

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149307.14 - phase: 0
(629 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
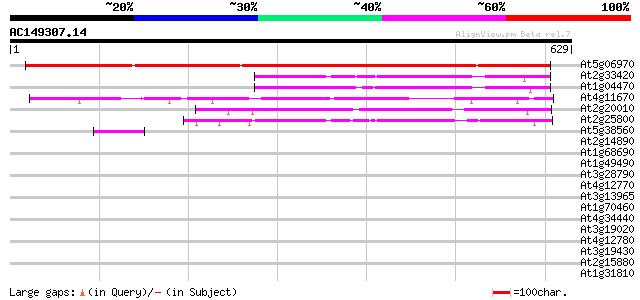
At5g06970 unknown protein 830 0.0
At2g33420 unknown protein 205 7e-53
At1g04470 unknown protein 201 1e-51
At4g11670 hypothetical protein 195 6e-50
At2g20010 unknown protein 195 6e-50
At2g25800 unknown protein 190 2e-48
At5g38560 putative protein 44 4e-04
At2g14890 arabinogalactan-protein AGP9 41 0.002
At1g68690 protein kinase, putative 40 0.003
At1g49490 hypothetical protein 39 0.007
At3g28790 unknown protein 39 0.009
At4g12770 auxilin-like protein 38 0.015
At3g13965 unknown protein 38 0.019
At1g70460 putative protein kinase 38 0.019
At4g34440 serine/threonine protein kinase - like 37 0.025
At3g19020 hypothetical protein 37 0.025
At4g12780 auxilin-like protein 37 0.033
At3g19430 putative late embryogenesis abundant protein 37 0.043
At2g15880 unknown protein 37 0.043
At1g31810 hypothetical protein 37 0.043
>At5g06970 unknown protein
Length = 1101
Score = 830 bits (2143), Expect = 0.0
Identities = 422/592 (71%), Positives = 497/592 (83%), Gaps = 10/592 (1%)
Query: 18 ENAIDLLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSE 77
ENA+++LQRYRRDRR LLDF+L+GSLIKKV+MPPGAVTLDDVDLDQVS+DYV+NCAKK
Sbjct: 4 ENAVEILQRYRRDRRKLLDFMLAGSLIKKVIMPPGAVTLDDVDLDQVSVDYVINCAKKGG 63
Query: 78 MLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVP--ISAVPPIA 135
MLEL+EAIRDYHDH GLP M+ G+ EF+L T PESSGSPPKRAPPP+P IS+ P+
Sbjct: 64 MLELAEAIRDYHDHIGLPYMNSVGTADEFFLATIPESSGSPPKRAPPPIPVLISSSSPMV 123
Query: 136 VSTPPPAYPTSPVASNISRSESLYSAQERELTVDDIEDFEDDDDTSMVEGLR-AKRTLND 194
+ P + SP A + RSES S + +ELTVDDI+DFEDDDD V R ++RT ND
Sbjct: 124 TN---PEWCESPSAPPLMRSESFDSPKAQELTVDDIDDFEDDDDLDEVGNFRISRRTAND 180
Query: 195 ASDLAVKLPPFSTGITDDDLRETAYEILLACAGATGGLIVPSKEKKKDRKSSSLIRKLGR 254
A+DL +LP F+TGITDDDLRETA+EILLACAGA+GGLIVPSKEKKK++ S LI+KLGR
Sbjct: 181 AADLVPRLPSFATGITDDDLRETAFEILLACAGASGGLIVPSKEKKKEKSRSRLIKKLGR 240
Query: 255 SKTGSIVSQSQNAPGLVGLLESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPL 314
VSQSQ++ GLV LLE MR Q+EISEAMDIRT+QGLLNAL GK GKRMD+LLVPL
Sbjct: 241 KSES--VSQSQSSSGLVSLLEMMRGQMEISEAMDIRTRQGLLNALAGKVGKRMDSLLVPL 298
Query: 315 ELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEES 374
ELLCCV+RTEFSDKKA++RWQKRQL +L EGL+N+PVVGFGESGRK +++ LL +IEES
Sbjct: 299 ELLCCVSRTEFSDKKAYLRWQKRQLNMLAEGLINNPVVGFGESGRKATDLKSLLLRIEES 358
Query: 375 EFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSV 434
E LPSS+GE+QR ECL+SLRE+AI LAERPARGDLTGE+CHWADGY NVRLYEKLLL V
Sbjct: 359 ESLPSSAGEVQRAECLKSLREVAISLAERPARGDLTGEVCHWADGYHLNVRLYEKLLLCV 418
Query: 435 FDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVLFRQYVITREHRILLHALE 494
FD+L++GKLTEEVEEILELLKSTWRVLGITETIH+TCYAWVLFRQYVIT E +L HA++
Sbjct: 419 FDILNDGKLTEEVEEILELLKSTWRVLGITETIHYTCYAWVLFRQYVITSERGLLRHAIQ 478
Query: 495 QLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEG 554
QL KIPL EQRG QERLHLK+L+ +V+ E ++SFL++FL+PI+ WADKQLGDYHLHF+EG
Sbjct: 479 QLKKIPLKEQRGPQERLHLKTLKCRVDNE-EISFLESFLSPIRSWADKQLGDYHLHFAEG 537
Query: 555 SAIMEKIVAVAMITRRLLLEEPDTSTQSLPISDRDQIEVYITSSIKHAFTRV 606
S +ME V VAMIT RLLLEE D + S SDR+QIE Y+ SSIK+ FTR+
Sbjct: 538 SLVMEDTVTVAMITWRLLLEESDRAMHS-NSSDREQIESYVLSSIKNTFTRM 588
>At2g33420 unknown protein
Length = 1039
Score = 205 bits (521), Expect = 7e-53
Identities = 123/344 (35%), Positives = 197/344 (56%), Gaps = 32/344 (9%)
Query: 275 ESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRW 334
E MR Q++++E D R ++ LL LVG+ G+R +T+++PLELL + +EF D + W
Sbjct: 179 EIMRQQMKVTEQSDSRLRKTLLRTLVGQTGRRAETIILPLELLRHLKTSEFGDIHEYQLW 238
Query: 335 QKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKI-EESEFLPSSSGELQRTECLRSL 393
Q+RQLKVLE GL+ HP + KTN + L ++ +SE P + + T +R+L
Sbjct: 239 QRRQLKVLEAGLLLHPSIPLD----KTNNFAMRLREVVRQSETKPIDTSKTSDT--MRTL 292
Query: 394 REIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILEL 453
+ + L+ R G+ T ++CHWADGY N+ LY LL S+FD+ DE + +E++E+LEL
Sbjct: 293 TNVVVSLSWRGTNGNPT-DVCHWADGYPLNIHLYVALLQSIFDVRDETLVLDEIDELLEL 351
Query: 454 LKSTWRVLGITETIHHTCYAWVLFRQYVIT--REHRILLHALEQLNKIPLMEQRGQQERL 511
+K TW LGIT IH+ C+ WVLF QYV+T E +L + L ++ ++ +E L
Sbjct: 352 MKKTWSTLGITRPIHNLCFTWVLFHQYVVTSQMEPDLLGASHAMLAEVANDAKKLDREAL 411
Query: 512 HLKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGS-AIMEKIVAVAMITRR 570
++K L S L +Q W +K+L YH +F G+ ++E ++ +A+ + R
Sbjct: 412 YVKLLNST-------------LASMQGWTEKRLLSYHDYFQRGNVGLIENLLPLALSSSR 458
Query: 571 LLLEE--------PDTSTQSLPISDRDQIEVYITSSIKHAFTRV 606
+L E+ + L D+++ YI SSIK+AF++V
Sbjct: 459 ILGEDVTISQGKGQEKGDVKLVDHSGDRVDYYIRSSIKNAFSKV 502
>At1g04470 unknown protein
Length = 1035
Score = 201 bits (510), Expect = 1e-51
Identities = 118/343 (34%), Positives = 194/343 (56%), Gaps = 31/343 (9%)
Query: 275 ESMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRW 334
E MR Q++++E D R ++ L+ LVG+ G+R +T+++PLELL V +EF D + W
Sbjct: 177 EIMRQQMKVTEQSDTRLRKTLMRTLVGQTGRRAETIILPLELLRHVKPSEFGDVHEYQIW 236
Query: 335 QKRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLR 394
Q+RQLKVLE GL+ HP + ++ +R ++ + E S + ++ T C
Sbjct: 237 QRRQLKVLEAGLLIHPSIPLEKTNNFAMRLREIIRQSETKAIDTSKNSDIMPTLC----- 291
Query: 395 EIAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELL 454
+ L+ R A T +ICHWADGY N+ LY LL S+FD+ DE + +E++E+LEL+
Sbjct: 292 NLVASLSWRNATP--TTDICHWADGYPLNIHLYVALLQSIFDIRDETLVLDEIDELLELM 349
Query: 455 KSTWRVLGITETIHHTCYAWVLFRQYVIT--REHRILLHALEQLNKIPLMEQRGQQERLH 512
K TW +LGIT IH+ C+ WVLF QY++T E +L + L ++ ++ +E L+
Sbjct: 350 KKTWIMLGITRAIHNLCFTWVLFHQYIVTSQMEPDLLGASHAMLAEVANDAKKSDREALY 409
Query: 513 LKSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGS-AIMEKIVAVAMITRRL 571
+K L S L +Q W +K+L YH +F G+ ++E ++ +A+ + ++
Sbjct: 410 VKLLTST-------------LASMQGWTEKRLLSYHDYFQRGNVGLIENLLPLALSSSKI 456
Query: 572 LLEEPDTSTQS--------LPISDRDQIEVYITSSIKHAFTRV 606
L E+ S + L S D+++ YI +SIK+AF++V
Sbjct: 457 LGEDVTISQMNGLEKGDVKLVDSSGDRVDYYIRASIKNAFSKV 499
>At4g11670 hypothetical protein
Length = 998
Score = 195 bits (496), Expect = 6e-50
Identities = 166/619 (26%), Positives = 267/619 (42%), Gaps = 130/619 (21%)
Query: 23 LLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVT-LDDVDLDQVSIDYVLNCAKKS----- 76
LLQRYR DRR L++F++S L+K++ P G+ T L DLD +S DYVL+C K
Sbjct: 4 LLQRYRNDRRKLMEFLMSSGLVKELRSPSGSPTSLSPADLDALSADYVLDCVKSEIAETL 63
Query: 77 --------EMLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPI 128
++++S+ Y+ + P + S ++LV+ P+ +GSPP R PPP
Sbjct: 64 HFVNTNAGGVVDVSKGREKYNFDSSYPVTIHSESGDSYFLVSSPDLAGSPPHRMPPP--- 120
Query: 129 SAVPPIAVSTPPPAYPTSPVASNISRSESLYSAQERELTVDDIEDFEDD----DDTSMVE 184
PV NI +S + + R + + D+ ++T ++
Sbjct: 121 ------------------PV--NIEKSSNNGADMSRHMDSSNTPSARDNYVFKEETPDIK 160
Query: 185 GLRAKRTLNDASDLAVKLPPFSTGITDDDLRETAYEILLAC----AGATGGLIVPSKEKK 240
++ + + + LPP TG++DDDLRE AYE+++A + T + ++
Sbjct: 161 PVKPIKII------PLGLPPLRTGLSDDDLREAAYELMIASMLLSSFLTNSVEAYPTHRR 214
Query: 241 KDRKSSSLIRKLGRSKTGSIVSQSQNAPGLVGLLESMRVQLEISEAMDIRTKQGLLNALV 300
K KSS L+ L R + Q N EIS MD ++ L+
Sbjct: 215 KIEKSSRLMLSLKRKDKPHLQPQISNTHS------------EISSKMDTCIRRNLVQLAT 262
Query: 301 GKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRK 360
+ G+++D + L LL + +++F ++K +++W+ RQ +LEE L P + E
Sbjct: 263 LRTGEQIDLPQLALGLLVGIFKSDFPNEKLYMKWKTRQANLLEEVLCFSPSLEKNERAT- 321
Query: 361 TNEMRILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGY 420
MR LA I +S+ R E L S+R++A L+ P R + E +W Y
Sbjct: 322 ---MRKCLATIRDSKEWDVVVSASLRIEVLSSIRQVASKLSSLPGRCGIEEETYYWTAIY 378
Query: 421 QFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWVLFRQY 480
N+RLYEKLL VFD LDEG +E++
Sbjct: 379 HLNIRLYEKLLFGVFDTLDEGSTIQELQ-------------------------------- 406
Query: 481 VITREHRILLHALEQLNKIPLMEQRGQQERLHLKSL---RSKVEGERDMSFLQAFLTPIQ 537
K+ E +E L+L L R + + + ++A LT +
Sbjct: 407 -----------------KVTSAESGNPKEDLYLSHLVCSRQTIGTDIHLGLVKAILTSVS 449
Query: 538 RWADKQLGDYHLHFSEGSAIMEKIVAVAMIT-------RRLLLEEPDTSTQSLPISDRDQ 590
W D +L DYHLHF + +V +A R L + DT + + D+
Sbjct: 450 AWCDDKLQDYHLHFGKKPRDFGMLVRLASTVGLPPADCTRTELIKLDTLSDDV----SDK 505
Query: 591 IEVYITSSIKHAFTRVRVF 609
I+ Y+ +SIK A R F
Sbjct: 506 IQSYVQNSIKGACARAAHF 524
>At2g20010 unknown protein
Length = 952
Score = 195 bits (496), Expect = 6e-50
Identities = 135/437 (30%), Positives = 224/437 (50%), Gaps = 57/437 (13%)
Query: 209 ITDDDLRETAYEILLACAGATGG--LIVPSKEKKKDRK------SSSLIRKLGRSKTGSI 260
+++ +LRETAYEIL+A +TG L + K DR S S L RS T +
Sbjct: 15 LSNSELRETAYEILVAACRSTGSRPLTYIPQSPKSDRSNGLTTASLSPSPSLHRSLTSTA 74
Query: 261 VSQSQNAPGL------------------------VGLLESMRVQLEISEAMDIRTKQGLL 296
S+ + A G+ V + E +RVQ+ ISE +D R ++ LL
Sbjct: 75 ASKVKKALGMKKRIGDGDGGAGESSSQPDRSKKSVTVGELVRVQMRISEQIDSRIRRALL 134
Query: 297 NALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGE 356
G+ G+R++ +++PLELL + ++F D++ + WQ+R LK+LE GL+ +P V +
Sbjct: 135 RIASGQLGRRVEMMVLPLELLQQLKASDFPDQEEYESWQRRNLKLLEAGLILYPCVPLSK 194
Query: 357 SGRKTNEMR-ILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICH 415
S + +++ I+ + +E +GE Q +LR + + LA R + E CH
Sbjct: 195 SDKSVQQLKQIIRSGLERPLDTGKITGETQ------NLRSLVMSLASRQNNNGIGSETCH 248
Query: 416 WADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLKSTWRVLGITETIHHTCYAWV 475
WADG+ N+R+Y+ LL S FD+ DE + EEV+E+LEL+K TW VLGI + IH+ C+ WV
Sbjct: 249 WADGFPLNLRIYQMLLESCFDVNDELLIVEEVDEVLELIKKTWPVLGINQMIHNVCFLWV 308
Query: 476 LFRQYVITRE-HRILLHALEQLNKIPLMEQRGQQERLHLKSLRSKVEGERDMSFLQAFLT 534
L +YV T + LL A L L +++ + L + L+
Sbjct: 309 LVNRYVSTGQVENDLLVAAHNL-------------ILEIENDAMETNDPEYSKILSSVLS 355
Query: 535 PIQRWADKQLGDYHLHFS-EGSAIMEKIVAVAMITRRLLLEEPDTS---TQSLPISDRDQ 590
+ W +K+L YH F+ + +E V++ ++ ++L E+ + + S RD+
Sbjct: 356 LVMDWGEKRLLAYHDTFNIDNVETLETTVSLGILVAKVLGEDISSEYRRKKKHVDSGRDR 415
Query: 591 IEVYITSSIKHAFTRVR 607
++ YI SS++ AF + +
Sbjct: 416 VDTYIRSSLRMAFQQTK 432
>At2g25800 unknown protein
Length = 993
Score = 190 bits (483), Expect = 2e-48
Identities = 147/460 (31%), Positives = 232/460 (49%), Gaps = 70/460 (15%)
Query: 195 ASDLAVKLPPFSTG-----ITDDDLRETAYEILLA-CAGATGGLI--------------- 233
+S ++ LPP G ++D DLR TAYEI +A C ATG +
Sbjct: 30 SSSMSSDLPPSPLGQLAVQLSDSDLRLTAYEIFVAACRSATGKPLSSAVSVAVLNQDSPN 89
Query: 234 ---------------VPSKEKKKDRKSSSLIRKLGRSKTGSIVSQSQNA---PGLVGLLE 275
SK KK SS G +K+ S S P VG E
Sbjct: 90 GSPASPAIQRSLTSTAASKMKKALGLRSSSSLSPGSNKSSGSASGSNGKSKRPTTVG--E 147
Query: 276 SMRVQLEISEAMDIRTKQGLLNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQ 335
MR+Q+ +SEA+D R ++ L + G++++++++PLELL + ++F+D++ + W
Sbjct: 148 LMRIQMRVSEAVDSRVRRAFLRIAASQVGRKIESVVLPLELLQQLKSSDFTDQQEYDAWL 207
Query: 336 KRQLKVLEEGLVNHPVVGFGESGRKTNEMRILLAKIEESEFLPSSSGELQRTECLRSLRE 395
KR LKVLE GL+ HP V KTN + L I + P +G + E ++SLR
Sbjct: 208 KRSLKVLEAGLLLHPRVPLD----KTNSSQRLRQIIHGALDRPLETG--RNNEQMQSLRS 261
Query: 396 IAIPLAERPARGDLTGEICHWADGYQFNVRLYEKLLLSVFDMLDEGKLTEEVEEILELLK 455
+ LA R + G + + CHWADG FN+RLYE LL + FD D + EEV++++E +K
Sbjct: 262 AVMSLATR-SDGSFS-DSCHWADGSPFNLRLYELLLEACFDSNDATSMVEEVDDLMEHIK 319
Query: 456 STWRVLGITETIHHTCYAWVLFRQYVITREHRI-LLHALE-QLNKIPLMEQRGQQERLHL 513
TW +LGI + +H+ C+ W+LF +YV+T + + LLHA + QL ++
Sbjct: 320 KTWVILGINQMLHNLCFTWLLFSRYVVTGQVEMDLLHACDSQLAEV-------------A 366
Query: 514 KSLRSKVEGERDMSFLQAFLTPIQRWADKQLGDYHLHFSEGSA-IMEKIVAVAMITRRLL 572
K ++ + E L + L+ I WA+K+L YH F G+ ME IV++ + R+L
Sbjct: 367 KDAKTTKDPEYS-QVLSSTLSAILGWAEKRLLAYHDTFDRGNIHTMEGIVSLGVSAARIL 425
Query: 573 LEEPDTSTQSLPISD----RDQIEVYITSSIKHAFTRVRV 608
+E+ + + R +IE YI SS++ +F + +
Sbjct: 426 VEDISNEYRRRRKGEVDVARTRIETYIRSSLRTSFAQASI 465
>At5g38560 putative protein
Length = 681
Score = 43.5 bits (101), Expect = 4e-04
Identities = 20/57 (35%), Positives = 30/57 (52%)
Query: 95 PQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P S++ + L T P + +PP PPP P + PP+ S+PPP +SP S+
Sbjct: 12 PPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSS 68
Score = 35.4 bits (80), Expect = 0.097
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 7/49 (14%)
Query: 108 LVTDPESSGSPPKRAP------PPVPISAVPPIAVSTPPPAYP-TSPVA 149
+V+ P S SPP P P V S PP+ +++PPP+ P T+P A
Sbjct: 60 VVSSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPA 108
Score = 33.9 bits (76), Expect = 0.28
Identities = 19/42 (45%), Positives = 23/42 (54%), Gaps = 3/42 (7%)
Query: 112 PESSGSPPKRAPPPVP--ISAVPP-IAVSTPPPAYPTSPVAS 150
P S PP +PPP P I++ PP +A S PPP SP S
Sbjct: 59 PVVSSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPS 100
Score = 31.2 bits (69), Expect = 1.8
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 4/44 (9%)
Query: 112 PESSGSPPKRAP-PPVPISAVPPIAVST---PPPAYPTSPVASN 151
P + SPP P PP P + P +T PPPA SP +SN
Sbjct: 142 PGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPPATSASPPSSN 185
Score = 30.8 bits (68), Expect = 2.4
Identities = 18/48 (37%), Positives = 22/48 (45%), Gaps = 12/48 (25%)
Query: 112 PESSGSPPKR--------APPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P +S SPP APPP P+ VP P A PT P ++N
Sbjct: 175 PATSASPPSSNPTDPSTLAPPPTPLPVVP----REKPIAKPTGPASNN 218
Score = 29.3 bits (64), Expect = 6.9
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query: 112 PESSGSPPKRAP-PPVPISAVPPIAVSTPPPAYPTSP 147
P+ S SPP P PP + P + STP P TSP
Sbjct: 135 PKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSP 171
Score = 29.3 bits (64), Expect = 6.9
Identities = 12/36 (33%), Positives = 19/36 (52%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P ++ PPK +P P + PP +PP P++P
Sbjct: 128 PTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTP 163
Score = 28.9 bits (63), Expect = 9.1
Identities = 15/41 (36%), Positives = 21/41 (50%), Gaps = 5/41 (12%)
Query: 112 PESSGSPPKRAPP-PVPISAV----PPIAVSTPPPAYPTSP 147
P + SPPK +P P P + PP ++PP + PT P
Sbjct: 149 PGETPSPPKPSPSTPTPTTTTSPPPPPATSASPPSSNPTDP 189
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 40.8 bits (94), Expect = 0.002
Identities = 20/41 (48%), Positives = 24/41 (57%), Gaps = 2/41 (4%)
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
T P ++ PP APPPV S PP + PPPA P PV+S
Sbjct: 38 TPPPAATPPPVSAPPPVTTS--PPPVTTAPPPANPPPPVSS 76
Score = 34.7 bits (78), Expect = 0.17
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 3/46 (6%)
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIA--VSTPPP-AYPTSPVAS 150
+ T P + P A PP P+S+ PP + +TPPP A P PVAS
Sbjct: 54 VTTSPPPVTTAPPPANPPPPVSSPPPASPPPATPPPVASPPPPVAS 99
Score = 33.9 bits (76), Expect = 0.28
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P + +PP A PP P+SA PP V+T PP T+P +N
Sbjct: 33 PPTPTTPPPAATPP-PVSAPPP--VTTSPPPVTTAPPPAN 69
Score = 32.7 bits (73), Expect = 0.63
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 8/46 (17%)
Query: 112 PESSGSPPKRAPPPV-------PISAVPPIAVSTPPPAYPTSPVAS 150
P ++ PP +PPP P A PP V++PPPA P PVA+
Sbjct: 66 PPANPPPPVSSPPPASPPPATPPPVASPPPPVASPPPATP-PPVAT 110
Score = 32.7 bits (73), Expect = 0.63
Identities = 22/77 (28%), Positives = 29/77 (37%), Gaps = 1/77 (1%)
Query: 109 VTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYSAQERELTV 168
V P + PP PPP P+ A PP V P P + + S S L S+ +
Sbjct: 97 VASPPPATPPPVATPPPAPL-ASPPAQVPAPAPTTKPDSPSPSPSSSPPLPSSDAPGPST 155
Query: 169 DDIEDFEDDDDTSMVEG 185
D I D + G
Sbjct: 156 DSISPAPSPTDVNDQNG 172
Score = 32.3 bits (72), Expect = 0.82
Identities = 21/41 (51%), Positives = 22/41 (53%), Gaps = 8/41 (19%)
Query: 117 SPPKRAPPPV----PISAVPPIA----VSTPPPAYPTSPVA 149
SPP PPPV P A PP A V+TPPPA SP A
Sbjct: 81 SPPPATPPPVASPPPPVASPPPATPPPVATPPPAPLASPPA 121
Score = 30.8 bits (68), Expect = 2.4
Identities = 18/50 (36%), Positives = 22/50 (44%), Gaps = 3/50 (6%)
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSES 157
+ + P SPP PPPV A PP A PPA +P + S S
Sbjct: 90 VASPPPPVASPPPATPPPV---ATPPPAPLASPPAQVPAPAPTTKPDSPS 136
>At1g68690 protein kinase, putative
Length = 708
Score = 40.4 bits (93), Expect = 0.003
Identities = 21/46 (45%), Positives = 26/46 (55%), Gaps = 3/46 (6%)
Query: 109 VTDP-ESSGSPPKRAPPPVP--ISAVPPIAVSTPPPAYPTSPVASN 151
VT P S PP RAPPP P ++ PP+A PPP P P +S+
Sbjct: 35 VTSPLPPSAPPPNRAPPPPPPVTTSPPPVANGAPPPPLPKPPESSS 80
Score = 35.4 bits (80), Expect = 0.097
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPV 148
++ P S SPP PP+P S PP +V P P+ P+ P+
Sbjct: 101 VIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPS-PSPPI 140
Score = 34.7 bits (78), Expect = 0.17
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 3/39 (7%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPP---PAYPTSP 147
P PP +PPP P+ PP + S PP P P+SP
Sbjct: 85 PVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSP 123
Score = 33.9 bits (76), Expect = 0.28
Identities = 18/35 (51%), Positives = 20/35 (56%), Gaps = 1/35 (2%)
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPA 142
LV SS PP PPP P S PPI V +PPP+
Sbjct: 115 LVPPLPSSPPPPASVPPPRP-SPSPPILVRSPPPS 148
Score = 33.1 bits (74), Expect = 0.48
Identities = 21/61 (34%), Positives = 30/61 (48%), Gaps = 7/61 (11%)
Query: 94 LPQMSDTGSVGEFYLVTDPESSGSPPKR----APPP---VPISAVPPIAVSTPPPAYPTS 146
LP+ ++ S ++ P S SPP + +PPP P + VPP+ S PPPA
Sbjct: 72 LPKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPP 131
Query: 147 P 147
P
Sbjct: 132 P 132
Score = 32.0 bits (71), Expect = 1.1
Identities = 16/49 (32%), Positives = 26/49 (52%), Gaps = 9/49 (18%)
Query: 112 PESSGSPP--------KRAPPPVPISAVPPIAVSTPP-PAYPTSPVASN 151
P ++G+PP +PPP P+ PP + S PP P P+ P +++
Sbjct: 62 PVANGAPPPPLPKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSAS 110
Score = 31.6 bits (70), Expect = 1.4
Identities = 19/53 (35%), Positives = 21/53 (38%), Gaps = 17/53 (32%)
Query: 112 PESSGSPPKRAPP-----------------PVPISAVPPIAVSTPPPAYPTSP 147
P S SPP +PP P+P SA PP PPP TSP
Sbjct: 8 PPVSNSPPVTSPPPPLNNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSP 60
Score = 31.6 bits (70), Expect = 1.4
Identities = 15/38 (39%), Positives = 20/38 (52%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVA 149
P S+ PP+ P P SA PP A+ P P+ P P +
Sbjct: 91 PPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPAS 128
Score = 30.8 bits (68), Expect = 2.4
Identities = 21/55 (38%), Positives = 27/55 (48%), Gaps = 8/55 (14%)
Query: 109 VTDPESSGSPP--KRAPPPV--PISAVPPIA----VSTPPPAYPTSPVASNISRS 155
V P S SPP R+PPP PI + PP +PPP P SP + ++S
Sbjct: 129 VPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQS 183
Score = 30.4 bits (67), Expect = 3.1
Identities = 16/44 (36%), Positives = 19/44 (42%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRS 155
P S SPP P P S +PPP P SP + S+S
Sbjct: 169 PPSPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQS 212
Score = 29.6 bits (65), Expect = 5.3
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 112 PESSGSPPKRA-PPPVPISAVPPIAVSTPPPAYPTSPVA 149
PESS PP+ P P P ++ PP V PP + P A
Sbjct: 76 PESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPA 114
>At1g49490 hypothetical protein
Length = 847
Score = 39.3 bits (90), Expect = 0.007
Identities = 19/53 (35%), Positives = 27/53 (50%)
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYS 160
+ + P S PP +PPP P+ + PP S PPP+ SP + S +YS
Sbjct: 579 VASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYS 631
Score = 36.6 bits (83), Expect = 0.043
Identities = 20/56 (35%), Positives = 23/56 (40%)
Query: 95 PQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
P M Y P S PP + PP P PP V++PPP P PV S
Sbjct: 538 PPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHS 593
Score = 36.2 bits (82), Expect = 0.057
Identities = 19/47 (40%), Positives = 24/47 (50%), Gaps = 3/47 (6%)
Query: 107 YLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAY---PTSPVAS 150
++ + P SPP +PPP S PP S PPP + P SPV S
Sbjct: 571 HVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYS 617
Score = 32.3 bits (72), Expect = 0.82
Identities = 16/37 (43%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 112 PESSGSPPKRAPPPV-PISAVPPIAVSTPPPAYPTSP 147
P S PP +PPP P+ + PP + S PPP Y P
Sbjct: 598 PVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPP 634
Score = 29.6 bits (65), Expect = 5.3
Identities = 17/45 (37%), Positives = 19/45 (41%), Gaps = 1/45 (2%)
Query: 107 YLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
Y P SPP P P S PP+ PPP + P PV S
Sbjct: 564 YSSPPPPHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFS 608
>At3g28790 unknown protein
Length = 608
Score = 38.9 bits (89), Expect = 0.009
Identities = 70/305 (22%), Positives = 117/305 (37%), Gaps = 29/305 (9%)
Query: 101 GSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPT--SPVASNISRSESL 158
GS G Y T SSG+ P +P P P + P STP P+ PT +P S + S
Sbjct: 257 GSSGNTYKDTTGSSSGASPSGSPTPTPSTPTP----STPTPSTPTPSTPTPSTPTPSTPA 312
Query: 159 YSAQERELTVDDIEDFEDDDDTSMVEGLRAKRTLNDASDLAV-KLPPFSTGITDDDLRET 217
S T + + + SM + +K A+ +V K + G + D ++T
Sbjct: 313 PSTPAAGKTSE-----KGSESASMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDT 367
Query: 218 AYEILLACAGATGGLIVPSKEKKKDRKSSSLIRKLGRSKTGSIVSQSQNAPGLVGLLESM 277
+ +G+ G PS D K+SS G + + S S +A ES
Sbjct: 368 TGTSSGSPSGSPSGSPTPS--TSTDGKASS----KGSASASAGASASASAGASASAEESA 421
Query: 278 RVQLEISEAMDIRTKQGL--LNALVGKAGKRMDTLLVPLELLCCVARTEFSDKKAFIRWQ 335
Q + S + + + + + +++ + LE T S+ K F
Sbjct: 422 ASQKKESNSKSSSSSSSTTSVKEVETQTSSEVNSFISNLE----KKYTGNSELKVFFEKL 477
Query: 336 KRQLKV---LEEGLVNHPVVGFGESGRKTNE-MRILLAKIEESEFLPSSSGELQRTECLR 391
K + L V G + K E M + ++ +SE +S Q+ E ++
Sbjct: 478 KTSMSASAKLSTSNAKELVTGMRSAASKIAEAMMFVSSRFSKSEETKTSMASCQQ-EVMQ 536
Query: 392 SLREI 396
SL+E+
Sbjct: 537 SLKEL 541
>At4g12770 auxilin-like protein
Length = 909
Score = 38.1 bits (87), Expect = 0.015
Identities = 42/157 (26%), Positives = 59/157 (36%), Gaps = 23/157 (14%)
Query: 102 SVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYSA 161
+V E L T P +S PP R PPP P PI P+ PTS S++ S
Sbjct: 356 TVSEIPLFTQP-TSAPPPTRPPPPRP---TRPIKKKVNEPSIPTSAYHSHVPSSGRASVN 411
Query: 162 QERELTVDDIEDFEDDDDTSMVEGLRAKRTLNDASDLAVKLPPFSTGITDDDLRETAYEI 221
+D+++DF + + G P S+G D D+ TA
Sbjct: 412 SPTASQMDELDDFSIGRNQTAANG----------------YPDPSSG-EDSDVFSTAAAS 454
Query: 222 LLACAGATGGLIVPSKEKKKDRKSSSLIRKLGRSKTG 258
A A + K+ R+ SL K RS+ G
Sbjct: 455 AAAMKDAMDKAEAKFRHAKERREKESL--KASRSREG 489
>At3g13965 unknown protein
Length = 209
Score = 37.7 bits (86), Expect = 0.019
Identities = 18/40 (45%), Positives = 19/40 (47%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P SPP PPP P PPI+ PPP SP SN
Sbjct: 94 PPPLPSPPPPPPPPTPSPPPPPISKPPPPPRAQASPPHSN 133
>At1g70460 putative protein kinase
Length = 710
Score = 37.7 bits (86), Expect = 0.019
Identities = 18/36 (50%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP APPP+PI PPI +PPP SP
Sbjct: 85 PPPSSPPPPDAPPPIPIVFPPPI--DSPPPESTNSP 118
Score = 32.0 bits (71), Expect = 1.1
Identities = 17/39 (43%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVP-PIAVSTPPPAYPTSPVA 149
P + S APPP + P P A S+PPPA P+ P A
Sbjct: 19 PPDTSSDGSAAPPPTDSAPPPSPPADSSPPPALPSLPPA 57
Score = 30.8 bits (68), Expect = 2.4
Identities = 18/41 (43%), Positives = 23/41 (55%), Gaps = 3/41 (7%)
Query: 112 PESSGSPPKR--APPPVPI-SAVPPIAVSTPPPAYPTSPVA 149
P + SPP +PPP P+ S+ PP TPPP+ P P A
Sbjct: 55 PPAVFSPPPTVSSPPPPPLDSSPPPPPDLTPPPSSPPPPDA 95
Score = 30.0 bits (66), Expect = 4.1
Identities = 15/40 (37%), Positives = 20/40 (49%), Gaps = 2/40 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P +PP +PPP A PPI + PPP P ++N
Sbjct: 79 PPPDLTPPPSSPPPP--DAPPPIPIVFPPPIDSPPPESTN 116
Score = 29.6 bits (65), Expect = 5.3
Identities = 17/46 (36%), Positives = 22/46 (46%), Gaps = 4/46 (8%)
Query: 110 TDPESSGSPPKR--APPPVPI--SAVPPIAVSTPPPAYPTSPVASN 151
T + S +PP APPP P S+ PP S PP + P S+
Sbjct: 22 TSSDGSAAPPPTDSAPPPSPPADSSPPPALPSLPPAVFSPPPTVSS 67
Score = 29.3 bits (64), Expect = 6.9
Identities = 14/37 (37%), Positives = 18/37 (47%), Gaps = 1/37 (2%)
Query: 112 PESSGSPPKRAPP-PVPISAVPPIAVSTPPPAYPTSP 147
P + SPP P P + + PP S PPP +SP
Sbjct: 41 PPADSSPPPALPSLPPAVFSPPPTVSSPPPPPLDSSP 77
Score = 29.3 bits (64), Expect = 6.9
Identities = 14/38 (36%), Positives = 19/38 (49%), Gaps = 1/38 (2%)
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
TD SPP + PP + ++PP S PPP + P
Sbjct: 33 TDSAPPPSPPADSSPPPALPSLPPAVFS-PPPTVSSPP 69
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 37.4 bits (85), Expect = 0.025
Identities = 23/52 (44%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query: 110 TDPESSGSPPK--RAPPPVPISAVPP--IAVSTPPPAYPTSPVASNISRSES 157
T P + SPP +PPP ISA PP A +PPPA PT + S S S
Sbjct: 24 TSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAPPTQETSPPTSPSSS 75
Score = 36.2 bits (82), Expect = 0.057
Identities = 14/32 (43%), Positives = 19/32 (58%)
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPP 141
T P + SP + PP P S+ PP ++S PPP
Sbjct: 18 TPPSNGTSPSNESSPPTPPSSPPPSSISAPPP 49
Score = 29.6 bits (65), Expect = 5.3
Identities = 21/59 (35%), Positives = 25/59 (41%), Gaps = 1/59 (1%)
Query: 93 GLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPP-VPISAVPPIAVSTPPPAYPTSPVAS 150
G P + T E T P S APPP + S PP A T + PTSP +S
Sbjct: 17 GTPPSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAPPTQETSPPTSPSSS 75
>At3g19020 hypothetical protein
Length = 951
Score = 37.4 bits (85), Expect = 0.025
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 750 PVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHS 789
Score = 37.0 bits (84), Expect = 0.033
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 668 PVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHS 707
Score = 35.4 bits (80), Expect = 0.097
Identities = 30/97 (30%), Positives = 45/97 (45%), Gaps = 7/97 (7%)
Query: 112 PESSGSPPKRAP---PPVPISAVPPIAVSTPPPAYPTSPVAS--NISRSESLYSAQEREL 166
P SPP +P PP P+ + PP V+ PPA TSP+A+ S SES + +
Sbjct: 791 PPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPA--TSPMANAPTPSSSESGEISTPVQA 848
Query: 167 TVDDIEDFEDDDDTSMVEGLRAKRTLNDASDLAVKLP 203
D ED E D++ ++ + S+ V+ P
Sbjct: 849 PTPDSEDIEAPSDSNHSPVFKSSPAPSPDSEPEVEAP 885
Score = 34.7 bits (78), Expect = 0.17
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYSAQERELT 167
P S PP +PPP P+ + PP S PPP + P + S ++S + +T
Sbjct: 765 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPVT 819
Score = 33.9 bits (76), Expect = 0.28
Identities = 19/43 (44%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Query: 110 TDPESSGSPPKRA-PPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
T S SPP + PPP P+ + PP S PPP + P PV S
Sbjct: 629 TKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYS 671
Score = 33.9 bits (76), Expect = 0.28
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 690 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVHSPPPPVQS 728
Score = 33.5 bits (75), Expect = 0.37
Identities = 15/38 (39%), Positives = 18/38 (46%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVA 149
P S PP +PPP S PP+ PPP + P A
Sbjct: 704 PVHSPPPPVHSPPPPVHSPPPPVQSPPPPPVFSPPPPA 741
Score = 33.5 bits (75), Expect = 0.37
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query: 112 PESSGSPPKRAPPPVPI-SAVPPIAVSTPPPAYPTSP 147
P S PP ++PPP P+ S PP + +PPP SP
Sbjct: 718 PVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSP 754
Score = 33.5 bits (75), Expect = 0.37
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 683 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVHSPPPPVHS 721
Score = 33.5 bits (75), Expect = 0.37
Identities = 17/40 (42%), Positives = 20/40 (49%), Gaps = 1/40 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP S PP+ PPP + P PV S
Sbjct: 654 PVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHS 693
Score = 33.1 bits (74), Expect = 0.48
Identities = 17/36 (47%), Positives = 18/36 (49%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP +PPP S PP S PPPA SP
Sbjct: 711 PVHSPPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSP 746
Score = 32.7 bits (73), Expect = 0.63
Identities = 18/45 (40%), Positives = 21/45 (46%), Gaps = 1/45 (2%)
Query: 107 YLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
Y P S PP PP P+ + PP S PPP + P PV S
Sbjct: 670 YSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHS 714
Score = 32.7 bits (73), Expect = 0.63
Identities = 16/49 (32%), Positives = 21/49 (42%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYS 160
P S PP PP P+ + PP S PPP + P + +YS
Sbjct: 757 PVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPSPIYS 805
Score = 32.7 bits (73), Expect = 0.63
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
PP +PPP P+ + PP S PPP Y P PV S
Sbjct: 646 PPVHSPPP-PVFSPPPPMHSPPPPVYSPPPPVHS 678
Score = 32.0 bits (71), Expect = 1.1
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 3/41 (7%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY--PTSPVAS 150
P S PP +PPP P+ + PP S PPP P PV S
Sbjct: 697 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVQSPPPPPVFS 736
Score = 32.0 bits (71), Expect = 1.1
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 3/41 (7%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY--PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 647 PVHSPPPPVFSPPP-PMHSPPPPVYSPPPPVHSPPPPPVHS 686
Score = 32.0 bits (71), Expect = 1.1
Identities = 16/38 (42%), Positives = 19/38 (49%), Gaps = 2/38 (5%)
Query: 112 PESSGSPPK--RAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP PPP PI + PP V +PPP + P
Sbjct: 725 PVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPP 762
Score = 32.0 bits (71), Expect = 1.1
Identities = 15/36 (41%), Positives = 20/36 (54%), Gaps = 1/36 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP +PPP P+ + PP V +PPP + P
Sbjct: 661 PMHSPPPPVYSPPP-PVHSPPPPPVHSPPPPVHSPP 695
Score = 30.8 bits (68), Expect = 2.4
Identities = 13/30 (43%), Positives = 18/30 (59%), Gaps = 1/30 (3%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PP +PPP P+ + PP V +PPP + P
Sbjct: 749 PPVHSPPP-PVHSPPPPPVHSPPPPVHSPP 777
Score = 30.0 bits (66), Expect = 4.1
Identities = 15/35 (42%), Positives = 18/35 (50%), Gaps = 4/35 (11%)
Query: 117 SPPKRAP----PPVPISAVPPIAVSTPPPAYPTSP 147
SPP AP PP P+ + PP S PPP + P
Sbjct: 736 SPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPP 770
>At4g12780 auxilin-like protein
Length = 904
Score = 37.0 bits (84), Expect = 0.033
Identities = 41/157 (26%), Positives = 59/157 (37%), Gaps = 23/157 (14%)
Query: 102 SVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYSA 161
+V E L T P +S PP R PPP P PI P+ PTS S++ S
Sbjct: 358 TVSEIPLFTQP-TSAPPPTRPPPPRP---TRPIKKKVNEPSIPTSAYHSHVPSSGRASVN 413
Query: 162 QERELTVDDIEDFEDDDDTSMVEGLRAKRTLNDASDLAVKLPPFSTGITDDDLRETAYEI 221
+D+++DF + + G P S+G D D+ TA
Sbjct: 414 SPTASQMDELDDFSIGRNQTAANG----------------YPDPSSG-EDSDVFSTAAAS 456
Query: 222 LLACAGATGGLIVPSKEKKKDRKSSSLIRKLGRSKTG 258
A A + K+ R+ +L K RS+ G
Sbjct: 457 AAAMKDAMDKAEAKFRHAKERREKENL--KASRSREG 491
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 36.6 bits (83), Expect = 0.043
Identities = 17/44 (38%), Positives = 22/44 (49%)
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNIS 153
TDP S PP PPP P +VP TP P P+ P +++
Sbjct: 171 TDPMPSPPPPVSPPPPTPTPSVPSPPDVTPTPPTPSVPSPPDVT 214
Score = 33.9 bits (76), Expect = 0.28
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 115 SGSPPKRAPPPVPISAVP-PIAVSTPPPAYPTSPVAS 150
S +PP PPP P +VP P +PPP PT V S
Sbjct: 92 SPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPS 128
Score = 33.9 bits (76), Expect = 0.28
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 115 SGSPPKRAPPPVPISAVP-PIAVSTPPPAYPTSPVAS 150
S +PP PPP P +VP P +PPP PT V S
Sbjct: 128 SPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPS 164
Score = 33.9 bits (76), Expect = 0.28
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 115 SGSPPKRAPPPVPISAVP-PIAVSTPPPAYPTSPVAS 150
S +PP PPP P +VP P +PPP PT V S
Sbjct: 110 SPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPS 146
Score = 31.6 bits (70), Expect = 1.4
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 5/36 (13%)
Query: 115 SGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
S +PP PPP P +VP +P P PT P+ S
Sbjct: 146 SPTPPVSPPPPTPTPSVP-----SPTPPVPTDPMPS 176
Score = 29.6 bits (65), Expect = 5.3
Identities = 16/38 (42%), Positives = 17/38 (44%), Gaps = 6/38 (15%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPA------YPTSPVA 149
PP PPP P P VS PPP PT PV+
Sbjct: 79 PPVSPPPPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVS 116
Score = 28.9 bits (63), Expect = 9.1
Identities = 16/43 (37%), Positives = 17/43 (39%), Gaps = 4/43 (9%)
Query: 109 VTDPESSGSPPKRAPPPVPISAVPPIAV----STPPPAYPTSP 147
V P SPP P P S PP+ S PPP P P
Sbjct: 144 VPSPTPPVSPPPPTPTPSVPSPTPPVPTDPMPSPPPPVSPPPP 186
Score = 28.9 bits (63), Expect = 9.1
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Query: 110 TDPESSGSPPKRAP-PPVPISAVPPIAVSTPP-PAYPTSP 147
T S SPP P PP P PP TPP P+ P+ P
Sbjct: 187 TPTPSVPSPPDVTPTPPTPSVPSPPDVTPTPPTPSVPSPP 226
>At2g15880 unknown protein
Length = 727
Score = 36.6 bits (83), Expect = 0.043
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 576 PVHSPPPPVYSPPPPPVHSPPPPVHSPPPPVHSPPPPVYS 615
Score = 35.0 bits (79), Expect = 0.13
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 3/41 (7%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY--PTSPVAS 150
P S PP +PPP P+ + PP S PPP Y P PV S
Sbjct: 555 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVYSPPPPPVHS 594
Score = 35.0 bits (79), Expect = 0.13
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 3/41 (7%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY--PTSPVAS 150
P S PP +PPP P+ + PP S PPP Y P PV S
Sbjct: 621 PVHSPPPPVFSPPP-PVHSPPPPVYSPPPPVYSPPPPPVKS 660
Score = 33.5 bits (75), Expect = 0.37
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 548 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVHSPPPPVYS 586
Score = 33.5 bits (75), Expect = 0.37
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 1/40 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP S PP S PPP + P PV S
Sbjct: 569 PVHSPPPPVHSPPPPVYSPPPPPVHSPPPPVHSPPPPVHS 608
Score = 33.5 bits (75), Expect = 0.37
Identities = 18/40 (45%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP P+ + PP S PPP + P PV S
Sbjct: 541 PVHSPPPPVHSPPP-PVHSPPPPVHSPPPPVHSPPPPVHS 579
Score = 33.1 bits (74), Expect = 0.48
Identities = 15/36 (41%), Positives = 17/36 (46%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP PP P+ + PP S PPP Y P
Sbjct: 583 PVYSPPPPPVHSPPPPVHSPPPPVHSPPPPVYSPPP 618
Score = 33.1 bits (74), Expect = 0.48
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
PP +PPP P+ + PP S PPP Y P PV S
Sbjct: 620 PPVHSPPP-PVFSPPPPVHSPPPPVYSPPPPVYS 652
Score = 32.7 bits (73), Expect = 0.63
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 2/41 (4%)
Query: 112 PESSGSPPKRA-PPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP + PPP P+ + PP S PPP + P PV S
Sbjct: 605 PVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSPPPPVYS 645
Score = 32.7 bits (73), Expect = 0.63
Identities = 18/41 (43%), Positives = 20/41 (47%), Gaps = 2/41 (4%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY--PTSPVAS 150
P S PP +PPP S PP V +PPP P PV S
Sbjct: 598 PVHSPPPPVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHS 638
Score = 32.7 bits (73), Expect = 0.63
Identities = 17/40 (42%), Positives = 20/40 (49%), Gaps = 1/40 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P S PP +PPP S PP+ PPP + P PV S
Sbjct: 562 PVHSPPPPVHSPPPPVHSPPPPVYSPPPPPVHSPPPPVHS 601
Score = 32.3 bits (72), Expect = 0.82
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP +PPP P+ + PP S PPP SP
Sbjct: 591 PVHSPPPPVHSPPP-PVHSPPPPVYSPPPPPPVHSP 625
Score = 32.0 bits (71), Expect = 1.1
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 2/38 (5%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPP--PAYPTSP 147
P S PP +PPP P+ + PP V +PP P +SP
Sbjct: 642 PVYSPPPPVYSPPPPPVKSPPPPPVYSPPLLPPKMSSP 679
Score = 32.0 bits (71), Expect = 1.1
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP +PPP P+ + PP V +PPP SP
Sbjct: 635 PVHSPPPPVYSPPP-PVYSPPPPPVKSPPPPPVYSP 669
Score = 32.0 bits (71), Expect = 1.1
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P S PP +PPP P+ + PP S PPP + P
Sbjct: 628 PVFSPPPPVHSPPP-PVYSPPPPVYSPPPPPVKSPP 662
Score = 31.6 bits (70), Expect = 1.4
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
PP +PPP P+ + PP S PPP + P PV S
Sbjct: 540 PPVHSPPP-PVHSPPPPVHSPPPPVHSPPPPVHS 572
Score = 30.8 bits (68), Expect = 2.4
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAY-PTSPVAS 150
P PPP P+ + PP S PPP + P PV S
Sbjct: 532 PVYSPPPPPPVHSPPPPVHSPPPPVHSPPPPVHS 565
Score = 30.4 bits (67), Expect = 3.1
Identities = 13/27 (48%), Positives = 15/27 (55%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYP 144
P PPP PI + PP V +PPP P
Sbjct: 497 PVHSPPPPSPIHSPPPPPVYSPPPPPP 523
Score = 30.0 bits (66), Expect = 4.1
Identities = 13/30 (43%), Positives = 16/30 (53%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PP +PPP P PP V +PPP + P
Sbjct: 531 PPVYSPPPPPPVHSPPPPVHSPPPPVHSPP 560
Score = 29.6 bits (65), Expect = 5.3
Identities = 13/31 (41%), Positives = 17/31 (53%), Gaps = 3/31 (9%)
Query: 120 KRAPPPVPISAVP---PIAVSTPPPAYPTSP 147
+R+PPP P+ + P PI PPP Y P
Sbjct: 490 RRSPPPPPVHSPPPPSPIHSPPPPPVYSPPP 520
>At1g31810 hypothetical protein
Length = 1201
Score = 36.6 bits (83), Expect = 0.043
Identities = 16/38 (42%), Positives = 19/38 (49%)
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRS 155
PP PPP+P ++PP PPP P P SRS
Sbjct: 574 PPPPPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSRS 611
Score = 30.4 bits (67), Expect = 3.1
Identities = 20/68 (29%), Positives = 29/68 (42%), Gaps = 10/68 (14%)
Query: 88 YHDHTGLPQMSDTGSVGEFYLVTDPESSGS-------PPKRAPPPVPISAVPPIAVSTPP 140
+H H +P + L +DP SSG PP PPP+ ++ + S PP
Sbjct: 449 HHHHHEIPAKDSVDN--PLNLPSDPPSSGDHVTLLPPPPPPPPPPL-FTSTTSFSPSQPP 505
Query: 141 PAYPTSPV 148
P P P+
Sbjct: 506 PPPPPPPL 513
Score = 30.0 bits (66), Expect = 4.1
Identities = 15/36 (41%), Positives = 18/36 (49%), Gaps = 3/36 (8%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
P + PP R PPP P PP + S P P+ P P
Sbjct: 589 PPLAQPPPPRPPPPPP---PPPSSRSIPSPSAPPPP 621
Score = 29.3 bits (64), Expect = 6.9
Identities = 14/44 (31%), Positives = 20/44 (44%)
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRS 155
P + +PP PPP S + + PP P P +NIS +
Sbjct: 652 PAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISNA 695
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,916,073
Number of Sequences: 26719
Number of extensions: 619250
Number of successful extensions: 6468
Number of sequences better than 10.0: 146
Number of HSP's better than 10.0 without gapping: 76
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 4039
Number of HSP's gapped (non-prelim): 1461
length of query: 629
length of database: 11,318,596
effective HSP length: 105
effective length of query: 524
effective length of database: 8,513,101
effective search space: 4460864924
effective search space used: 4460864924
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149307.14


