

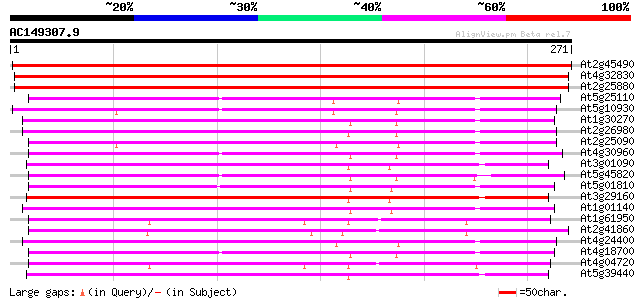
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149307.9 - phase: 0
(271 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45490 putative protein kinase 454 e-128
At4g32830 putative serine/threonine protein kinase 376 e-105
At2g25880 putative protein kinase 374 e-104
At5g25110 serine/threonine protein kinase-like protein 190 6e-49
At5g10930 serine/threonine protein kinase -like protein 188 2e-48
At1g30270 serine/threonine kinase, putative 187 5e-48
At2g26980 putative protein kinase (At2g26980) 184 4e-47
At2g25090 CBL-interacting protein kinase 16 (CIPK16) 183 7e-47
At4g30960 CBL-interacting protein kinase 6 (CIPK6) 182 2e-46
At3g01090 putative SNF1-related protein kinase 179 1e-45
At5g45820 CBL-interacting protein kinase 20 (CIPK20) 178 2e-45
At5g01810 serine/threonine protein kinase ATPK10 178 2e-45
At3g29160 SNF1-like protein kinase (AKin11) 177 4e-45
At1g01140 SOS2-like protein kinase PKS6/CBL-interacting protein ... 177 4e-45
At1g61950 hypothetical protein 176 9e-45
At2g41860 putative Ca2+ dependent protein kinase 176 2e-44
At4g24400 serine/threonine kinase-like protein 175 2e-44
At4g18700 putative protein kinase 174 5e-44
At4g04720 putative calcium dependent protein kinase 174 6e-44
At5g39440 serine/threonine-specific protein kinase -like protein 173 8e-44
>At2g45490 putative protein kinase
Length = 288
Score = 454 bits (1169), Expect = e-128
Identities = 211/270 (78%), Positives = 243/270 (89%)
Query: 2 DREWSINDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEI 61
+++WS+ DFEIG+PLGKGKFGRVY+ARE KSK++VALKVIFKEQ+EKYK HHQLRREMEI
Sbjct: 14 EKQWSLADFEIGRPLGKGKFGRVYLAREAKSKYIVALKVIFKEQIEKYKIHHQLRREMEI 73
Query: 62 QISLKHPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEAL 121
Q SL+HPN+L L+GWFHD+ER+FLILEYAH GELY L + GH +E+QAATYI SL++AL
Sbjct: 74 QTSLRHPNILRLFGWFHDNERIFLILEYAHGGELYGVLKQNGHLTEQQAATYIASLSQAL 133
Query: 122 TYCHENHVIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRKTMCGTLDYLAPEMVENKT 181
YCH VIHRDIKPENLLLDHEGRLKIADFGWSVQS +KRKTMCGTLDYLAPEMVEN+
Sbjct: 134 AYCHGKCVIHRDIKPENLLLDHEGRLKIADFGWSVQSSNKRKTMCGTLDYLAPEMVENRD 193
Query: 182 HDYAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRL 241
HDYAVDNWTLGILCYEFLYG PPFEAESQ DTF+RI K+DLSFP +P VS +AKNLIS+L
Sbjct: 194 HDYAVDNWTLGILCYEFLYGNPPFEAESQKDTFKRILKIDLSFPLTPNVSEEAKNLISQL 253
Query: 242 LVKDSSRRLSLQKIMEHPWIIKNANRMGVC 271
LVKD S+RLS++KIM+HPWI+KNA+ GVC
Sbjct: 254 LVKDPSKRLSIEKIMQHPWIVKNADPKGVC 283
>At4g32830 putative serine/threonine protein kinase
Length = 294
Score = 376 bits (965), Expect = e-105
Identities = 172/268 (64%), Positives = 222/268 (82%)
Query: 3 REWSINDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQ 62
+ W+++DF+IGKPLG+GKFG VY+ARE +S VVALKV+FK QL++ + HQLRRE+EIQ
Sbjct: 24 KRWTLSDFDIGKPLGRGKFGHVYLAREKRSNHVVALKVLFKSQLQQSQVEHQLRREVEIQ 83
Query: 63 ISLKHPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALT 122
L+HPN+L LYG+F+D +RV+LILEYA GELYK+L K +FSE++AATY+ SL AL
Sbjct: 84 SHLRHPNILRLYGYFYDQKRVYLILEYAARGELYKDLQKCKYFSERRAATYVASLARALI 143
Query: 123 YCHENHVIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRKTMCGTLDYLAPEMVENKTH 182
YCH HVIHRDIKPENLL+ +G LKIADFGWSV + ++R+TMCGTLDYL PEMVE+ H
Sbjct: 144 YCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEMVESVEH 203
Query: 183 DYAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLL 242
D +VD W+LGILCYEFLYGVPPFEA +DT+ RI +VDL FPP P++S+ AK+LIS++L
Sbjct: 204 DASVDIWSLGILCYEFLYGVPPFEAMEHSDTYRRIVQVDLKFPPKPIISASAKDLISQML 263
Query: 243 VKDSSRRLSLQKIMEHPWIIKNANRMGV 270
VK+SS+RL L K++EHPWI++NA+ G+
Sbjct: 264 VKESSQRLPLHKLLEHPWIVQNADPSGI 291
>At2g25880 putative protein kinase
Length = 282
Score = 374 bits (961), Expect = e-104
Identities = 172/268 (64%), Positives = 223/268 (83%)
Query: 3 REWSINDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQ 62
+ W+ +DF+IGKPLG+GKFG VY+ARE +S +VALKV+FK QL++ + HQLRRE+EIQ
Sbjct: 12 KRWTTSDFDIGKPLGRGKFGHVYLAREKRSDHIVALKVLFKAQLQQSQVEHQLRREVEIQ 71
Query: 63 ISLKHPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALT 122
L+HPN+L LYG+F+D +RV+LILEYA GELYKEL K +FSE++AATY+ SL AL
Sbjct: 72 SHLRHPNILRLYGYFYDQKRVYLILEYAVRGELYKELQKCKYFSERRAATYVASLARALI 131
Query: 123 YCHENHVIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRKTMCGTLDYLAPEMVENKTH 182
YCH HVIHRDIKPENLL+ +G LKIADFGWSV + ++R+TMCGTLDYL PEMVE+ H
Sbjct: 132 YCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEMVESVEH 191
Query: 183 DYAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLL 242
D +VD W+LGILCYEFLYGVPPFEA ++T++RI +VDL FPP P+VSS AK+LIS++L
Sbjct: 192 DASVDIWSLGILCYEFLYGVPPFEAREHSETYKRIVQVDLKFPPKPIVSSSAKDLISQML 251
Query: 243 VKDSSRRLSLQKIMEHPWIIKNANRMGV 270
VK+S++RL+L K++EHPWI++NA+ G+
Sbjct: 252 VKESTQRLALHKLLEHPWIVQNADPSGL 279
>At5g25110 serine/threonine protein kinase-like protein
Length = 488
Score = 190 bits (483), Expect = 6e-49
Identities = 102/263 (38%), Positives = 154/263 (57%), Gaps = 9/263 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHPN 69
+E+G+ LGKG FG+VY +E+ + VA+K+I K+Q+++ Q++RE+ I ++HPN
Sbjct: 43 YEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIKREISIMRLVRHPN 102
Query: 70 VLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENHV 129
++ L ++F I+EY GEL+ ++ K G E A Y L A+ +CH V
Sbjct: 103 IVELKEVMATKTKIFFIMEYVKGGELFSKIVK-GKLKEDSARKYFQQLISAVDFCHSRGV 161
Query: 130 IHRDIKPENLLLDHEGRLKIADFGWS-----VQSKDKRKTMCGTLDYLAPEMVENKTHDY 184
HRD+KPENLL+D G LK++DFG S + T CGT Y+APE++ K +D
Sbjct: 162 SHRDLKPENLLVDENGDLKVSDFGLSALPEQILQDGLLHTQCGTPAYVAPEVLRKKGYDG 221
Query: 185 AV-DNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLV 243
A D W+ GI+ Y L G PF+ E+ + +I K + +P P S ++K LIS+LLV
Sbjct: 222 AKGDIWSCGIILYVLLAGFLPFQDENLMKMYRKIFKSEFEYP--PWFSPESKRLISKLLV 279
Query: 244 KDSSRRLSLQKIMEHPWIIKNAN 266
D ++R+S+ IM PW KN N
Sbjct: 280 VDPNKRISIPAIMRTPWFRKNIN 302
>At5g10930 serine/threonine protein kinase -like protein
Length = 445
Score = 188 bits (478), Expect = 2e-48
Identities = 106/270 (39%), Positives = 155/270 (57%), Gaps = 10/270 (3%)
Query: 2 DREWSINDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYK-FHHQLRREME 60
+R +E+G+ LGKG F +VY +E+ VA+KVI K+Q+ K Q++RE+
Sbjct: 4 ERRVLFGKYEMGRLLGKGTFAKVYYGKEIIGGECVAIKVINKDQVMKRPGMMEQIKREIS 63
Query: 61 IQISLKHPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEA 120
I ++HPN++ L ++F ++E+ GEL+ ++SK G E A Y L A
Sbjct: 64 IMKLVRHPNIVELKEVMATKTKIFFVMEFVKGGELFCKISK-GKLHEDAARRYFQQLISA 122
Query: 121 LTYCHENHVIHRDIKPENLLLDHEGRLKIADFGWS-----VQSKDKRKTMCGTLDYLAPE 175
+ YCH V HRD+KPENLLLD G LKI+DFG S + T CGT Y+APE
Sbjct: 123 VDYCHSRGVSHRDLKPENLLLDENGDLKISDFGLSALPEQILQDGLLHTQCGTPAYVAPE 182
Query: 176 MVENKTHDYA-VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDA 234
+++ K +D A D W+ G++ Y L G PF+ E+ + + +I + D FP P S +A
Sbjct: 183 VLKKKGYDGAKADIWSCGVVLYVLLAGCLPFQDENLMNMYRKIFRADFEFP--PWFSPEA 240
Query: 235 KNLISRLLVKDSSRRLSLQKIMEHPWIIKN 264
+ LIS+LLV D RR+S+ IM PW+ KN
Sbjct: 241 RRLISKLLVVDPDRRISIPAIMRTPWLRKN 270
>At1g30270 serine/threonine kinase, putative
Length = 482
Score = 187 bits (475), Expect = 5e-48
Identities = 96/263 (36%), Positives = 155/263 (58%), Gaps = 8/263 (3%)
Query: 7 INDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLK 66
+ +E+G+ LG+G F +V AR V++ VA+KVI KE++ K K Q++RE+ +K
Sbjct: 28 VGKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMIAQIKREISTMKLIK 87
Query: 67 HPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHE 126
HPNV+ ++ +++ +LE+ GEL+ ++S G E +A Y L A+ YCH
Sbjct: 88 HPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARKYFQQLINAVDYCHS 147
Query: 127 NHVIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRK-----TMCGTLDYLAPEMVENKT 181
V HRD+KPENLLLD G LK++DFG S + R+ T CGT +Y+APE++ NK
Sbjct: 148 RGVYHRDLKPENLLLDANGALKVSDFGLSALPQQVREDGLLHTTCGTPNYVAPEVINNKG 207
Query: 182 HDYA-VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISR 240
+D A D W+ G++ + + G PFE + +++I K + + P P S+ AK LI R
Sbjct: 208 YDGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEFTCP--PWFSASAKKLIKR 265
Query: 241 LLVKDSSRRLSLQKIMEHPWIIK 263
+L + + R++ +++E+ W K
Sbjct: 266 ILDPNPATRITFAEVIENEWFKK 288
>At2g26980 putative protein kinase (At2g26980)
Length = 441
Score = 184 bits (467), Expect = 4e-47
Identities = 98/264 (37%), Positives = 154/264 (58%), Gaps = 8/264 (3%)
Query: 7 INDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLK 66
+ +E+G+ +G+G F +V AR ++ VALK++ KE++ K+K Q+RRE+ +K
Sbjct: 11 VGKYEVGRTIGEGTFAKVKFARNSETGEPVALKILDKEKVLKHKMAEQIRREIATMKLIK 70
Query: 67 HPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHE 126
HPNV+ LY ++F+ILEY GEL+ ++ G E +A Y L A+ YCH
Sbjct: 71 HPNVVQLYEVMASKTKIFIILEYVTGGELFDKIVNDGRMKEDEARRYFQQLIHAVDYCHS 130
Query: 127 NHVIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKR-----KTMCGTLDYLAPEMVENKT 181
V HRD+KPENLLLD G LKI+DFG S S+ R T CGT +Y+APE++ ++
Sbjct: 131 RGVYHRDLKPENLLLDSYGNLKISDFGLSALSQQVRDDGLLHTSCGTPNYVAPEVLNDRG 190
Query: 182 HDYA-VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISR 240
+D A D W+ G++ Y L G PF+ + + +++I + + P P +S A LI+R
Sbjct: 191 YDGATADMWSCGVVLYVLLAGYLPFDDSNLMNLYKKISSGEFNCP--PWLSLGAMKLITR 248
Query: 241 LLVKDSSRRLSLQKIMEHPWIIKN 264
+L + R++ Q++ E W K+
Sbjct: 249 ILDPNPMTRVTPQEVFEDEWFKKD 272
>At2g25090 CBL-interacting protein kinase 16 (CIPK16)
Length = 469
Score = 183 bits (465), Expect = 7e-47
Identities = 105/269 (39%), Positives = 150/269 (55%), Gaps = 16/269 (5%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYK-FHHQLRREMEIQISLKHP 68
+ IG+ LG G F +VY E+ + VA+KVI K+ + K + Q+ RE+ + L+HP
Sbjct: 15 YNIGRLLGTGNFAKVYHGTEISTGDDVAIKVIKKDHVFKRRGMMEQIEREIAVMRLLRHP 74
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
NV+ L +++F ++EY + GEL++ + + G E A Y L A+ +CH
Sbjct: 75 NVVELREVMATKKKIFFVMEYVNGGELFEMIDRDGKLPEDLARKYFQQLISAVDFCHSRG 134
Query: 129 VIHRDIKPENLLLDHEGRLKIADFGWSV------------QSKDKRKTMCGTLDYLAPEM 176
V HRDIKPENLLLD EG LK+ DFG S S D T CGT Y+APE+
Sbjct: 135 VFHRDIKPENLLLDGEGDLKVTDFGLSALMMPEGLGGRRGSSDDLLHTRCGTPAYVAPEV 194
Query: 177 VENKTHDYAV-DNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAK 235
+ NK +D A+ D W+ GI+ Y L G PF E+ + +I K + FP P S ++K
Sbjct: 195 LRNKGYDGAMADIWSCGIVLYALLAGFLPFIDENVMTLYTKIFKAECEFP--PWFSLESK 252
Query: 236 NLISRLLVKDSSRRLSLQKIMEHPWIIKN 264
L+SRLLV D +R+S+ +I PW KN
Sbjct: 253 ELLSRLLVPDPEQRISMSEIKMIPWFRKN 281
>At4g30960 CBL-interacting protein kinase 6 (CIPK6)
Length = 441
Score = 182 bits (462), Expect = 2e-46
Identities = 94/264 (35%), Positives = 157/264 (58%), Gaps = 9/264 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHPN 69
+E+G+ LG G F +VY AR +++ VA+KV+ KE++ K Q++RE+ + +KHPN
Sbjct: 24 YELGRLLGHGTFAKVYHARNIQTGKSVAMKVVGKEKVVKVGMVDQIKREISVMRMVKHPN 83
Query: 70 VLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENHV 129
++ L+ +++ +E GEL+ +++K G E A Y L A+ +CH V
Sbjct: 84 IVELHEVMASKSKIYFAMELVRGGELFAKVAK-GRLREDVARVYFQQLISAVDFCHSRGV 142
Query: 130 IHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRK-----TMCGTLDYLAPEMVENKTHDY 184
HRD+KPENLLLD EG LK+ DFG S ++ ++ T CGT Y+APE++ K +D
Sbjct: 143 YHRDLKPENLLLDEEGNLKVTDFGLSAFTEHLKQDGLLHTTCGTPAYVAPEVILKKGYDG 202
Query: 185 A-VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLV 243
A D W+ G++ + L G PF+ ++ + + +I + D P +SSDA+ L+++LL
Sbjct: 203 AKADLWSCGVILFVLLAGYLPFQDDNLVNMYRKIYRGDFKCP--GWLSSDARRLVTKLLD 260
Query: 244 KDSSRRLSLQKIMEHPWIIKNANR 267
+ + R++++K+M+ PW K A R
Sbjct: 261 PNPNTRITIEKVMDSPWFKKQATR 284
>At3g01090 putative SNF1-related protein kinase
Length = 512
Score = 179 bits (455), Expect = 1e-45
Identities = 89/255 (34%), Positives = 154/255 (59%), Gaps = 5/255 (1%)
Query: 9 DFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHP 68
++++G+ LG G FGRV +A + VA+K++ + +++ + ++RRE++I HP
Sbjct: 18 NYKLGRTLGIGSFGRVKIAEHALTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFMHP 77
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
+++ LY ++L++EY ++GEL+ + ++G E +A + + + YCH N
Sbjct: 78 HIIRLYEVIETPTDIYLVMEYVNSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCHRNM 137
Query: 129 VIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKR--KTMCGTLDYLAPEMVENKTH-DYA 185
V+HRD+KPENLLLD + +KIADFG S +D KT CG+ +Y APE++ K +
Sbjct: 138 VVHRDLKPENLLLDSKCNVKIADFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLYAGPE 197
Query: 186 VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLVKD 245
VD W+ G++ Y L G PF+ E+ + F++IK + P +S A++LI R+LV D
Sbjct: 198 VDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSH--LSPGARDLIPRMLVVD 255
Query: 246 SSRRLSLQKIMEHPW 260
+R+++ +I +HPW
Sbjct: 256 PMKRVTIPEIRQHPW 270
>At5g45820 CBL-interacting protein kinase 20 (CIPK20)
Length = 439
Score = 178 bits (452), Expect = 2e-45
Identities = 98/269 (36%), Positives = 155/269 (57%), Gaps = 17/269 (6%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHPN 69
+E+G+ LG+G F +VY AR +K+ VA+KVI K+++ K Q++RE+ + ++HP+
Sbjct: 12 YELGRLLGQGTFAKVYHARNIKTGESVAIKVIDKQKVAKVGLIDQIKREISVMRLVRHPH 71
Query: 70 VLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENHV 129
V+ L+ +++ +EY GEL+ ++SK G E A Y L A+ YCH V
Sbjct: 72 VVFLHEVMASKTKIYFAMEYVKGGELFDKVSK-GKLKENIARKYFQQLIGAIDYCHSRGV 130
Query: 130 IHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRK-----TMCGTLDYLAPEMVENKTHDY 184
HRD+KPENLLLD G LKI+DFG S + K++ T CGT Y+APE++ K +D
Sbjct: 131 YHRDLKPENLLLDENGDLKISDFGLSALRESKQQDGLLHTTCGTPAYVAPEVIGKKGYDG 190
Query: 185 A-VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLS----FPPSPLVSSDAKNLIS 239
A D W+ G++ Y L G PF ++ + + +I K + FPP + K L+S
Sbjct: 191 AKADVWSCGVVLYVLLAGFLPFHEQNLVEMYRKITKGEFKCPNWFPP------EVKKLLS 244
Query: 240 RLLVKDSSRRLSLQKIMEHPWIIKNANRM 268
R+L + + R+ ++KIME+ W K ++
Sbjct: 245 RILDPNPNSRIKIEKIMENSWFQKGFKKI 273
>At5g01810 serine/threonine protein kinase ATPK10
Length = 421
Score = 178 bits (452), Expect = 2e-45
Identities = 95/260 (36%), Positives = 151/260 (57%), Gaps = 9/260 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHPN 69
+E+GK LG+G F +VY AR +K+ VA+KVI KE++ K Q++RE+ L+HPN
Sbjct: 12 YEVGKFLGQGTFAKVYHARHLKTGDSVAIKVIDKERILKVGMTEQIKREISAMRLLRHPN 71
Query: 70 VLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENHV 129
++ L+ +++ ++E+ GEL+ ++S G E A Y L A+ +CH V
Sbjct: 72 IVELHEVMATKSKIYFVMEHVKGGELFNKVS-TGKLREDVARKYFQQLVRAVDFCHSRGV 130
Query: 130 IHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRK-----TMCGTLDYLAPEMVENKTHD- 183
HRD+KPENLLLD G LKI+DFG S S +R+ T CGT Y+APE++ +D
Sbjct: 131 CHRDLKPENLLLDEHGNLKISDFGLSALSDSRRQDGLLHTTCGTPAYVAPEVISRNGYDG 190
Query: 184 YAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLV 243
+ D W+ G++ + L G PF + + +++I K ++ FP ++ AK L+ R+L
Sbjct: 191 FKADVWSCGVILFVLLAGYLPFRDSNLMELYKKIGKAEVKFP--NWLAPGAKRLLKRILD 248
Query: 244 KDSSRRLSLQKIMEHPWIIK 263
+ + R+S +KIM+ W K
Sbjct: 249 PNPNTRVSTEKIMKSSWFRK 268
>At3g29160 SNF1-like protein kinase (AKin11)
Length = 512
Score = 177 bits (450), Expect = 4e-45
Identities = 87/255 (34%), Positives = 154/255 (60%), Gaps = 5/255 (1%)
Query: 9 DFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHP 68
++++GK LG G FG+V +A V + VA+K++ + +++ + ++RRE++I HP
Sbjct: 19 NYKLGKTLGIGSFGKVKIAEHVVTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFMHP 78
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
+++ Y + +++++EY +GEL+ + ++G E +A + + + YCH N
Sbjct: 79 HIIRQYEVIETTSDIYVVMEYVKSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCHRNM 138
Query: 129 VIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKR--KTMCGTLDYLAPEMVENKTH-DYA 185
V+HRD+KPENLLLD +KIADFG S +D KT CG+ +Y APE++ K +
Sbjct: 139 VVHRDLKPENLLLDSRCNIKIADFGLSNVMRDGHFLKTSCGSPNYAAPEVISGKLYAGPE 198
Query: 186 VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLVKD 245
VD W+ G++ Y L G PF+ E+ + F++IK + P +SS+A++LI R+L+ D
Sbjct: 199 VDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSH--LSSEARDLIPRMLIVD 256
Query: 246 SSRRLSLQKIMEHPW 260
+R+++ +I +H W
Sbjct: 257 PVKRITIPEIRQHRW 271
>At1g01140 SOS2-like protein kinase PKS6/CBL-interacting protein
kinase 9 (CIPK9)
Length = 447
Score = 177 bits (450), Expect = 4e-45
Identities = 89/263 (33%), Positives = 155/263 (58%), Gaps = 8/263 (3%)
Query: 7 INDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLK 66
+ ++E+G+ LG+G F +V A+ + A+K++ +E++ ++K QL+RE+ +K
Sbjct: 16 VGNYEMGRTLGEGSFAKVKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREISTMKLIK 75
Query: 67 HPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHE 126
HPNV+ + +++++LE + GEL+ +++++G E +A Y L A+ YCH
Sbjct: 76 HPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINAVDYCHS 135
Query: 127 NHVIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRK-----TMCGTLDYLAPEMVENKT 181
V HRD+KPENL+LD G LK++DFG S S+ R+ T CGT +Y+APE++ +K
Sbjct: 136 RGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPEVLSDKG 195
Query: 182 HD-YAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISR 240
+D A D W+ G++ + + G PF+ + ++RI K + S P P S AK +I R
Sbjct: 196 YDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRICKAEFSCP--PWFSQGAKRVIKR 253
Query: 241 LLVKDSSRRLSLQKIMEHPWIIK 263
+L + R+S+ +++E W K
Sbjct: 254 ILEPNPITRISIAELLEDEWFKK 276
>At1g61950 hypothetical protein
Length = 547
Score = 176 bits (447), Expect = 9e-45
Identities = 96/261 (36%), Positives = 153/261 (57%), Gaps = 10/261 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLK-HP 68
+ +G+ LG+G+FG Y+ E+ S A K I K +L + K +RRE++I L P
Sbjct: 98 YSLGRELGRGQFGITYICTEISSGKNFACKSILKRKLIRTKDREDVRREIQIMHYLSGQP 157
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
N++ + G + D + V L++E GEL+ +++KRGH+SEK AA I S+ + + CH
Sbjct: 158 NIVEIKGAYEDRQSVHLVMELCEGGELFDKITKRGHYSEKAAAEIIRSVVKVVQICHFMG 217
Query: 129 VIHRDIKPENLLL----DHEGRLKIADFGWSVQSKDKR--KTMCGTLDYLAPEMVENKTH 182
VIHRD+KPEN LL + LK DFG SV ++ + + + G+ Y+APE+++ + +
Sbjct: 218 VIHRDLKPENFLLSSKDEASSMLKATDFGVSVFIEEGKVYEDIVGSAYYVAPEVLK-RNY 276
Query: 183 DYAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKK--VDLSFPPSPLVSSDAKNLISR 240
A+D W+ G++ Y L G PPF AE+ FE I + +D P P +S AK+L+
Sbjct: 277 GKAIDIWSAGVILYILLCGNPPFWAETDKGIFEEILRGEIDFESEPWPSISESAKDLVRN 336
Query: 241 LLVKDSSRRLSLQKIMEHPWI 261
+L D +R + +++EHPWI
Sbjct: 337 MLKYDPKKRFTAAQVLEHPWI 357
>At2g41860 putative Ca2+ dependent protein kinase
Length = 530
Score = 176 bits (445), Expect = 2e-44
Identities = 95/269 (35%), Positives = 162/269 (59%), Gaps = 9/269 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISL-KHP 68
+++G+ LG+G+FG Y+ E+++ + A K I K++L+ ++RE+EI + +HP
Sbjct: 54 YKLGRELGRGEFGVTYLCTEIETGEIFACKSILKKKLKTSIDIEDVKREVEIMRQMPEHP 113
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
N++ L + D + V L++E GEL+ + RGH++E+ AA+ I ++ E + CH++
Sbjct: 114 NIVTLKETYEDDKAVHLVMELCEGGELFDRIVARGHYTERAAASVIKTIIEVVQMCHKHG 173
Query: 129 VIHRDIKPENLLLDHE---GRLKIADFGWSVQSK--DKRKTMCGTLDYLAPEMVENKTHD 183
V+HRD+KPEN L ++ LK DFG SV K ++ + G+ Y+APE++ +++
Sbjct: 174 VMHRDLKPENFLFANKKETASLKAIDFGLSVFFKPGERFNEIVGSPYYMAPEVL-RRSYG 232
Query: 184 YAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKK--VDLSFPPSPLVSSDAKNLISRL 241
+D W+ G++ Y L GVPPF AE++ + I K +D P P VS +AK+LI ++
Sbjct: 233 QEIDIWSAGVILYILLCGVPPFWAETEHGVAKAILKSVIDFKRDPWPKVSDNAKDLIKKM 292
Query: 242 LVKDSSRRLSLQKIMEHPWIIKNANRMGV 270
L D RRL+ Q++++HPWI N V
Sbjct: 293 LHPDPRRRLTAQQVLDHPWIQNGKNASNV 321
>At4g24400 serine/threonine kinase-like protein
Length = 445
Score = 175 bits (444), Expect = 2e-44
Identities = 91/262 (34%), Positives = 153/262 (57%), Gaps = 6/262 (2%)
Query: 7 INDFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLK 66
+ +E+G+ +G+G F +V A+ ++ VA+K++ + + K K Q++RE+ I ++
Sbjct: 6 VGKYELGRTIGEGTFAKVKFAQNTETGESVAMKIVDRSTIIKRKMVDQIKREISIMKLVR 65
Query: 67 HPNVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHE 126
HP V+ LY ++++ILEY GEL+ ++ + G SE +A Y L + + YCH
Sbjct: 66 HPCVVRLYEVLASRTKIYIILEYITGGELFDKIVRNGRLSESEARKYFHQLIDGVDYCHS 125
Query: 127 NHVIHRDIKPENLLLDHEGRLKIADFGWSV---QSKDKRKTMCGTLDYLAPEMVENKTHD 183
V HRD+KPENLLLD +G LKI+DFG S Q KT CGT +Y+APE++ +K ++
Sbjct: 126 KGVYHRDLKPENLLLDSQGNLKISDFGLSALPEQGVTILKTTCGTPNYVAPEVLSHKGYN 185
Query: 184 YAV-DNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLL 242
AV D W+ G++ Y + G PF+ + +I K + S P + AK+LI+R+L
Sbjct: 186 GAVADIWSCGVILYVLMAGYLPFDEMDLPTLYSKIDKAEFSCP--SYFALGAKSLINRIL 243
Query: 243 VKDSSRRLSLQKIMEHPWIIKN 264
+ R+++ +I + W +K+
Sbjct: 244 DPNPETRITIAEIRKDEWFLKD 265
>At4g18700 putative protein kinase
Length = 489
Score = 174 bits (441), Expect = 5e-44
Identities = 95/260 (36%), Positives = 151/260 (57%), Gaps = 9/260 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHPN 69
+E+GK LG G F +VY+AR VK+ VA+KVI KE++ K ++RE+ I ++HPN
Sbjct: 26 YEMGKLLGHGTFAKVYLARNVKTNESVAIKVIDKEKVLKGGLIAHIKREISILRRVRHPN 85
Query: 70 VLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENHV 129
++ L+ +++ ++EY GEL+ +++K G E+ A Y L A+T+CH V
Sbjct: 86 IVQLFEVMATKAKIYFVMEYVRGGELFNKVAK-GRLKEEVARKYFQQLISAVTFCHARGV 144
Query: 130 IHRDIKPENLLLDHEGRLKIADFGWSVQSKDKRK-----TMCGTLDYLAPEMVENKTHDY 184
HRD+KPENLLLD G LK++DFG S S R+ T CGT Y+APE++ K +D
Sbjct: 145 YHRDLKPENLLLDENGNLKVSDFGLSAVSDQIRQDGLFHTFCGTPAYVAPEVLARKGYDA 204
Query: 185 A-VDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLV 243
A VD W+ G++ + + G PF + +++I + + P S++ L+S+LL
Sbjct: 205 AKVDIWSCGVILFVLMAGYLPFHDRNVMAMYKKIYRGEFRCP--RWFSTELTRLLSKLLE 262
Query: 244 KDSSRRLSLQKIMEHPWIIK 263
+ +R + +IME+ W K
Sbjct: 263 TNPEKRFTFPEIMENSWFKK 282
>At4g04720 putative calcium dependent protein kinase
Length = 531
Score = 174 bits (440), Expect = 6e-44
Identities = 90/260 (34%), Positives = 154/260 (58%), Gaps = 9/260 (3%)
Query: 10 FEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLK-HP 68
+ +GK LG+G+FG Y+ +E+ + A K I K +L + ++RE++I L P
Sbjct: 80 YSLGKELGRGQFGITYMCKEIGTGNTYACKSILKRKLISKQDKEDVKREIQIMQYLSGQP 139
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
N++ + G + D + + L++E GEL+ + +GH+SE+ AA I S+ + CH
Sbjct: 140 NIVEIKGAYEDRQSIHLVMELCAGGELFDRIIAQGHYSERAAAGIIRSIVNVVQICHFMG 199
Query: 129 VIHRDIKPENLLL---DHEGRLKIADFGWSVQSKDKR--KTMCGTLDYLAPEMVENKTHD 183
V+HRD+KPEN LL + LK DFG SV ++ + + + G+ Y+APE++ +++
Sbjct: 200 VVHRDLKPENFLLSSKEENAMLKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVL-RRSYG 258
Query: 184 YAVDNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSF--PPSPLVSSDAKNLISRL 241
+D W+ G++ Y L GVPPF AE++ F+ + K ++ F P P +S AK+L+ ++
Sbjct: 259 KEIDIWSAGVILYILLSGVPPFWAENEKGIFDEVIKGEIDFVSEPWPSISESAKDLVRKM 318
Query: 242 LVKDSSRRLSLQKIMEHPWI 261
L KD RR++ +++EHPWI
Sbjct: 319 LTKDPKRRITAAQVLEHPWI 338
>At5g39440 serine/threonine-specific protein kinase -like protein
Length = 494
Score = 173 bits (439), Expect = 8e-44
Identities = 86/254 (33%), Positives = 149/254 (57%), Gaps = 4/254 (1%)
Query: 9 DFEIGKPLGKGKFGRVYVAREVKSKFVVALKVIFKEQLEKYKFHHQLRREMEIQISLKHP 68
++ IGK LG G F +V +A V + VA+K++ + +++ +++RE++I L HP
Sbjct: 18 NYRIGKTLGHGSFAKVKLALHVATGHKVAIKILNRSKIKNMGIEIKVQREIKILRFLMHP 77
Query: 69 NVLHLYGWFHDSERVFLILEYAHNGELYKELSKRGHFSEKQAATYILSLTEALTYCHENH 128
+++ Y +++++EY +GEL+ + ++G E +A + + YCH N
Sbjct: 78 HIIRQYEVIETPNDIYVVMEYVKSGELFDYIVEKGKLQEDEARHLFQQIISGVEYCHRNM 137
Query: 129 VIHRDIKPENLLLDHEGRLKIADFGWSVQSKDKR--KTMCGTLDYLAPEMVENKTHDYAV 186
++HRD+KPEN+LLD + +KI DFG S D KT CG+ +Y APE++ K + V
Sbjct: 138 IVHRDLKPENVLLDSQCNIKIVDFGLSNVMHDGHFLKTSCGSPNYAAPEVISGKPYGPDV 197
Query: 187 DNWTLGILCYEFLYGVPPFEAESQADTFERIKKVDLSFPPSPLVSSDAKNLISRLLVKDS 246
D W+ G++ Y L G PF+ E+ + FE+IK+ + P +S A++LI R+L+ D
Sbjct: 198 DIWSCGVILYALLCGTLPFDDENIPNVFEKIKRGMYTLPNH--LSHFARDLIPRMLMVDP 255
Query: 247 SRRLSLQKIMEHPW 260
+ R+S+ +I +HPW
Sbjct: 256 TMRISITEIRQHPW 269
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,193,825
Number of Sequences: 26719
Number of extensions: 261557
Number of successful extensions: 3532
Number of sequences better than 10.0: 1006
Number of HSP's better than 10.0 without gapping: 922
Number of HSP's successfully gapped in prelim test: 84
Number of HSP's that attempted gapping in prelim test: 673
Number of HSP's gapped (non-prelim): 1112
length of query: 271
length of database: 11,318,596
effective HSP length: 98
effective length of query: 173
effective length of database: 8,700,134
effective search space: 1505123182
effective search space used: 1505123182
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149307.9


