Y ++ +P
Sbjct: 179 FFVGMRFYVYVKPEGSVFSGIARVIVAARKKRDLKISLVDDGTEEYYEPPVKPGVLSKLP 238
Query: 290 -TDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAIT--M 346
TD+ +FL+KA VI D D+ S+G N+W LC++ +VEE+K +I+V+P+WS I +
Sbjct: 239 LTDQFKFLDKAAVILDG--DLTSEGVPANKWRLCSIQEVEEVKCLIRVVPVWSAGIISIV 296
Query: 347 SINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIR 406
++ +F + QA +DRH+ +FE+PA S +VI + I IW+ IY+ +L+P ++R
Sbjct: 297 AMTTQATFMVFQATKMDRHM--GPHFEIPAASITVISYITIGIWVPIYEHLLVPFLWRMR 354
Query: 407 GKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQ 466
V + +RMGIG+ F L + TA E VRR A + + +MS WLA
Sbjct: 355 KFRVTLL--QRMGIGIVFAILSMFTAGFVEGVRRTRATE---------MTQMSVFWLALP 403
Query: 467 LCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSG 526
L L G+ E FN IG EF+ +FP+ M S+A SL L+ N +SSL+++ + + +
Sbjct: 404 LILMGLCESFNFIGLIEFFNSQFPEHMRSIANSLFPLSFAAANYLSSLLVTTVHKVSGTK 463
Query: 527 GNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEE 586
+ W++ ++++G D +Y++I + +NL+Y+ C+ Y + GS++E+
Sbjct: 464 DHPDWLNKDLDRGKLDYFYYLIAVLGVVNLVYFWYCAHRY----------QYKAGSQIED 513
Query: 587 STEFKHM 593
E K +
Sbjct: 514 FNEEKSL 520
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 317 bits (811), Expect = 2e-86
Identities = 187/538 (34%), Positives = 293/538 (53%), Gaps = 21/538 (3%)
Query: 36 FIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPVIGAFIADSY 95
FI+G E ++A G+ N+I YL + +++ + S TP+IGAFIAD+Y
Sbjct: 34 FILGTECCERLAYYGMSTNLINYLEKQMNMENVSASKSVSNWSGTCYATPLIGAFIADAY 93
Query: 96 LGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQLAMLFSALILI 155
LGR+ + I GM+LL ++A +P P+ E C AT GQ A+ F AL LI
Sbjct: 94 LGRYWTIASFVVIYIAGMTLLTISASVPGLTPTCSG---ETCH-ATAGQTAITFIALYLI 149
Query: 156 AIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGIVYIQDHL 214
A+G GGI C +FGADQ + D FF+W+Y I + +IA + +V+IQ ++
Sbjct: 150 ALGTGGIKPCVSSFGADQFDDTDEKEK-ESKSSFFNWFYFVINVGAMIASSVLVWIQMNV 208
Query: 215 GWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKLSLPPKTS 274
GW G GVP M I+ V FF S Y S LT QV VA+ + K+ +P S
Sbjct: 209 GWGWGLGVPTVAMAIAVVFFFAGSNFYRLQKPGGSPLTRMLQVIVASCRKSKVKIPEDES 268
Query: 275 PEFYHHKKDSDLVIP-----TDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEE 329
+ + +S ++ T L F +KA V + + A+ S+ W LCTV QVEE
Sbjct: 269 LLYENQDAESSIIGSRKLEHTKILTFFDKAAVETESDNKGAAKSSS---WKLCTVTQVEE 325
Query: 330 LKAIIKVIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAI 387
LKA+I+++P+W+T I + G+ +LQ +LD+H+ NF++P+ S S+ +++
Sbjct: 326 LKALIRLLPIWATGIVFASVYSQMGTVFVLQGNTLDQHM--GPNFKIPSASLSLFDTLSV 383
Query: 388 LIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEG 447
L W +YD++++P A K G + +R+GIGL + +++A I E R
Sbjct: 384 LFWAPVYDKLIVPFARKYTGHERGFTQLQRIGIGLVISIFSMVSAGILEVARLNYVQTHN 443
Query: 448 YLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGV 507
N+ + M+ W PQ L G AE+F IGQ EF+Y + P +M S+ ++LS A+
Sbjct: 444 LYNEE--TIPMTIFWQVPQYFLVGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTAIAF 501
Query: 508 GNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYL-VCSW 564
GN +S+ +++++ T SGG GW++ N+N GH D ++W++ G++ LN L YL + W
Sbjct: 502 GNYLSTFLVTLVTKVTRSGGRPGWIAKNLNNGHLDYFFWLLAGLSFLNFLVYLWIAKW 559
>At3g45660 putative protein
Length = 557
Score = 314 bits (805), Expect = 8e-86
Identities = 191/556 (34%), Positives = 301/556 (53%), Gaps = 27/556 (4%)
Query: 14 DEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQI 73
D E + S R GG +T PF+I +A++G L N+I+YL+ Y + + +I
Sbjct: 8 DAEALKSADSSTRRSGGRITFPFMIVTLFGLTLATLGWLQNLIVYLIEEYNMKSIAAAKI 67
Query: 74 LLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHP 133
L + S + P IGA ADS+ G + + S IS +G+ LL LT + + RP AC
Sbjct: 68 LNIFSGFTFMFPAIGAIAADSFFGTIPVILVSSFISLVGVVLLALTTLFDSLRPQACETA 127
Query: 134 SEGCESATPGQLAMLFSALILIAIGNGGITCSLA-FGADQVNRKDNPNNYRVLEIFFSWY 192
S+ C++ T QL +L++A+ L +G GG+ +LA GA+Q + + + FF+W+
Sbjct: 128 SKLCQAPTNIQLGVLYTAITLGCVGAGGLRFTLATAGANQYEKTKDQGS------FFNWF 181
Query: 193 YAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLT 252
+ +A I+ T IVY ++++ W GFG+ A L+ ++F Y S T
Sbjct: 182 FFTWYLAASISATAIVYAEENISWSFGFGLCVAANLLGLIVFISGKKFYKHDKPLGSPFT 241
Query: 253 GFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASD 312
+V AA + RK + T+ + YH + PT RF N+A + +D E + SD
Sbjct: 242 SLLRVIFAAIRKRKAVV--STNEKDYHSESKK---TPTKSFRFFNRAALKQDDE--VNSD 294
Query: 313 GSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGSFGL--LQAKSLDRHIISSS 370
G+ N+W LC+V QVE+ KA+I++IPL + +S I GL LQ +DR +
Sbjct: 295 GTIHNQWRLCSVQQVEDFKAVIRIIPLVLAILFLSTPIAMQLGLTVLQGLVMDRRL--GP 352
Query: 371 NFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLI 430
+F++PAGS VI +++ ++II+ DR L P K+ GK +P +R+GIG FN L +
Sbjct: 353 HFKIPAGSLQVITLLSTCLFIIVNDRFLYPFYQKLTGK--FPTPIQRVGIGHVFNILSMA 410
Query: 431 TAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFP 490
AI E +R + +++G+ + V MS +WL P L + GI E F+ G Y+EFP
Sbjct: 411 VTAIVE-AKRLKIVQKGHFLGSSSVADMSVLWLFPPLVIVGIGEAFHFPGNVALCYQEFP 469
Query: 491 KSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVG 550
+SM S A S++ + +G+ S+ ++ +I+ TT W+ D+IN G D YW++V
Sbjct: 470 ESMRSTATSITSVLIGICFYTSTALIDLIQKTT------AWLPDDINHGRVDNVYWILVI 523
Query: 551 INALNLLYYLVCSWAY 566
LNL Y+LVCSW Y
Sbjct: 524 GGVLNLGYFLVCSWFY 539
>At2g26690 putative nitrate transporter
Length = 586
Score = 311 bits (796), Expect = 9e-85
Identities = 184/573 (32%), Positives = 314/573 (54%), Gaps = 17/573 (2%)
Query: 13 ADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQ 72
AD + + + GG +T I+G E + +++++G+ N++ YLM + L S
Sbjct: 19 ADAVDYKGRPADKSKTGGWITAALILGIEVVERLSTMGIAVNLVTYLMETMHLPSSTSAN 78
Query: 73 ILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNH 132
I+ S ++G F+ADS+LGRF +GI S+I LG L + +P RP C+H
Sbjct: 79 IVTDFMGTSFLLCLLGGFLADSFLGRFKTIGIFSTIQALGTGALAVATKLPELRPPTCHH 138
Query: 133 PSEGCESATPGQLAMLFSALILIAIGNGGITCSLA-FGADQVNRKDNPNNYRVLEIFFSW 191
E C AT Q+ +L+ +L LIA+G GG+ S++ FG+DQ + KD P + FF+
Sbjct: 139 -GEACIPATAFQMTILYVSLYLIALGTGGLKSSISGFGSDQFDDKD-PKEKAHMAFFFNR 196
Query: 192 YYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLL 251
++ FI++ ++A+T +VY+QD +G +G+ M I+ V+F + Y + S +
Sbjct: 197 FFFFISMGTLLAVTVLVYMQDEVGRSWAYGICTVSMAIAIVIFLCGTKRYRYKKSQGSPV 256
Query: 252 TGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIAS 311
QV AA++ RK+ LP ++ Y + + TD+ L+KA ++ + + +
Sbjct: 257 VQIFQVIAAAFRKRKMELP-QSIVYLYEDNPEGIRIEHTDQFHLLDKAAIVAEGDFEQTL 315
Query: 312 DGSAI-NRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG--SFGLLQAKSLDRHIIS 368
DG AI N W L +V +VEE+K +++++P+W+T I +F + QA ++ R+I
Sbjct: 316 DGVAIPNPWKLSSVTKVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQASTMRRNI-- 373
Query: 369 SSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLH 428
+F++PAGS +V + AILI + +YDR ++P K +GKP S +R+ IGL +
Sbjct: 374 -GSFKIPAGSLTVFFVAAILITLAVYDRAIMPFWKKWKGKPGF-SSLQRIAIGLVLSTAG 431
Query: 429 LITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKE 488
+ AA+ E R A + + L +S L PQ L G E F GQ +F+ +
Sbjct: 432 MAAAALVEQKRLSVA-----KSSSQKTLPISVFLLVPQFFLVGAGEAFIYTGQLDFFITQ 486
Query: 489 FPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVI 548
PK M +++ L + +G VSS ++SI++ T + + GW++DNIN G D +YW++
Sbjct: 487 SPKGMKTMSTGLFLTTLSLGFFVSSFLVSIVKRVTSTSTDVGWLADNINHGRLDYFYWLL 546
Query: 549 VGINALNLLYYLVCSWAYGPTVDEVSNVSKENG 581
V ++ +N + Y++C+ + PT + +V KENG
Sbjct: 547 VILSGINFVVYIICALWFKPTKGK-DSVEKENG 578
>At1g69860 putative peptide transporter
Length = 555
Score = 310 bits (794), Expect = 2e-84
Identities = 184/552 (33%), Positives = 308/552 (55%), Gaps = 31/552 (5%)
Query: 25 QRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFT 84
QR G MP+IIGNE L ++A+ GL+ N ++Y++ Y + + ++ SA +NF
Sbjct: 16 QRKPLGWKAMPYIIGNETLERLATFGLMANFMVYMVREYHMDQVQAVTLINTWSALTNFA 75
Query: 85 PVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSAC--NHPSEGCESATP 142
P+IGAFI+DSY G+F + GS LGM +L T+++P RP C + + C +
Sbjct: 76 PIIGAFISDSYTGKFNTIVFGSIAELLGMLVLTFTSLVPNLRPPPCTADQITGQCIPYSY 135
Query: 143 GQLAMLFSALILIAIGNGGI-TCSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVI 201
QL +L S L L+++G GGI +CS+ F DQ + FFSWYY TI +
Sbjct: 136 SQLYVLLSGLFLLSVGTGGIRSCSVPFSLDQFD-DSTEEGREGSRSFFSWYYTTHTIVQL 194
Query: 202 IALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAA 261
+++T ++Y+Q+++ W IGF +P L + +L F+ + YV + S+ +G +V VAA
Sbjct: 195 VSMTLVLYVQNNISWGIGFAIPTVLNFFALLLLFVGTRYYVFVKPEGSVFSGVFKVLVAA 254
Query: 262 YKNRKLSLPPKTSPEFYHH-----KKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAI 316
YK RK TS YH S+ ++ TD+ RFLNKA ++ + +D +
Sbjct: 255 YKKRKARF---TSGIDYHQPLLETDLQSNKLVLTDQFRFLNKAVIVMN------NDEAGN 305
Query: 317 NRWSLCTVDQVEELKAIIKVIPLWSTAIT--MSINIGGSFGLLQAKSLDRHIISSSNFEV 374
W CTV Q+E++K+II +IP+++++I +++N +F + QA +D +S + +
Sbjct: 306 EEWRTCTVRQIEDIKSIISIIPIFASSIIGFLAMNQQQTFTVSQALKMDLQFPGTS-YLI 364
Query: 375 PAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAI 434
P S +VI ++ I IW+ Y+ VL+ I + IS +++GIG F+ ++ + I
Sbjct: 365 PPASITVISLLNIGIWLPFYETVLVRHIENITKQNGGISLLQKVGIGNIFSISTMLISGI 424
Query: 435 FETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMS 494
E RR ++ +GV KMS WL PQ L G ++F ++G EF+ K+ P +M
Sbjct: 425 VERKRRDLSL--------NGV-KMSVFWLTPQQVLMGFYQVFTIVGLTEFFNKQVPINMR 475
Query: 495 SVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINAL 554
S+ SL L + + + +SS ++SI+ S T GG + W++D+I+K D +Y+ I ++ L
Sbjct: 476 SIGNSLLYLGLSLASYLSSAMVSIVHSVTARGG-QSWLTDDIDKSKLDCFYYFIAALSTL 534
Query: 555 NLLYYLVCSWAY 566
N +++ C+ Y
Sbjct: 535 NFIFFFWCARRY 546
>At3g45650 unknown protein
Length = 558
Score = 307 bits (786), Expect = 1e-83
Identities = 187/561 (33%), Positives = 298/561 (52%), Gaps = 27/561 (4%)
Query: 9 SVDAADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLG 68
SV E + + +R GG +T PF+I +A+ G L N+I+YL+ + +
Sbjct: 4 SVTGDAETAISADSSTKRRGGGWITFPFMIATLLGLTIAAWGWLLNLIVYLIEEFNVKSI 63
Query: 69 ISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPS 128
+ QI + S P + A +DS+ G + + + IS +G++LL LTA + RP
Sbjct: 64 AAAQIANIVSGCICMVPAVAAIASDSFFGTIPVISVSAFISLMGVALLTLTASLDTLRPR 123
Query: 129 ACNHPSEGCESATPGQLAMLFSALILIAIGNGGITCSLA-FGADQVNRKDNPNNYRVLEI 187
C S C+S + QL +L++A+ L +IG GG +LA GA+Q + + +
Sbjct: 124 PCETASILCQSPSKTQLGVLYTAITLASIGTGGTRFTLATAGANQYEKTKDQGS------ 177
Query: 188 FFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQR 247
FF+W++ +A I+ T IVY +D++ W +GFG+ A S ++F Y
Sbjct: 178 FFNWFFFTTYLAGAISATAIVYTEDNISWTLGFGLSVAANFFSFLVFVSGKRFYKHDKPL 237
Query: 248 TSLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQ 307
S T V AA + RK + T+ + YH++ + +PT RF N+A + E
Sbjct: 238 GSPFTSLLCVIFAALRKRKAVV--STNEKDYHNE---SITMPTKSFRFFNRAAL--KQED 290
Query: 308 DIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGSFGL--LQAKSLDRH 365
++ DG+ N W LC+V QVE+ KA+I++IPL I +S I L LQ +DR
Sbjct: 291 EVKPDGTIRNPWRLCSVQQVEDFKAVIRIIPLALATIFLSTPIAMQLSLTVLQGLVMDRR 350
Query: 366 IISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFN 425
+ +F++PAGS VI +++ ++II+ DRVL P K+ GK ++P +R+GIG FN
Sbjct: 351 L--GPSFKIPAGSLQVITLLSTCLFIIVNDRVLYPFYQKLTGKH--LTPLQRVGIGHAFN 406
Query: 426 FLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFY 485
L + AI E +R + +++G+ + V MS +WL P L + GI E F+ G
Sbjct: 407 ILSMAVTAIVE-AKRLKIVQKGHFLGSSSVADMSVLWLFPPLVIVGIGEAFHFPGNVALC 465
Query: 486 YKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYY 545
Y+EFP+SM S A S++ + +G+ S+ ++ +I+ TT W+ D+IN G D Y
Sbjct: 466 YQEFPESMRSTATSITSVVIGICFYTSTALIDLIQRTT------AWLPDDINHGRVDNVY 519
Query: 546 WVIVGINALNLLYYLVCSWAY 566
W++V LNL Y+LVCSW Y
Sbjct: 520 WILVIGGVLNLGYFLVCSWLY 540
>At3g21670 nitrate transporter
Length = 590
Score = 293 bits (749), Expect = 3e-79
Identities = 184/586 (31%), Positives = 316/586 (53%), Gaps = 26/586 (4%)
Query: 11 DAADEEEMVSQQKPQRSK-GGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGI 69
D ++E P +SK GG + I+G+E ++ +G+ N++ YL+G +
Sbjct: 12 DGSEEAYDYRGNPPDKSKTGGWLGAGLILGSELSERICVMGISMNLVTYLVGDLHISSAK 71
Query: 70 STQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSA 129
S I+ N ++G F+AD+ LGR+ V I +S++ LG+ LL + I + RP
Sbjct: 72 SATIVTNFMGTLNLLGLLGGFLADAKLGRYKMVAISASVTALGVLLLTVATTISSMRPPI 131
Query: 130 CN---HPSEGCESATPGQLAMLFSALILIAIGNGGITCSLA-FGADQVNRKDNPNNYRVL 185
C+ C A QLA+L+ AL IA+G GGI +++ FG+DQ + D P + +
Sbjct: 132 CDDFRRLHHQCIEANGHQLALLYVALYTIALGGGGIKSNVSGFGSDQFDTSD-PKEEKQM 190
Query: 186 EIFFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKIT 245
FF+ +Y I++ + A+ +VY+QD++G G+G+ AA M+++ ++ + Y
Sbjct: 191 IFFFNRFYFSISVGSLFAVIALVYVQDNVGRGWGYGISAATMVVAAIVLLCGTKRYRFKK 250
Query: 246 QRTSLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDH 305
+ S T +V A+K RK S P S + D+ V T+ L+ L+KA + K+
Sbjct: 251 PKGSPFTTIWRVGFLAWKKRKESYPAHPS---LLNGYDNTTVPHTEMLKCLDKAAISKNE 307
Query: 306 EQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SINIGGSFGLLQAKSLD 363
+ D + W + TV QVEE+K ++K++P+W+T I + +F + QA +D
Sbjct: 308 SSPSSKDFEEKDPWIVSTVTQVEEVKLVMKLVPIWATNILFWTIYSQMTTFTVEQATFMD 367
Query: 364 RHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLF 423
R + +F VPAGS+S LI+ IL++ + +RV +PL ++ KP I+ +R+G+GL
Sbjct: 368 RKL---GSFTVPAGSYSAFLILTILLFTSLNERVFVPLTRRLTKKPQGITSLQRIGVGLV 424
Query: 424 FNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNE 483
F+ + AA+ E RR+ A+ ND K+SA WL PQ L G E F +GQ E
Sbjct: 425 FSMAAMAVAAVIENARREAAVN----NDK----KISAFWLVPQYFLVGAGEAFAYVGQLE 476
Query: 484 FYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDK 543
F+ +E P+ M S++ L + +G VSSL++S+++ T ++ W+ N+NK +
Sbjct: 477 FFIREAPERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVT----DKSWLRSNLNKARLNY 532
Query: 544 YYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEESTE 589
+YW++V + ALN L ++V + + D ++ V ++ S +E T+
Sbjct: 533 FYWLLVVLGALNFLIFIVFAMKHQYKADVITVVVTDDDSVEKEVTK 578
>At3g45720 putative transporter protein
Length = 555
Score = 288 bits (738), Expect = 5e-78
Identities = 176/545 (32%), Positives = 286/545 (52%), Gaps = 33/545 (6%)
Query: 28 KGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPVI 87
+GG +T PF+I + S G + N+I++L+ Y + + QI + + + PV+
Sbjct: 20 RGGCITFPFMIATLLGISVTSYGWVLNLIVFLIEEYNIKSIAAAQISNIVNGCLSMLPVV 79
Query: 88 GAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQLAM 147
A +ADS+ G + + IS LG+ LL L + RP C S C+S + L +
Sbjct: 80 TAILADSFFGNIPVISASAFISLLGIFLLTLISSFENLRPRPCETGSILCQSPSKLHLGV 139
Query: 148 LFSALILIAIGNGGITCSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGI 207
L++AL L+ G G +LA N+ D P R FF+WY+ + II+ T I
Sbjct: 140 LYAALALVTAGTSGTRVALASAG--ANQYDKP---RDKGSFFNWYFLTVNTGAIISATAI 194
Query: 208 VYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKL 267
VY Q++ WR+GFG+ AA LIS ++F Y S T +V VAA K+
Sbjct: 195 VYTQENASWRLGFGLCAAANLISFIVFISGKRFYKHDKPMGSPFTSLIRVLVAAI--LKI 252
Query: 268 SLPPKTSPEFYHHKKDSD----LVIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCT 323
+ + E YH + + + + +P+ RFLN+A +K + DG N W LC+
Sbjct: 253 KVVTSSKEEDYHREVEKESKTCIGMPSKSFRFLNRA-ALKSEKDLNQEDGLCHNPWRLCS 311
Query: 324 VDQVEELKAIIKVIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSV 381
V++VE+ K++++V+PLW + + +IG S +LQA DR + S F+VPAGS V
Sbjct: 312 VEEVEDFKSVLRVLPLWLAILFVGTSIGVQASMTVLQALVTDRGL--DSKFKVPAGSLQV 369
Query: 382 ILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRK 441
I++++ +++++ + + P+ KI K ++P +++GIG FN L + +AI E R K
Sbjct: 370 IVLISSCVFLVLNNWTIYPIYQKITHKQ--LTPLQQVGIGQVFNILSMAISAIVEAKRLK 427
Query: 442 EAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLS 501
E MS +WL P L + GI + F+ + +Y EFP+SM + A S++
Sbjct: 428 TVENEH---------PMSVLWLLPPLVIVGIGDAFHYMANVAVFYGEFPESMRNTATSVT 478
Query: 502 GLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLV 561
+A G+ +S+ ++++I+ TT W+ D+IN G D YWV+V LNL Y+ V
Sbjct: 479 SVAFGISFYLSTALINLIQRTT------AWLPDDINHGRVDNVYWVLVIGGVLNLGYFFV 532
Query: 562 CSWAY 566
CSW +
Sbjct: 533 CSWYF 537
>At3g45710 putative transporter protein
Length = 556
Score = 288 bits (737), Expect = 6e-78
Identities = 183/563 (32%), Positives = 291/563 (51%), Gaps = 39/563 (6%)
Query: 12 AADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGIST 71
+ D E P +GG +T+PF++G + S G N+I++L+ + + +
Sbjct: 6 SGDTEVAHRSSDPSEKRGGWITLPFMLG----MSITSFGWGMNLIVFLIEEFHIKNIAAA 61
Query: 72 QILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACN 131
QI + + N PV+ A +ADS+ G + + IS G SLL L + P C
Sbjct: 62 QISNVVNGVVNMLPVVAAILADSFFGNIPVISTSTFISLAGTSLLTLITSLNYLMPRPCE 121
Query: 132 HPSEGCESATPGQLAMLFSALILIAIGNGGITCSLAF-GADQVNRKDNPNNYRVLEIFFS 190
S C+S + QL +L+ AL L+ IG+ G +LA GA+Q + FF+
Sbjct: 122 TGSILCQSPSKLQLGILYVALALVIIGSAGTRFTLAAAGANQYKKPKEQGR------FFN 175
Query: 191 WYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSL 250
W++ + I I T IVY QD+ W++GFG+ A LIS ++F Y S
Sbjct: 176 WFFLALYIGAITGTTAIVYTQDNASWKLGFGLCAVANLISFIVFIAGVRFYKHDKPLGSP 235
Query: 251 LTGFAQVSVAAYKNRKLSLPPKTSPEFYHH-----KKDSDLVIPTDKLRFLNKACVIKDH 305
T +V VAA RK + K E YH + + +P+ RFLN+A +K+
Sbjct: 236 YTSLIRVLVAATMKRKAVISSKD--EDYHQYGLGKEAKTYTTMPSKSFRFLNRAA-LKNK 292
Query: 306 EQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMS--INIGGSFGLLQAKSLD 363
E S S+ N W LC+V +VE+ KAI++++PLW+ + +S + + S +LQA +D
Sbjct: 293 EDLNTSGDSSNNMWRLCSVQEVEDFKAILRLVPLWAAVMFLSTPVAVQMSMTVLQALVMD 352
Query: 364 RHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLF 423
R + S +FEV AGS VI++V ++I++ + ++ P+ K+ GKP ++P +++GIG
Sbjct: 353 RKL--SPHFEVSAGSLQVIVLVFGCVFIMLNNWIIYPMYQKLIGKP--LTPLQQVGIGHV 408
Query: 424 FNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNE 483
F L + +A+ E R K G+ MS +WL P L + GI E F+
Sbjct: 409 FTILSMAISAVVEAKRLKTVENGGH--------PMSVLWLVPALVMVGIGEAFHFPANVA 460
Query: 484 FYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDK 543
+Y EFP+S+ + A SL+ + +G+ +S+ V+ +I+ TT W+ ++IN G D
Sbjct: 461 VFYGEFPESLKNTATSLTSVVIGISFYLSTAVIDVIQRTT------SWLPNDINHGRVDN 514
Query: 544 YYWVIVGINALNLLYYLVCSWAY 566
YWV+V LNL Y+LVCSW Y
Sbjct: 515 VYWVVVIGGVLNLGYFLVCSWFY 537
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,085,718
Number of Sequences: 26719
Number of extensions: 597624
Number of successful extensions: 2174
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1783
Number of HSP's gapped (non-prelim): 77
length of query: 638
length of database: 11,318,596
effective HSP length: 106
effective length of query: 532
effective length of database: 8,486,382
effective search space: 4514755224
effective search space used: 4514755224
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149489.10


