

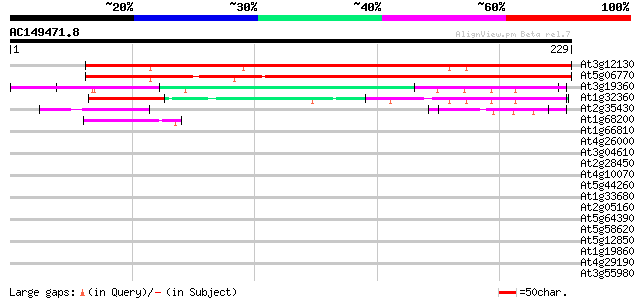
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149471.8 + phase: 0
(229 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g12130 unknown protein 181 3e-46
At5g06770 unknown protein 176 1e-44
At3g19360 unknown protein 51 6e-07
At1g32360 RING-H2 zinc finger like protein ATL6 47 6e-06
At2g35430 hypothetical protein 44 5e-05
At1g68200 putative zinc finger protein 40 8e-04
At1g66810 hypothetical protein 40 0.001
At4g26000 nucleic acid binding protein like 38 0.004
At3g04610 putative RNA-binding protein 38 0.005
At2g28450 RNA methyltransferase like protein 37 0.011
At4g10070 putative DNA-directed RNA polymerase 36 0.015
At5g44260 zinc finger like protein 35 0.025
At1g33680 single-strand nucleic acid-binding protein, putative 35 0.025
At2g05160 hypothetical protein 35 0.033
At5g64390 HEN4 35 0.043
At5g58620 unknown protein 33 0.16
At5g12850 zinc finger transcription factor -like protein 33 0.16
At1g19860 unknown protein 33 0.16
At4g29190 unknown protein 32 0.21
At3g55980 unknown protein 32 0.21
>At3g12130 unknown protein
Length = 248
Score = 181 bits (459), Expect = 3e-46
Identities = 100/211 (47%), Positives = 132/211 (62%), Gaps = 13/211 (6%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH----GEWELGRPTVPAYEDTRAMGQMQSSSVGGRI 87
+K++ C KF + GC FG+ CHF H G + + T + MQ S GGR
Sbjct: 37 SKSKPCTKFFSTSGCPFGENCHFLHYVPGGYNAVSQMTNMGPPIPQVSRNMQGSGNGGRF 96
Query: 88 EPPPPAH-GAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDP 146
+ G + FG SATA S++A+LAGAIIGK V+SKQIC TG KLSI++H+ DP
Sbjct: 97 SGRGESGPGHVSNFGDSATARFSVDASLAGAIIGKGGVSSKQICRQTGVKLSIQDHERDP 156
Query: 147 NLRNIELEGSFDQIKQASAMVHDLILNVSSVS-GPPGKNI-------TSQTSAPANNFKT 198
NL+NI LEG+ +QI +ASAMV DLI ++S + PPG + + P +NFKT
Sbjct: 157 NLKNIVLEGTLEQISEASAMVKDLIGRLNSAAKKPPGGGLGGGGGMGSEGKPHPGSNFKT 216
Query: 199 KLCENFTKGSCTFGERCHFAHGTDELRKPGM 229
K+CE F+KG+CTFG+RCHFAHG ELRK G+
Sbjct: 217 KICERFSKGNCTFGDRCHFAHGEAELRKSGI 247
>At5g06770 unknown protein
Length = 240
Score = 176 bits (445), Expect = 1e-44
Identities = 93/206 (45%), Positives = 128/206 (61%), Gaps = 11/206 (5%)
Query: 32 AKTRLCNKFNTAEGCKFGDKCHFAH-------GEWELGRPTVPAYEDTRAMGQMQSSSVG 84
+K++ C KF + GC FGD CHF H ++ P + +R M S G
Sbjct: 37 SKSKPCTKFFSTSGCPFGDNCHFLHYVPGGYNAAAQMTNLRPPVSQVSRNM--QGSGGPG 94
Query: 85 GRIEPP-PPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHD 143
GR P G + FG S T+ +S++A+LAGAIIGK ++SKQIC TGAKLSI++H+
Sbjct: 95 GRFSGRGDPGSGPVSIFGAS-TSKISVDASLAGAIIGKGGIHSKQICRETGAKLSIKDHE 153
Query: 144 SDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPANNFKTKLCEN 203
DPNL+ IELEG+F+QI AS MV +LI + SV P G P +N+KTK+C+
Sbjct: 154 RDPNLKIIELEGTFEQINVASGMVRELIGRLGSVKKPQGIGGPEGKPHPGSNYKTKICDR 213
Query: 204 FTKGSCTFGERCHFAHGTDELRKPGM 229
++KG+CT+G+RCHFAHG ELR+ G+
Sbjct: 214 YSKGNCTYGDRCHFAHGESELRRSGI 239
>At3g19360 unknown protein
Length = 386
Score = 50.8 bits (120), Expect = 6e-07
Identities = 30/74 (40%), Positives = 38/74 (50%), Gaps = 13/74 (17%)
Query: 1 MINVGSSPAIPPIGRNPNVPQSFPDGSSPPVA-------------KTRLCNKFNTAEGCK 47
+I+VG++ A P N+ + GS P A KTRLC KF+ C
Sbjct: 226 VISVGATAADQPSDTASNLIEVNRQGSIPVPAPMNNGGVVKTVYWKTRLCMKFDITGQCP 285
Query: 48 FGDKCHFAHGEWEL 61
FGDKCHFAHG+ EL
Sbjct: 286 FGDKCHFAHGQAEL 299
Score = 50.8 bits (120), Expect = 6e-07
Identities = 59/224 (26%), Positives = 80/224 (35%), Gaps = 28/224 (12%)
Query: 20 PQSFPDGSSPPVAK--------TRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYE- 70
P P + PPV K TR+C KF A C+ G+ C+FAHG +L +P E
Sbjct: 85 PWMVPSLNPPPVNKGTANIFYKTRMCAKFR-AGTCRNGELCNFAHGIEDLRQPPSNWQEI 143
Query: 71 -DTRAMGQMQSSSVGGRIEPPPPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQI 129
GQ + E P+ + + L +
Sbjct: 144 VGPPPAGQDRERERERERERERPSLAPVVNNNWEDDQKIILRMKLCRKFCFGEECPYGDR 203
Query: 130 CHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQT 189
C+ LS DS + + Q S +LI V+ P
Sbjct: 204 CNFIHEDLSKFREDSGKLRESSVISVGATAADQPSDTASNLI-EVNRQGSIP-------V 255
Query: 190 SAPANN--------FKTKLCENFT-KGSCTFGERCHFAHGTDEL 224
AP NN +KT+LC F G C FG++CHFAHG EL
Sbjct: 256 PAPMNNGGVVKTVYWKTRLCMKFDITGQCPFGDKCHFAHGQAEL 299
Score = 46.2 bits (108), Expect = 1e-05
Identities = 26/72 (36%), Positives = 37/72 (51%), Gaps = 10/72 (13%)
Query: 166 MVHDLILN-VSSVSGPPGKN---ITSQTSAPANN------FKTKLCENFTKGSCTFGERC 215
+V D + N SS P N + S P N +KT++C F G+C GE C
Sbjct: 65 LVDDNLFNPASSFPQPSSSNPWMVPSLNPPPVNKGTANIFYKTRMCAKFRAGTCRNGELC 124
Query: 216 HFAHGTDELRKP 227
+FAHG ++LR+P
Sbjct: 125 NFAHGIEDLRQP 136
>At1g32360 RING-H2 zinc finger like protein ATL6
Length = 384
Score = 47.4 bits (111), Expect = 6e-06
Identities = 35/99 (35%), Positives = 47/99 (47%), Gaps = 20/99 (20%)
Query: 146 PNLRNIELEGSFDQIKQASAMVHDLILNVSSVS-GPPGKNI--------TSQTSAPANN- 195
PNL + + + D +A AM D N S + GPP K + TSA +N
Sbjct: 28 PNLGDSAVWATEDDYNRAWAMNPD---NTSGDNNGPPNKKTRGSPSSSSATTTSAASNRT 84
Query: 196 -------FKTKLCENFTKGSCTFGERCHFAHGTDELRKP 227
FKTKLC F G+C + C+FAH +ELR+P
Sbjct: 85 KAIGKMFFKTKLCCKFRAGTCPYITNCNFAHTVEELRRP 123
Score = 44.7 bits (104), Expect = 4e-05
Identities = 19/31 (61%), Positives = 21/31 (67%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGR 63
KTR+CNK+ C FG KCHFAHG EL R
Sbjct: 262 KTRICNKWEITGYCPFGAKCHFAHGAAELHR 292
Score = 42.7 bits (99), Expect = 2e-04
Identities = 54/215 (25%), Positives = 86/215 (39%), Gaps = 32/215 (14%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGGRIEPPPP 92
KT+LC KF A C + C+FAH EL RP P +++ A + + S G + P
Sbjct: 93 KTKLCCKFR-AGTCPYITNCNFAHTVEELRRPP-PNWQEIVAAHEEERS---GGMGTPTV 147
Query: 93 AHGAAAGFGVSATATVSINATLAGAIIGKN--DVNSKQICHITGAKLSIREHDSDPNLRN 150
+ + VS A + G++ +++ C G + ++ N +
Sbjct: 148 SVVEIPREEFQIPSLVSSTAESGRSFKGRHCKKFYTEEGCPY-GESCTFLHDEASRNRES 206
Query: 151 IELE----------------GSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPAN 194
+ + GS +S +V V G G Q P+N
Sbjct: 207 VAISLGPGGYGSGGGGGSGGGSVGGGGSSSNVV------VLGGGGGSGSGSGIQILKPSN 260
Query: 195 NFKTKLCENFT-KGSCTFGERCHFAHGTDELRKPG 228
+KT++C + G C FG +CHFAHG EL + G
Sbjct: 261 -WKTRICNKWEITGYCPFGAKCHFAHGAAELHRFG 294
Score = 32.0 bits (71), Expect = 0.28
Identities = 39/179 (21%), Positives = 65/179 (35%), Gaps = 31/179 (17%)
Query: 61 LGRPTVPAYED--TRAMGQMQSSSVGGRIEPPPPAHGAAAGFGVSATATVSINATLAGAI 118
LG V A ED RA ++ G PP + + T + + N T A
Sbjct: 30 LGDSAVWATEDDYNRAWAMNPDNTSGDNNGPPNKKTRGSPSSSSATTTSAASNRTKA--- 86
Query: 119 IGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLIL------ 172
IGK +K C P + N + +++++ +++
Sbjct: 87 IGKMFFKTKLCCKFRAGTC--------PYITNCNFAHTVEELRRPPPNWQEIVAAHEEER 138
Query: 173 -------NVSSVSGPPGK----NITSQTSAPANNFKTKLCENF-TKGSCTFGERCHFAH 219
VS V P + ++ S T+ +FK + C+ F T+ C +GE C F H
Sbjct: 139 SGGMGTPTVSVVEIPREEFQIPSLVSSTAESGRSFKGRHCKKFYTEEGCPYGESCTFLH 197
>At2g35430 hypothetical protein
Length = 180
Score = 44.3 bits (103), Expect = 5e-05
Identities = 23/58 (39%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query: 172 LNVSSVSGPPGKNITSQTSAPANNF-KTKLCENFTKGSCTFG-ERCHFAHGTDELRKP 227
+N P K +S +S +F KTKLC F G+C + CHFAH +ELR P
Sbjct: 47 INSDGAESPSKKTRSSSSSEIGKSFFKTKLCFKFRAGTCPYSASSCHFAHSAEELRLP 104
Score = 43.9 bits (102), Expect = 7e-05
Identities = 21/45 (46%), Positives = 25/45 (54%), Gaps = 4/45 (8%)
Query: 13 IGRNPNVPQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHG 57
+G NV Q+ P KTR+CNK+ T C FG CHFAHG
Sbjct: 129 LGPRGNVAQTLKS----PNWKTRICNKWQTTGYCPFGSHCHFAHG 169
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query: 176 SVSGPPGKNITSQTSAPANNFKTKLCENF-TKGSCTFGERCHFAHG 220
+VS P N+ +P N+KT++C + T G C FG CHFAHG
Sbjct: 126 AVSLGPRGNVAQTLKSP--NWKTRICNKWQTTGYCPFGSHCHFAHG 169
Score = 31.2 bits (69), Expect = 0.48
Identities = 15/35 (42%), Positives = 16/35 (44%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVP 67
KT+LC KF CHFAH EL P P
Sbjct: 73 KTKLCFKFRAGTCPYSASSCHFAHSAEELRLPPPP 107
>At1g68200 putative zinc finger protein
Length = 308
Score = 40.4 bits (93), Expect = 8e-04
Identities = 20/42 (47%), Positives = 25/42 (58%), Gaps = 3/42 (7%)
Query: 31 VAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTV--PAYE 70
+ KT LCNK+ C +GD C FAHG EL RP + P Y+
Sbjct: 222 MTKTELCNKWQETGTCPYGDHCQFAHGIKEL-RPVIRHPRYK 262
Score = 37.0 bits (84), Expect = 0.009
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 1/30 (3%)
Query: 197 KTKLCENFTK-GSCTFGERCHFAHGTDELR 225
KT+LC + + G+C +G+ C FAHG ELR
Sbjct: 224 KTELCNKWQETGTCPYGDHCQFAHGIKELR 253
Score = 32.7 bits (73), Expect = 0.16
Identities = 13/32 (40%), Positives = 18/32 (55%), Gaps = 1/32 (3%)
Query: 196 FKTKLCENFTKG-SCTFGERCHFAHGTDELRK 226
+KT++C G +C +G RCHF H E K
Sbjct: 261 YKTEVCRMVLAGDNCPYGHRCHFRHSLSEQEK 292
Score = 30.4 bits (67), Expect = 0.81
Identities = 10/27 (37%), Positives = 14/27 (51%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAH 56
P KT +C + C +G +CHF H
Sbjct: 259 PRYKTEVCRMVLAGDNCPYGHRCHFRH 285
>At1g66810 hypothetical protein
Length = 310
Score = 40.0 bits (92), Expect = 0.001
Identities = 20/40 (50%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTV--PAYE 70
KT LCNK+ C +GD C FAHG EL RP + P Y+
Sbjct: 234 KTELCNKWQETGACCYGDNCQFAHGIDEL-RPVIRHPRYK 272
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/30 (53%), Positives = 22/30 (73%), Gaps = 1/30 (3%)
Query: 197 KTKLCENFTK-GSCTFGERCHFAHGTDELR 225
KT+LC + + G+C +G+ C FAHG DELR
Sbjct: 234 KTELCNKWQETGACCYGDNCQFAHGIDELR 263
Score = 32.0 bits (71), Expect = 0.28
Identities = 14/32 (43%), Positives = 20/32 (61%), Gaps = 2/32 (6%)
Query: 196 FKTKLCENFTKGS-CTFGERCHFAHG-TDELR 225
+KT++C G+ C +G RCHF H TD+ R
Sbjct: 271 YKTEVCRMMVTGAMCPYGHRCHFRHSLTDQER 302
Score = 30.4 bits (67), Expect = 0.81
Identities = 11/27 (40%), Positives = 14/27 (51%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAH 56
P KT +C T C +G +CHF H
Sbjct: 269 PRYKTEVCRMMVTGAMCPYGHRCHFRH 295
>At4g26000 nucleic acid binding protein like
Length = 495
Score = 38.1 bits (87), Expect = 0.004
Identities = 31/119 (26%), Positives = 55/119 (46%), Gaps = 11/119 (9%)
Query: 71 DTRAMGQMQSSSVGGRIEPP-------PPAHGAAAGFGVSATATVSINATLAGAIIGKND 123
D+R ++Q S V + P P ++ F + T+ I + A IIG
Sbjct: 301 DSRVDSRVQPSGVSIYSQDPVLSARHSPGLARVSSAFVTQVSQTMQIPFSYAEDIIGVEG 360
Query: 124 VNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVS---SVSG 179
N I +GA ++I+E P+ +E++G+ Q++ A ++ + I+N SVSG
Sbjct: 361 ANIAYIRRRSGATITIKE-SPHPDQITVEIKGTTSQVQTAEQLIQEFIINHKEPVSVSG 418
>At3g04610 putative RNA-binding protein
Length = 577
Score = 37.7 bits (86), Expect = 0.005
Identities = 35/116 (30%), Positives = 54/116 (46%), Gaps = 8/116 (6%)
Query: 79 QSSSVGGRIEPPPPAHGAAAGFGVSATAT--VSINATLAGAIIGKNDVNSKQICHITGAK 136
Q S GR EPP H ++A V+ T + I + A A+IG + N ++GA
Sbjct: 432 QGISAYGR-EPPMNVHVSSAPPMVAQQVTQQMQIPLSYADAVIGTSGSNISYTRRLSGAT 490
Query: 137 LSIREHDSDPNLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAP 192
++I+E P +E+ G+ Q++ A LI N + +G P QT AP
Sbjct: 491 VTIQETRGVPGEMTVEVSGTGSQVQTAV----QLIQNFMAEAGAPAP-AQPQTVAP 541
>At2g28450 RNA methyltransferase like protein
Length = 809
Score = 36.6 bits (83), Expect = 0.011
Identities = 17/48 (35%), Positives = 24/48 (49%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQ 77
P KT LC+ F C G++C +AHGE EL ++ T G+
Sbjct: 78 PWWKTSLCSYFRREASCSHGNECKYAHGEAELRMKPDNTWDPTSERGK 125
Score = 35.4 bits (80), Expect = 0.025
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query: 187 SQTSAPANNFKTKLCENFTK-GSCTFGERCHFAHGTDELR 225
++ + P+ +KT LC F + SC+ G C +AHG ELR
Sbjct: 71 TKITTPSPWWKTSLCSYFRREASCSHGNECKYAHGEAELR 110
>At4g10070 putative DNA-directed RNA polymerase
Length = 748
Score = 36.2 bits (82), Expect = 0.015
Identities = 22/81 (27%), Positives = 42/81 (51%), Gaps = 3/81 (3%)
Query: 103 SATATVSINATLAGAIIGKNDVNSKQICHITGAKLSI-REHDSDPN--LRNIELEGSFDQ 159
S T + + ++ G +IGK + + +GAK+ I R+ ++DP+ LR +E+ GS
Sbjct: 198 STTRRIDVPSSKVGVLIGKGGETIRYLQFNSGAKIQILRDSEADPSSALRPVEIIGSVAC 257
Query: 160 IKQASAMVHDLILNVSSVSGP 180
I+ A ++ +I + P
Sbjct: 258 IESAEKLISAVIAEAEAGGSP 278
>At5g44260 zinc finger like protein
Length = 381
Score = 35.4 bits (80), Expect = 0.025
Identities = 39/154 (25%), Positives = 70/154 (45%), Gaps = 17/154 (11%)
Query: 36 LCNKFNTAEGCKFGDKCHFAHGEWEL----GRPTVPAYED----TRAMGQMQSSSVGGRI 87
+C +F+ C GD+C FAHG +E R A +D R + S R+
Sbjct: 105 VCPEFSRHGDCSRGDECGFAHGVFECWLHPSRYRTEACKDGKHCKRKVCFFAHSPRQLRV 164
Query: 88 EPPPPAHGAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHI-TGAKLSI-REHDSD 145
PP P + + G G S +++ A + KN+ H T L++ R S
Sbjct: 165 LPPSPENHISGGCGGSPSSSP------ASVLSNKNNRCCLFCSHSPTSTLLNLSRSPSSS 218
Query: 146 PNLRNIELEGSFDQI-KQASAMVHDLILNVSSVS 178
P L + +F ++ ++ +A++++LI ++ S+S
Sbjct: 219 PPLSPADKADAFSRLSRRRTAVLNELISSLDSLS 252
Score = 28.5 bits (62), Expect = 3.1
Identities = 11/30 (36%), Positives = 19/30 (62%), Gaps = 1/30 (3%)
Query: 195 NFKTKLCENFTK-GSCTFGERCHFAHGTDE 223
++ ++C F++ G C+ G+ C FAHG E
Sbjct: 100 HYTGEVCPEFSRHGDCSRGDECGFAHGVFE 129
>At1g33680 single-strand nucleic acid-binding protein, putative
Length = 407
Score = 35.4 bits (80), Expect = 0.025
Identities = 22/81 (27%), Positives = 45/81 (55%), Gaps = 3/81 (3%)
Query: 103 SATATVSINATLAGAIIGKNDVNSKQICHITGAKLSI-REHDSDPN--LRNIELEGSFDQ 159
S T + + ++ G +IGK + + +GAK+ I R+ ++DP+ LR +E+ G+
Sbjct: 231 STTRRIDVPSSKVGTLIGKGGEMVRYLQVNSGAKIQIRRDAEADPSSALRPVEIIGTVSC 290
Query: 160 IKQASAMVHDLILNVSSVSGP 180
I++A +++ +I V + P
Sbjct: 291 IEKAEKLINAVIAEVEAGGVP 311
>At2g05160 hypothetical protein
Length = 536
Score = 35.0 bits (79), Expect = 0.033
Identities = 14/35 (40%), Positives = 17/35 (48%)
Query: 186 TSQTSAPANNFKTKLCENFTKGSCTFGERCHFAHG 220
TS+ S F K+C F KG C G C + HG
Sbjct: 147 TSRRSPSLPEFPVKICHYFNKGFCKHGNNCRYFHG 181
Score = 27.3 bits (59), Expect = 6.9
Identities = 14/44 (31%), Positives = 24/44 (53%), Gaps = 11/44 (25%)
Query: 15 RNPNVPQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGE 58
R+P++P+ FP ++C+ FN CK G+ C + HG+
Sbjct: 150 RSPSLPE-FP---------VKICHYFNKGF-CKHGNNCRYFHGQ 182
>At5g64390 HEN4
Length = 857
Score = 34.7 bits (78), Expect = 0.043
Identities = 20/78 (25%), Positives = 37/78 (46%)
Query: 95 GAAAGFGVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELE 154
G + + T + + A + G+ N +Q+ I+GA++ I E + R I +
Sbjct: 767 GHKSAIVTNTTVEIRVPANAMSFVYGEQGYNLEQLRQISGARVIIHEPPLGTSDRIIVIS 826
Query: 155 GSFDQIKQASAMVHDLIL 172
G+ DQ + A ++H IL
Sbjct: 827 GTPDQTQAAQNLLHAFIL 844
Score = 32.0 bits (71), Expect = 0.28
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 4/67 (5%)
Query: 115 AGAIIGKNDVNSKQICHITGAKLSIREHD----SDPNLRNIELEGSFDQIKQASAMVHDL 170
AGA+IGK I TG K+SIR + +D + +E+EG+ +K+A +
Sbjct: 161 AGAVIGKGGQMVGSIRKETGCKISIRIENLPICADTDDEMVEVEGNAIAVKKALVSISRC 220
Query: 171 ILNVSSV 177
+ N S+
Sbjct: 221 LQNCQSI 227
Score = 29.6 bits (65), Expect = 1.4
Identities = 16/66 (24%), Positives = 34/66 (51%), Gaps = 5/66 (7%)
Query: 103 SATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDP-----NLRNIELEGSF 157
S TA + + + G ++GK V ++ TGA + I + + +P N + +++ G F
Sbjct: 541 SITARLVVPTSQIGCVLGKGGVIVSEMRKTTGAAIQILKVEQNPKCISENDQVVQITGEF 600
Query: 158 DQIKQA 163
+++A
Sbjct: 601 PNVREA 606
>At5g58620 unknown protein
Length = 607
Score = 32.7 bits (73), Expect = 0.16
Identities = 12/23 (52%), Positives = 15/23 (65%)
Query: 201 CENFTKGSCTFGERCHFAHGTDE 223
C F KGSC+ G+ C +AHG E
Sbjct: 255 CPEFRKGSCSRGDTCEYAHGIFE 277
>At5g12850 zinc finger transcription factor -like protein
Length = 706
Score = 32.7 bits (73), Expect = 0.16
Identities = 12/23 (52%), Positives = 15/23 (65%)
Query: 201 CENFTKGSCTFGERCHFAHGTDE 223
C +F KGSC G+ C +AHG E
Sbjct: 303 CPDFKKGSCKQGDMCEYAHGVFE 325
>At1g19860 unknown protein
Length = 413
Score = 32.7 bits (73), Expect = 0.16
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Query: 11 PPIGRNPNVPQSFPDGSSPPVAKTRL---CNKFNTAEGCKFGDKCHFAH 56
P G NP + S + P +K ++ C FN+A GC+ G C + H
Sbjct: 334 PGGGPNPEMVNSSNNNQRPRDSKPKIMKACMYFNSARGCRHGANCMYQH 382
>At4g29190 unknown protein
Length = 356
Score = 32.3 bits (72), Expect = 0.21
Identities = 12/23 (52%), Positives = 14/23 (60%)
Query: 201 CENFTKGSCTFGERCHFAHGTDE 223
C +F KG C G+ C FAHG E
Sbjct: 125 CPDFRKGGCKKGDSCEFAHGVFE 147
Score = 30.0 bits (66), Expect = 1.1
Identities = 11/16 (68%), Positives = 12/16 (74%)
Query: 45 GCKFGDKCHFAHGEWE 60
GCK GD C FAHG +E
Sbjct: 132 GCKKGDSCEFAHGVFE 147
>At3g55980 unknown protein
Length = 580
Score = 32.3 bits (72), Expect = 0.21
Identities = 12/23 (52%), Positives = 14/23 (60%)
Query: 201 CENFTKGSCTFGERCHFAHGTDE 223
C F KGSC G+ C +AHG E
Sbjct: 257 CPEFRKGSCPKGDSCEYAHGVFE 279
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,718,446
Number of Sequences: 26719
Number of extensions: 242009
Number of successful extensions: 831
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 695
Number of HSP's gapped (non-prelim): 147
length of query: 229
length of database: 11,318,596
effective HSP length: 96
effective length of query: 133
effective length of database: 8,753,572
effective search space: 1164225076
effective search space used: 1164225076
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149471.8


