

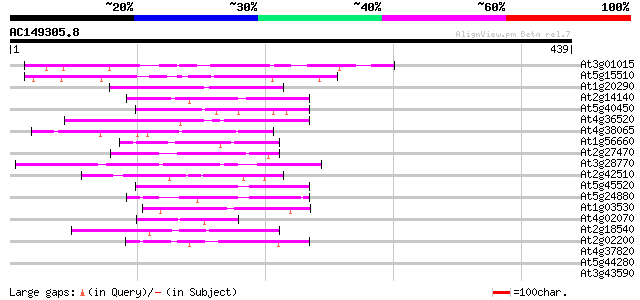
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.8 - phase: 0 /pseudo
(439 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g01015 hypothetical protein 125 6e-29
At5g15510 putative protein 118 7e-27
At1g20290 hypothetical protein 50 3e-06
At2g14140 putative transposase of FARE2.8 (cds1) 49 4e-06
At5g40450 unknown protein 48 1e-05
At4g36520 trichohyalin like protein 47 2e-05
At4g38065 hypothetical protein 45 8e-05
At1g56660 hypothetical protein 44 1e-04
At2g27470 putative CCAAT-binding transcription factor subunit 44 2e-04
At3g28770 hypothetical protein 43 3e-04
At2g42510 hypothetical protein 43 3e-04
At5g45520 putative protein 42 5e-04
At5g24880 glutamic acid-rich protein 42 5e-04
At1g03530 unknown protein 42 7e-04
At4g02070 G/T DNA mismatch repair enzyme 42 9e-04
At2g18540 putative vicilin storage protein (globulin-like) 42 9e-04
At2g02200 putative protein on transposon FARE2.10 (cds2) 42 9e-04
At4g37820 unknown protein 41 0.001
At5g44280 putative protein 40 0.002
At3g43590 putative protein 40 0.002
>At3g01015 hypothetical protein
Length = 488
Score = 125 bits (313), Expect = 6e-29
Identities = 106/302 (35%), Positives = 146/302 (48%), Gaps = 61/302 (20%)
Query: 12 ENLNPNEFSNRTPLQK----SLTKGDNNCVNVV--VVSPQKRIRQRKFVVAKKK-KNDQS 64
EN NPN S+ TPL+K S K N V V SP+ RIR+R+FVV KK + +++
Sbjct: 33 ENSNPNFVSHSTPLEKPSKSSAQKNPKWKPNPVPAVFSPRNRIRERRFVVVKKNSRKEKN 92
Query: 65 PRKTVLCKCGEN--DDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEV 122
+V CKCG + KCVC AY LR SQE FF + E+
Sbjct: 93 DSASVDCKCGAKTISNMKKCVCIAYETLRASQEEFFNNRR--------------ESVSEI 138
Query: 123 IEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
E++ + D GNE+ + + + S++KRRREKV+EEAR S+PE GKVMH
Sbjct: 139 GESSQNLED-GNEQVEFGDSDETRV---------SLMKRRREKVLEEARMSIPEFGKVMH 188
Query: 183 LVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDD 242
LVKAFE+L K ++EEE D+ + +KW LPG+ S+ S +
Sbjct: 189 LVKAFEKLTCFPLSKVTSKEEE--DQIKQPLKWELPGM------------SQPKCSESET 234
Query: 243 DDFDF*TTWIGSKG--FSYIFLGLWKWKKCV*QEFFWW*KKQKKSW-NHLVLLEEGGGRR 299
D F + +++ S G + LGL + V SW N + L GGRR
Sbjct: 235 DQFTWSSSFYPSSGLILTATNLGLEQPHASV-----------SSSWDNSVSSLNSNGGRR 283
Query: 300 SR 301
R
Sbjct: 284 GR 285
>At5g15510 putative protein
Length = 494
Score = 118 bits (295), Expect = 7e-27
Identities = 96/270 (35%), Positives = 132/270 (48%), Gaps = 45/270 (16%)
Query: 12 ENLNPN---EFSNRTPLQKSLTKGDNNCVNV------VVVSPQKRIRQRKFVVAKKKKND 62
EN NPN +++PL KS K + V SP+ RIR+RKFVV KK +
Sbjct: 32 ENANPNISQASPSKSPLMKSAKKSAQKNPSPSPKPTQAVFSPRNRIRERKFVVVAKKNSR 91
Query: 63 QSPRKTVL---------CKCGENDDGS-KCVCEAYRNLRESQEGFFEKENFFDGEEKNDE 112
+ ++ + CKCGE G+ KCVC AY LR SQE FF+ + +
Sbjct: 92 KGKKEPAITESKAAEIDCKCGERKKGNMKCVCVAYETLRASQEEFFKN--------RIES 143
Query: 113 EKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARN 172
E+ + LEE ++GN DE EG E EK S+M KR R KV+EEAR
Sbjct: 144 EEDKEALEECC-------NLGNG----DELEGKESGEPEKTGVSTM-KRSRAKVVEEARQ 191
Query: 173 SVPENGKVMHLVKAFERLLSIKKEKEKNEEEE---ENDKKNKVMKWALPGLQFQQPVKDG 229
SVP +GKVM+LV+AFE+L K N+ EE E D K V L G + + P
Sbjct: 192 SVPVSGKVMNLVEAFEKLTCFSNSKTANKIEENQTEEDTKKPVKLEFLEGEKEKNPWSSS 251
Query: 230 DEQSEVVSSCCD---DDDFDF*TTWIGSKG 256
SE+V + + D + ++W ++G
Sbjct: 252 FCPSEMVLTAKNLGLDPNASISSSWDSTRG 281
>At1g20290 hypothetical protein
Length = 497
Score = 49.7 bits (117), Expect = 3e-06
Identities = 39/136 (28%), Positives = 62/136 (44%), Gaps = 3/136 (2%)
Query: 79 GSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERK 138
G + +A E +E E+E + EE+ +EE+ E EE E + EE +
Sbjct: 356 GEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 415
Query: 139 IDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKE 198
+EEE EEE EE+ + E+ EE E + + E ++E+E
Sbjct: 416 EEEEEEEEEEEEEE---EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 472
Query: 199 KNEEEEENDKKNKVMK 214
+ EEEEE +KK ++K
Sbjct: 473 EEEEEEEEEKKYMIVK 488
Score = 38.9 bits (89), Expect = 0.006
Identities = 25/74 (33%), Positives = 40/74 (53%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
E +E E+E + EE+ +EE+ E EE E + EE + +EEE EEE EE
Sbjct: 420 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Query: 152 KGNCSSMVKRRREK 165
+ +VK++++K
Sbjct: 480 EEKKYMIVKKKKKK 493
Score = 35.8 bits (81), Expect = 0.048
Identities = 23/74 (31%), Positives = 39/74 (52%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
E +E E+E + EE+ +EE+ E EE E + EE + +EEE EEE EE
Sbjct: 421 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 480
Query: 152 KGNCSSMVKRRREK 165
+ + K+++++
Sbjct: 481 EKKYMIVKKKKKKR 494
Score = 32.0 bits (71), Expect = 0.69
Identities = 23/72 (31%), Positives = 35/72 (47%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
E +E E+E + EE+ +EE+ E EE E + EE + +EEE EEE ++
Sbjct: 424 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKK 483
Query: 152 KGNCSSMVKRRR 163
K+RR
Sbjct: 484 YMIVKKKKKKRR 495
>At2g14140 putative transposase of FARE2.8 (cds1)
Length = 783
Score = 49.3 bits (116), Expect = 4e-06
Identities = 42/148 (28%), Positives = 69/148 (46%), Gaps = 13/148 (8%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKI-----DEEEGVE 146
E QE + + +GEE+ EE+G+ EE +E D G E+++I +E EG E
Sbjct: 402 EKQEIPKQGDEEMEGEEEKQEEEGKEEEEEKVEYR---GDEGTEKQEIPKQGDEEMEGEE 458
Query: 147 EENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEEN 206
E+ EE+G K E + +P+ G + E ++E+ K EEEE+
Sbjct: 459 EKQEEEGKEKEEEKVEYRGDEETEKQEIPKQGD-----EEMEGEEEKQEEEGKEEEEEKV 513
Query: 207 DKKNKVMKWALPGLQFQQPVKDGDEQSE 234
+ K+ + + Q+ K GDE+ E
Sbjct: 514 EYKDHHSTCNVEETEKQENPKQGDEEME 541
Score = 33.9 bits (76), Expect = 0.18
Identities = 29/109 (26%), Positives = 47/109 (42%), Gaps = 16/109 (14%)
Query: 131 DIGNEERKI-----DEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVK 185
D G E+++I +E EG EE+ EE+G K + +P+ G
Sbjct: 398 DEGTEKQEIPKQGDEEMEGEEEKQEEEGKEEEEEKVEYRGDEGTEKQEIPKQGDE----- 452
Query: 186 AFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSE 234
++ E+EK EEE + ++ KV + Q+ K GDE+ E
Sbjct: 453 ------EMEGEEEKQEEEGKEKEEEKVEYRGDEETEKQEIPKQGDEEME 495
>At5g40450 unknown protein
Length = 2910
Score = 47.8 bits (112), Expect = 1e-05
Identities = 42/148 (28%), Positives = 73/148 (48%), Gaps = 13/148 (8%)
Query: 99 EKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSM 158
+KE EEK + E + +E+ I+ + + + N ID E E E EE G +++
Sbjct: 2654 QKEETSSIEEKREVEHVKAEMEDAIKHEVSVEEKNNTSENIDHEAAKEIEQEE-GKQTNI 2712
Query: 159 VK---RRREK-VMEEARNSVPEN----GKVMHLVKAFERLLSIKKEKEKNEEEEE--NDK 208
VK R EK + +E+ N+V E K ++ E L S+ K ++K E E E N +
Sbjct: 2713 VKEEIREEEKEINQESFNNVKETDDAIDKTQPEIRDIESLSSVSKTQDKPEPEYEVPNQQ 2772
Query: 209 KNKVMKW--ALPGLQFQQPVKDGDEQSE 234
K ++ +L + ++ ++ DE+SE
Sbjct: 2773 KREITNEVPSLENSKIEEELQKKDEESE 2800
Score = 32.7 bits (73), Expect = 0.41
Identities = 46/244 (18%), Positives = 92/244 (36%), Gaps = 57/244 (23%)
Query: 8 PLEIENLNPNEFSNRTPLQKSLTKGD----NNCVNVVVVSPQKRIRQRKFVVAKKKKNDQ 63
PL+ E+ PNE T L+K + + + V+ SP K + V A+ +N +
Sbjct: 984 PLQEESSQPNEQEKETKLEKHEPTNEEVKSDEVIEVLSASPSKELEGETVVEAENIENIK 1043
Query: 64 SPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVI 123
EN++ + + ++L Q F EE++ E +++E
Sbjct: 1044 -----------ENEE-EQAAEKIQKSLETVQTVESPSSLLFSSEEQDHVTVAEEIVDEKA 1091
Query: 124 EANLIIHDIGNE-------ERKIDEEEGV-------------EEENEEKGNCSSMVKRRR 163
+ + + I NE E ++++ + + E + K CS K +
Sbjct: 1092 KEEVPMLQIKNEDDATKIHETRVEQARDIGPSLTEICSINQNQPEEQVKEACS---KEEQ 1148
Query: 164 EKVMEEARNSVPENGKVMHLVKAFER------------------LLSIKKEKEKNEEEEE 205
EK + ++ +H V+A E LL ++KE+E+ E + +
Sbjct: 1149 EKEISTNSENIVNETYALHSVEAAEEETATNGESLDDVETTKSVLLEVRKEEEEAEMKTD 1208
Query: 206 NDKK 209
+ +
Sbjct: 1209 AEPR 1212
Score = 32.3 bits (72), Expect = 0.53
Identities = 43/177 (24%), Positives = 77/177 (43%), Gaps = 42/177 (23%)
Query: 111 DEEKGENVLE--------EVIEANLIIHDIGNEERKIDEEEG------------VEEENE 150
+E+KG++V+E +++EA + D E KI EE G ++EE++
Sbjct: 2248 EEKKGDDVVESNEKDFVSDILEAKRLHGDKSGEAEKIKEESGLAGKSLPIEEINLQEEHK 2307
Query: 151 EKGNCS------SMVKRRREKVMEEARNSVPENGKVMHLVKAFER------------LLS 192
E+ + V R E ++ + S E V+ K ER +S
Sbjct: 2308 EEVKVQEETREIAQVLPREEILISSSPLSAEEQEHVISDEKQEEREPQQDFNGSTSEKIS 2367
Query: 193 IKKEKEKNEE---EEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDDDDFD 246
++ E K+ E +E+ D+ ++ +K + + KD DE+ E+VSS D+ D
Sbjct: 2368 LQVEHLKDFETSKKEQKDETHETVKEEDQIVDIKDKKKD-DEEQEIVSSEVKKDNKD 2423
Score = 31.6 bits (70), Expect = 0.91
Identities = 38/163 (23%), Positives = 72/163 (43%), Gaps = 25/163 (15%)
Query: 56 AKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKE-NFFDGEEKNDEEK 114
A K+ + ++T + K ++ + E++ N++E+ + + + D E + K
Sbjct: 2698 AAKEIEQEEGKQTNIVKEEIREEEKEINQESFNNVKETDDAIDKTQPEIRDIESLSSVSK 2757
Query: 115 GENVLEEVIEA-NLIIHDIGNE-----ERKIDEE-EGVEEENEEKGNCSSMVKRRREKVM 167
++ E E N +I NE KI+EE + +EE+E + S+VK +
Sbjct: 2758 TQDKPEPEYEVPNQQKREITNEVPSLENSKIEEELQKKDEESENTKDLFSVVKETEPTLK 2817
Query: 168 EEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKN 210
E AR S+ + H+ +KE EE+END ++
Sbjct: 2818 EPARKSLSD-----HI------------QKEPKTEEDENDDED 2843
Score = 30.4 bits (67), Expect = 2.0
Identities = 40/177 (22%), Positives = 71/177 (39%), Gaps = 43/177 (24%)
Query: 65 PRKTVLCKCG--ENDDGSKCVCEAYRNLRESQEGF---------FEKENFFDGEEKNDEE 113
PR+ +L ++ + + + RE Q+ F + E+ D E E+
Sbjct: 2324 PREEILISSSPLSAEEQEHVISDEKQEEREPQQDFNGSTSEKISLQVEHLKDFETSKKEQ 2383
Query: 114 KGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKR------------ 161
K E H+ EE +I + + ++++EE+ SS VK+
Sbjct: 2384 KDET------------HETVKEEDQIVDIKDKKKDDEEQEIVSSEVKKDNKDARELEVGN 2431
Query: 162 ---RREKVMEEARNSVPENGKVMHLVKAFERLLS-----IKKEKEKNEEEEENDKKN 210
R+ EE ++ EN + M+ V A E+ +S IKK E E+ +D K+
Sbjct: 2432 DFVSRDGEKEEVPHNALENEEEMNEVVASEKQISDPVGVIKKASEAEHEDPVDDIKS 2488
>At4g36520 trichohyalin like protein
Length = 1432
Score = 47.0 bits (110), Expect = 2e-05
Identities = 41/194 (21%), Positives = 85/194 (43%), Gaps = 11/194 (5%)
Query: 44 PQKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENF 103
P KR+ + + + + + R+ V + EN+ K E R+ +E + EN
Sbjct: 628 PLKRMEEETRIKEARLREENDRRERVAVEKAENEKRLKAALEQEEKERKIKEAREKAENE 687
Query: 104 FDGEEKNDEEKGENVLEEVIEANLIIHDI---GNEERKIDEEEGVEEENEEKGNCSSMVK 160
E ++ + E ++E E L + + E R++ E +E+E E + +K
Sbjct: 688 RRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFALEQEKERR------IK 741
Query: 161 RRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGL 220
REK EE + E + L + + L ++++ + +E +E ++ + K L
Sbjct: 742 EAREK--EENERRIKEAREKAELEQRLKATLEQEEKERQIKERQEREENERRAKEVLEQA 799
Query: 221 QFQQPVKDGDEQSE 234
+ ++ +K+ EQ E
Sbjct: 800 ENERKLKEALEQKE 813
Score = 41.2 bits (95), Expect = 0.001
Identities = 45/200 (22%), Positives = 92/200 (45%), Gaps = 30/200 (15%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
++ + R+ ++K +Q + L + E ++ ++ + EA+ +E + E
Sbjct: 688 RRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFALEQEKERRIKEAREKE 747
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRRE 164
+ E + E + + LE+ ++A L + +ER+I E + EENE RR +
Sbjct: 748 ENERRIKEAREKAELEQRLKATL---EQEEKERQIKERQE-REENE----------RRAK 793
Query: 165 KVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEE---------ENDKKNKVMKW 215
+V+E+A N L +A E+ + ++ KE E+EE E ++K K +
Sbjct: 794 EVLEQAENE-------RKLKEALEQKENERRLKETREKEENKKKLREAIELEEKEKRLIE 846
Query: 216 ALPGLQFQQPVKDGDEQSEV 235
A + ++ +K+ EQ E+
Sbjct: 847 AFERAEIERRLKEDLEQEEM 866
Score = 34.7 bits (78), Expect = 0.11
Identities = 39/143 (27%), Positives = 61/143 (42%), Gaps = 29/143 (20%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGN 134
EN+ +K V E N R+ +E +KEN +E ++E+ + L E IE
Sbjct: 787 ENERRAKEVLEQAENERKLKEALEQKENERRLKETREKEENKKKLREAIELE------EK 840
Query: 135 EERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIK 194
E+R I+ E E E K + R R ++EA K ERL
Sbjct: 841 EKRLIEAFERAEIERRLKEDLEQEEMRMR---LQEA--------------KERERLHREN 883
Query: 195 KEKEKNEEE------EENDKKNK 211
+E ++NE + EE+D+K +
Sbjct: 884 QEHQENERKQHEYSGEESDEKER 906
>At4g38065 hypothetical protein
Length = 1050
Score = 45.1 bits (105), Expect = 8e-05
Identities = 51/203 (25%), Positives = 93/203 (45%), Gaps = 18/203 (8%)
Query: 18 EFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQSPRKTV---LCKC- 73
++ N+T L ++L K N ++ + + + ++ ++K + + K L +C
Sbjct: 21 DYRNKTELLENLKKVQNE--QLIEIREARLVNEKHGFEIEEKSREIAELKRANEELQRCL 78
Query: 74 GENDDGSKCVCEAYRNLRESQEGFF-----EKENFFDG----EEKN-DEEKGENVLEEVI 123
E D K V + LR + E + EK N G EKN D E+ NV I
Sbjct: 79 REKDSVVKRVNDVNDKLRANGEDKYREFEEEKRNMMSGLDEASEKNIDLEQKNNVYRAEI 138
Query: 124 EANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHL 183
E + + E ++I+ E+ V+ E +G +VK EK E + + + HL
Sbjct: 139 EGLKGLLAVA-ETKRIEAEKTVKGMKEMRGRDDVVVKMEEEKSQVEEKLKWKKE-QFKHL 196
Query: 184 VKAFERLLSIKKEKEKNEEEEEN 206
+A+E+L ++ K+ +K EEE++
Sbjct: 197 EEAYEKLKNLFKDSKKEWEEEKS 219
>At1g56660 hypothetical protein
Length = 522
Score = 44.3 bits (103), Expect = 1e-04
Identities = 33/129 (25%), Positives = 69/129 (52%), Gaps = 12/129 (9%)
Query: 87 YRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVE 146
+ L E +EG +K+N + +E EEK + +E D+ E+ +++EE+G +
Sbjct: 123 HEELEEEKEGK-KKKNKKEKDESGPEEKNKKADKEKKH-----EDVSQEKEELEEEDGKK 176
Query: 147 EENEEKGNCSSMVKRRR----EKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEE 202
+ +EK + K+++ +K EE++++ E+ KV + E+ K+++EK +E
Sbjct: 177 NKKKEKDESGTEEKKKKPKKEKKQKEESKSN--EDKKVKGKKEKGEKGDLEKEDEEKKKE 234
Query: 203 EEENDKKNK 211
+E D++ K
Sbjct: 235 HDETDQEMK 243
Score = 37.0 bits (84), Expect = 0.022
Identities = 43/177 (24%), Positives = 77/177 (43%), Gaps = 27/177 (15%)
Query: 57 KKKKNDQSPRK--TVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEK 114
KKKKN +K TV+ + VCE ++ EG +++ E+K+ EK
Sbjct: 325 KKKKNKDKAKKKETVIDE----------VCEKETKDKDDDEGETKQKKNKKKEKKS--EK 372
Query: 115 GE-NVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNS 173
GE +V E+ + N + ++ + + K++E E ++E ++ EK +
Sbjct: 373 GEKDVKEDKKKENPLETEVMSRDIKLEEPEAEKKEEDDT----------EEKKKSKVEGG 422
Query: 174 VPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGD 230
E GK K ++ K KE E+E +KK+ + G + ++ KD D
Sbjct: 423 ESEEGKKKK--KKDKKKNKKKDTKEPKMTEDEEEKKDDSKDVKIEGSKAKEEKKDKD 477
Score = 36.6 bits (83), Expect = 0.028
Identities = 36/137 (26%), Positives = 67/137 (48%), Gaps = 10/137 (7%)
Query: 88 RNLRESQE-GFFEKENFFDGEEKNDE--EKGENVLEEVIEANLIIH--DIGNEERKIDEE 142
+N +E E G EK D E+K+++ ++ E + EE + N + G EE+K +
Sbjct: 136 KNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPK 195
Query: 143 EGVEEENEEKGNCSSMVKRRREK-----VMEEARNSVPENGKVMHLVKAFERLLSIKKEK 197
+ +++ E K N VK ++EK + +E E+ + +K + + KKEK
Sbjct: 196 KEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEK 255
Query: 198 EKNEEEEENDKKNKVMK 214
+++ EE+ K +K K
Sbjct: 256 DESCAEEKKKKPDKEKK 272
Score = 35.4 bits (80), Expect = 0.063
Identities = 54/244 (22%), Positives = 101/244 (41%), Gaps = 31/244 (12%)
Query: 4 EKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQ 63
EKKK + + E ++ +K + D +C P K +++ K+ K +
Sbjct: 230 EKKKEHDETDQEMKEKDSKKNKKK---EKDESCAEEKKKKPDKEKKEKDESTEKEDKKLK 286
Query: 64 SPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKG---ENVLE 120
+K K + D+G K + QE E + +G++K +++K E V++
Sbjct: 287 G-KKGKGEKPEKEDEGKK----TKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKETVID 341
Query: 121 EVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKV 180
EV E +++ DE E +++N++K S ++ + V E+ + P +V
Sbjct: 342 EVCEKE-------TKDKDDDEGETKQKKNKKKEKKS---EKGEKDVKEDKKKENPLETEV 391
Query: 181 MHLVKAFERLLSIKKEKEKNEEE----------EENDKKNKVMKWALPGLQFQQPVKDGD 230
M E + KKE++ EE+ EE KK K K ++P D
Sbjct: 392 MSRDIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTED 451
Query: 231 EQSE 234
E+ +
Sbjct: 452 EEEK 455
Score = 31.6 bits (70), Expect = 0.91
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 18/124 (14%)
Query: 107 EEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKG---NCSSMVKRRR 163
+E + +EKGENV E+ I + + +K +E G ++++EK N S VK +
Sbjct: 20 QELDPKEKGENVEVEMEVKAKSIEKV--KAKKDEESSGKSKKDKEKKKGKNVDSEVKEDK 77
Query: 164 EKVMEEARNSVPENGKVMHL--------VKAFERLLSIKKEKEKN-----EEEEENDKKN 210
+ ++ V + + H VK E KK KEK EE+E KKN
Sbjct: 78 DDDKKKDGKMVSKKHEEGHGDLEVKESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKN 137
Query: 211 KVMK 214
K K
Sbjct: 138 KKEK 141
>At2g27470 putative CCAAT-binding transcription factor subunit
Length = 275
Score = 43.9 bits (102), Expect = 2e-04
Identities = 40/140 (28%), Positives = 63/140 (44%), Gaps = 23/140 (16%)
Query: 80 SKCVCEAYRNLRESQEGFFEKENFFDGEEKN-DEEKGENVLEEVIEANLIIHDIGNEERK 138
SK E RN E++ E+EN D E++N ++E+ EN D N E
Sbjct: 151 SKIDEETKRNDEETENDNTEEENGNDEEDENGNDEEDEN-------------DDENTEEN 197
Query: 139 IDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKE 198
++EE +E EE GN K E MEE N E+G H ++ E + ++ E
Sbjct: 198 GNDEENDDENTEENGNDEENEKEDEENSMEENGNESEESGNEDHSME--ENGSGVGEDNE 255
Query: 199 KNE-------EEEENDKKNK 211
+ EE E+D++++
Sbjct: 256 NEDGSVSGSGEEVESDEEDE 275
>At3g28770 hypothetical protein
Length = 2081
Score = 43.1 bits (100), Expect = 3e-04
Identities = 48/240 (20%), Positives = 98/240 (40%), Gaps = 24/240 (10%)
Query: 5 KKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQS 64
K K E E E N +K K + ++ +K ++ + ++KK+ D+
Sbjct: 1059 KAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKK 1118
Query: 65 PRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIE 124
+ + D S E ++SQ K+ E+K +EEK E +E+
Sbjct: 1119 DMEKL------EDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSET--KEIES 1170
Query: 125 ANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLV 184
+ +++ +E+K +++ ++E E K + +K+ E + + SV EN K
Sbjct: 1171 SKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEED--RKKQTSVEENKK----- 1223
Query: 185 KAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDDDD 244
+KE +K + + ++DKKN + + K+ + Q + ++ D D
Sbjct: 1224 ---------QKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSD 1274
Score = 38.5 bits (88), Expect = 0.007
Identities = 39/172 (22%), Positives = 76/172 (43%), Gaps = 15/172 (8%)
Query: 67 KTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEAN 126
++V K E +G+K + N Q+G +K+ + + N ++K E+ +E +
Sbjct: 912 ESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDK-KEYVNNE 970
Query: 127 LIIHDIGNEERKIDEEEGVEEEN----EEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
L + +E E ++EEN E+K + S K R +K EE ++ E K
Sbjct: 971 LKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEK 1030
Query: 183 LVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSE 234
K + +K EE++ ++K+K K L+ ++ ++ E+ E
Sbjct: 1031 K----------KSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKE 1072
Score = 37.7 bits (86), Expect = 0.013
Identities = 39/176 (22%), Positives = 72/176 (40%), Gaps = 9/176 (5%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
Q + R+ K +K K ++ + + K E + K E +++ ++ + E
Sbjct: 1034 QDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSM 1093
Query: 105 DGEE-----KNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMV 159
EE K EE EE + + D + ++K D+ E + ++ + S
Sbjct: 1094 KKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDK 1153
Query: 160 KRRREKV----MEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
K ++E +E +S + +V K + KKEKE E EE+ KKN+
Sbjct: 1154 KEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNE 1209
Score = 37.4 bits (85), Expect = 0.017
Identities = 45/211 (21%), Positives = 83/211 (39%), Gaps = 24/211 (11%)
Query: 4 EKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQ 63
+KKK + N+ E + + L K ++N K + K KK+ D
Sbjct: 946 KKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDS 1005
Query: 64 SPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVI 123
+ + K + SK EA + ++SQ+ +K D EE+ +++ E
Sbjct: 1006 ASKNRE--KKEYEEKKSKTKEEAKKEKKKSQD---KKREEKDSEERKSKKEKE------- 1053
Query: 124 EANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHL 183
E R + ++ EE E+K + + K++ +K E S+ +
Sbjct: 1054 -----------ESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEK 1102
Query: 184 VKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
K E S KKE++K + E+ D+ + K
Sbjct: 1103 -KKHEESKSRKKEEDKKDMEKLEDQNSNKKK 1132
Score = 36.6 bits (83), Expect = 0.028
Identities = 39/168 (23%), Positives = 77/168 (45%), Gaps = 16/168 (9%)
Query: 55 VAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEK 114
V K+KN+ T G +D + ++ ++E +K+ + ++ND+
Sbjct: 837 VEAKEKNENGGVDT---NVGNKEDSKDLKDDRSVEVKANKEESMKKKR--EEVQRNDKSS 891
Query: 115 GENVLEEVIEANLIIHDIGNEE---RKIDEEEGVEEENEEKGNCSSMVK-RRREKVMEEA 170
+ V + ++ + E +K +++EG +EEN++ N SS K + ++K +E+
Sbjct: 892 TKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKES 951
Query: 171 RNS---VPENGKVMHLVKAFERLLSIKKEKEKNE----EEEENDKKNK 211
+NS E K ++ ++ KKE K+E +EE D K K
Sbjct: 952 KNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEK 999
Score = 33.1 bits (74), Expect = 0.31
Identities = 28/77 (36%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE-KGNCSSMVKRRR 163
D +++N EE EN E V NL + GNEE E + NEE KGN S K
Sbjct: 414 DHKKENKEETHENNGESVKGENLE-NKAGNEESMKGENLENKVGNEELKGNASVEAKTNN 472
Query: 164 EKVMEEARNSVPENGKV 180
E EE R + +V
Sbjct: 473 ESSKEEKREESQRSNEV 489
Score = 31.2 bits (69), Expect = 1.2
Identities = 39/151 (25%), Positives = 59/151 (38%), Gaps = 26/151 (17%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDG----EEKNDEEKG--ENVLEEVIEANLI 128
E+ + K EA +N S G E+ +G E N E KG E+ ++++ A
Sbjct: 352 ESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGESTNDKMVNATTN 411
Query: 129 IHDIGNEERKIDEE---EGVEEENEEK--GNCSSMVKRRREKVM--EEARNSVPENGKVM 181
D E ++ E E V+ EN E GN SM E + EE + + K
Sbjct: 412 DEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGNEELKGNASVEAKT- 470
Query: 182 HLVKAFERLLSIKKEKEKNEEEEENDKKNKV 212
E K E+ EE+ + N+V
Sbjct: 471 ------------NNESSKEEKREESQRSNEV 489
Score = 31.2 bits (69), Expect = 1.2
Identities = 32/124 (25%), Positives = 53/124 (41%), Gaps = 13/124 (10%)
Query: 99 EKENFFDGEEKNDEE-KGENVL-----EEVIEANLIIHDIGNEERKID-------EEEGV 145
+KEN + E N E KGEN+ EE ++ + + +GNEE K + E
Sbjct: 416 KKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGNEELKGNASVEAKTNNESS 475
Query: 146 EEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEE 205
+EE E+ S+ V +E E N E+ + E +K + + NE +
Sbjct: 476 KEEKREESQRSNEVYMNKETTKGENVNIQGESIGDSTKDNSLENKEDVKPKVDANESDGN 535
Query: 206 NDKK 209
+ K+
Sbjct: 536 STKE 539
Score = 30.8 bits (68), Expect = 1.5
Identities = 45/249 (18%), Positives = 99/249 (39%), Gaps = 26/249 (10%)
Query: 4 EKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQ 63
+K+K + E + +K L K + + V K+ ++ K K+K +
Sbjct: 1180 KKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETK----KEKNKPK 1235
Query: 64 SPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVI 123
+K + G G K E+ ES+E ++++ + +DE K E +++
Sbjct: 1236 DDKKNTTKQSG----GKKESMES-----ESKEAENQQKSQATTQADSDESKNEILMQADS 1286
Query: 124 EANLIIHDIGNEER-------KIDEEEGVEEENEE-KGNCSSMVKRRREKVMEEARNSVP 175
+A+ + + + D + + NEE + +S+ + +++K +E +N
Sbjct: 1287 QADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPK 1346
Query: 176 ENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEV 235
++ K ++ K+ E +E EN +K++ A + + D Q++
Sbjct: 1347 DDKK-----NTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADS 1401
Query: 236 VSSCCDDDD 244
S D D
Sbjct: 1402 HSDSQADSD 1410
Score = 29.6 bits (65), Expect = 3.4
Identities = 44/179 (24%), Positives = 75/179 (41%), Gaps = 18/179 (10%)
Query: 74 GENDDGSKCVCEAYRNLR------ESQEGFFEKENFFDGEEKNDEE--KGENVLEEVIEA 125
G+ D+G + E + + ES++G K N G+E + EE K N++E
Sbjct: 1590 GKEDNGDEVGKENSKTIEVKGRHEESKDG---KTNENGGKEVSTEEGSKDSNIVERNGGK 1646
Query: 126 NLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVK 185
I + G+E+ K E G EE + E+G+ + E+ E NS E K + +
Sbjct: 1647 EDSIKE-GSEDGKTVEINGGEELSTEEGSKDGKI----EEGKEGKENSTKEGSKDDKIEE 1701
Query: 186 AFERLLSIKKEKEKNEE--EEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDD 242
E + KE K+ + E DK+ + + + G D +S ++ DD
Sbjct: 1702 GMEGKENSTKESSKDGKINEIHGDKEATMEEGSKDGGTNSTGKDSKDSKSVEINGVKDD 1760
Score = 29.3 bits (64), Expect = 4.5
Identities = 46/200 (23%), Positives = 72/200 (36%), Gaps = 33/200 (16%)
Query: 57 KKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQ-----EGFFEKENFFDGEEKND 111
++KKND+S T + D + E N ES E +K+ D E
Sbjct: 564 EQKKNDKSVEVTT----NDGDHTKEKREETQGNNGESVKNENLENKEDKKELKDDESVGA 619
Query: 112 EEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEAR 171
+ E LEE E HD + +D + G + N+EK E
Sbjct: 620 KTNNETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEK----------------EVH 663
Query: 172 NSVPENGKVMHLVKAFERLLSIKK---EKEKNEEEEENDK-----KNKVMKWALPGLQFQ 223
N M + + + +KK EK EE +EN+K K K + +
Sbjct: 664 VGDSTNDNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDD 723
Query: 224 QPVKDGDEQSEVVSSCCDDD 243
+ V D E++++ DD
Sbjct: 724 KSVDDKQEEAQIYGGESKDD 743
>At2g42510 hypothetical protein
Length = 646
Score = 43.1 bits (100), Expect = 3e-04
Identities = 50/188 (26%), Positives = 79/188 (41%), Gaps = 39/188 (20%)
Query: 57 KKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGE 116
KK D R + + K +N +GSK + RE E K+N F G N + +
Sbjct: 254 KKNDGDTLARASDIHKIEKNGNGSK-------DQREKVERVRVKDNAFVGRSVNIDLVDD 306
Query: 117 NVLEEVI----------EANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKV 166
L +V+ + + H + RK ++ GVE+ + KGN SS+V+R
Sbjct: 307 TALFDVVPFYKKGKDHSKRPVTAHTDKDAPRK-HKKVGVEKPID-KGNASSIVERNASTK 364
Query: 167 MEEARNSVPENGKVM----------------HLVKAFERLLSIKKEKE----KNEEEEEN 206
+ + RNS NGK + LV +E L + K E K E+ E+N
Sbjct: 365 VSDFRNSGEMNGKQLRIMYSRNQMESMRLLPELVTEYEGLKNHKSILEDYVVKEEKTEDN 424
Query: 207 DKKNKVMK 214
D N +++
Sbjct: 425 DDYNSILR 432
>At5g45520 putative protein
Length = 1167
Score = 42.4 bits (98), Expect = 5e-04
Identities = 31/136 (22%), Positives = 65/136 (47%), Gaps = 8/136 (5%)
Query: 99 EKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSM 158
EK N + ++ D ++G+ + EE + HD G EER + +E VEEE + + +
Sbjct: 600 EKGNVEETGKQEDGDQGDGINEEANLEDGKKHDEGKEERSLKSDEVVEEEKKTSPSEEAT 659
Query: 159 VKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALP 218
K + + ++ +++V +G + +++EK+++E E E K +++++
Sbjct: 660 EKFQNKPGDQKGKSNVEGDGD--------KGKADLEEEKKQDEVEAEKSKSDEIVEGEKK 711
Query: 219 GLQFQQPVKDGDEQSE 234
+ K GD E
Sbjct: 712 PDDKSKVEKKGDGDKE 727
Score = 37.7 bits (86), Expect = 0.013
Identities = 40/145 (27%), Positives = 71/145 (48%), Gaps = 18/145 (12%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKND--EEKGENVLEEVIEANLIIHDI 132
++D+G + V +A + RES + EN D ++ D EEKG+ E+V NL
Sbjct: 805 KSDNGVEGVDKASPS-RESTDAI---ENKPDDHQRGDKQEEKGDGEKEKV---NL----- 852
Query: 133 GNEERKIDE--EEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFE-R 189
E +K DE EE +++N G+ + + ME R+ EN KV + +
Sbjct: 853 -EEWKKHDEIKEESSKQDNVTGGDVKKSPPKESKDTMESKRDDQKENSKVQEKGDVDKGK 911
Query: 190 LLSIKKEKEKNEEEEENDKKNKVMK 214
+ + K++N+ + E+ K +KV++
Sbjct: 912 AADLDEGKKENDVKAESSKSDKVIE 936
Score = 30.0 bits (66), Expect = 2.6
Identities = 41/174 (23%), Positives = 71/174 (40%), Gaps = 22/174 (12%)
Query: 85 EAYRNLRES---QEGFFEKENFFDGEEKNDE---EKGENVLEEVIEANLIIHDIGNEERK 138
EA+ N +S Q G + ++KN++ K +N + ++I A + + K
Sbjct: 65 EAFNNASDSGGSQGGSRRSVQNIEEDQKNEDLEIRKLQNDIRQMIAAFKNLTQFQTDMSK 124
Query: 139 IDEEE-------GVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLL 191
E + + ++ G+ S +VK R KV + +P ++H K R +
Sbjct: 125 NLERDLRSTKLIAILQKKNSFGSRSHVVKEIRRKV-SALKCQIPS---LLH--KQSSRKI 178
Query: 192 SIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVVSSCCDDDDF 245
SI + E +E ++ + LPGL F + K EVV DDF
Sbjct: 179 SITDSQTAEENDEISETGIDIY---LPGLHFSEKFKTSSAFEEVVEKFQGLDDF 229
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 42.4 bits (98), Expect = 5e-04
Identities = 41/148 (27%), Positives = 67/148 (44%), Gaps = 31/148 (20%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGV-----E 146
E +E + E+ D EEK +E K ++ N+ K +EEE V E
Sbjct: 269 ELEEKLIKNED--DIEEKTEEMKEQD---------------NNQANKSEEEEDVKKKIDE 311
Query: 147 EENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEEN 206
E EK + S E+ +E V E GK ER+ +KEKEK +E+++
Sbjct: 312 NETPEKVDTESKEVESVEETTQEKEEEVKEEGK--------ERVEEEEKEKEKVKEDDQK 363
Query: 207 DKKNKVMKWALPGLQFQQPVKDGDEQSE 234
+K + K + G + ++ VK+ +E +E
Sbjct: 364 EKVEEEEKEKVKGDEEKEKVKE-EESAE 390
Score = 40.8 bits (94), Expect = 0.001
Identities = 50/200 (25%), Positives = 88/200 (44%), Gaps = 22/200 (11%)
Query: 18 EFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIR-----QRKFVVAKKKKNDQSPR----KT 68
E S TP+ + K +C VV +K I+ + K K++ N+Q+ + +
Sbjct: 248 ESSGPTPVASPVGK---DCNAVVAELEEKLIKNEDDIEEKTEEMKEQDNNQANKSEEEED 304
Query: 69 VLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF--DGEEKNDEEKGENVLEEVIEAN 126
V K EN+ K E+ + + +E EKE +G+E+ +EE+ E + +
Sbjct: 305 VKKKIDENETPEKVDTES-KEVESVEETTQEKEEEVKEEGKERVEEEEKEKEKVKEDDQK 363
Query: 127 LIIHDIGNEERKIDEE-EGVEEENEEKGNCSSMVKRRREK------VMEEARNSVPENGK 179
+ + E+ K DEE E V+EE +G +VK ++E V+ P K
Sbjct: 364 EKVEEEEKEKVKGDEEKEKVKEEESAEGKKKEVVKGKKESPSAYNDVIASKMQENPRKNK 423
Query: 180 VMHLVKAFERLLSIKKEKEK 199
V+ L AF+ ++ + K
Sbjct: 424 VLALAGAFQTVIDYETAASK 443
Score = 39.3 bits (90), Expect = 0.004
Identities = 44/184 (23%), Positives = 80/184 (42%), Gaps = 33/184 (17%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
+K ++ + V K + + +P + + K D + V E L ++++ EK
Sbjct: 234 EKVVQANESVEEKAESSGPTPVASPVGK-----DCNAVVAELEEKLIKNEDDIEEKTEEM 288
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEG---VEEENEEKGNCSSMVKR 161
++ N K E EE ++ +KIDE E V+ E++E + +
Sbjct: 289 KEQDNNQANKSEE--EEDVK------------KKIDENETPEKVDTESKEVESVEETTQE 334
Query: 162 RREKVMEEARNSVPENGKVMHLVKAFERLLSIKKE----------KEKNEEEEEND-KKN 210
+ E+V EE + V E K VK ++ +++E KEK +EEE + KK
Sbjct: 335 KEEEVKEEGKERVEEEEKEKEKVKEDDQKEKVEEEEKEKVKGDEEKEKVKEEESAEGKKK 394
Query: 211 KVMK 214
+V+K
Sbjct: 395 EVVK 398
Score = 35.4 bits (80), Expect = 0.063
Identities = 45/157 (28%), Positives = 68/157 (42%), Gaps = 19/157 (12%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDE-EKGENVLEEVIEANLIIHDIG 133
+N+D + E + +Q E+E D ++K DE E E V E E +
Sbjct: 276 KNEDDIEEKTEEMKEQDNNQANKSEEEE--DVKKKIDENETPEKVDTESKEVESVEETTQ 333
Query: 134 NEERKIDEEEGVEE-ENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLS 192
+E ++ +EEG E E EEK ++EKV EE + V + + VK E
Sbjct: 334 EKEEEV-KEEGKERVEEEEKEKEKVKEDDQKEKVEEEEKEKV-KGDEEKEKVKEEESAEG 391
Query: 193 IKKEKEKNEEE-------------EENDKKNKVMKWA 216
KKE K ++E +EN +KNKV+ A
Sbjct: 392 KKKEVVKGKKESPSAYNDVIASKMQENPRKNKVLALA 428
Score = 33.1 bits (74), Expect = 0.31
Identities = 37/157 (23%), Positives = 70/157 (44%), Gaps = 35/157 (22%)
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHD-------------IGN---------EERKIDEE 142
D EE E++ + V E+V++AN + + +G EE+ I E
Sbjct: 219 DIEEPKYEKEEKEVQEKVVQANESVEEKAESSGPTPVASPVGKDCNAVVAELEEKLIKNE 278
Query: 143 EGVEEENEE-KGNCSSMVKRRREKVMEEARNSVPENG---KVMHLVKAFERLLSIKKEKE 198
+ +EE+ EE K ++ + E+ E+ + + EN KV K E + +EKE
Sbjct: 279 DDIEEKTEEMKEQDNNQANKSEEE--EDVKKKIDENETPEKVDTESKEVESVEETTQEKE 336
Query: 199 KNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEV 235
+ +EE ++ + K ++ VK+ D++ +V
Sbjct: 337 EEVKEEGKERVEEEEK-------EKEKVKEDDQKEKV 366
>At1g03530 unknown protein
Length = 801
Score = 42.0 bits (97), Expect = 7e-04
Identities = 32/141 (22%), Positives = 70/141 (48%), Gaps = 15/141 (10%)
Query: 105 DGEEKNDEEKGE-----NVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMV 159
D +EK+DE KGE + E + +EE + DE+E +EEN+++ MV
Sbjct: 230 DDDEKSDEAKGEMDSAESESETSSSSASSSDSSSSEEEESDEDESDKEENKKEEKFEHMV 289
Query: 160 KRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALP- 218
+ + + E E G++ +L + ++ + +++++++D N+++ W+
Sbjct: 290 VGKEDDLAGEL-----EEGEIENLDEENGDDDIEDEDDDDDDDDDDDDDVNEMVAWSNDE 344
Query: 219 ----GLQFQQPVKDGDEQSEV 235
GLQ ++P++ +E E+
Sbjct: 345 DDDLGLQTKEPIRSKNELKEL 365
>At4g02070 G/T DNA mismatch repair enzyme
Length = 1324
Score = 41.6 bits (96), Expect = 9e-04
Identities = 28/82 (34%), Positives = 48/82 (58%), Gaps = 3/82 (3%)
Query: 100 KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE--KGNCSS 157
+E+ DG++ +DE+ G+NV +EV E+ ++ +E ++DEEE VEE++EE K N S
Sbjct: 212 EEDKSDGDDSSDEDWGKNVGKEVCESEEDDVEL-VDENEMDEEELVEEKDEETSKVNRVS 270
Query: 158 MVKRRREKVMEEARNSVPENGK 179
R+ K E ++ + K
Sbjct: 271 KTDSRKRKTSEVTKSGGEKKSK 292
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 41.6 bits (96), Expect = 9e-04
Identities = 38/166 (22%), Positives = 71/166 (41%), Gaps = 9/166 (5%)
Query: 49 RQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEE 108
R+ + +KK+ ++ RK + E ++ + E + E+++ +E EE
Sbjct: 461 RREEEETERKKREEEEARKREEERKREEEEAKRREEERKKREEEAEQARKREEEREKEEE 520
Query: 109 ---KNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREK 165
K +EE+ EEV +ERK EEE + E E K M KRR ++
Sbjct: 521 MAKKREEERQRKEREEVERKRR-----EEQERKRREEEARKREEERKRE-EEMAKRREQE 574
Query: 166 VMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
+ R V + K E + ++++ + +E EE ++K +
Sbjct: 575 RQRKEREEVERKIREEQERKREEEMAKRREQERQKKEREEMERKKR 620
Score = 40.0 bits (92), Expect = 0.003
Identities = 44/180 (24%), Positives = 80/180 (44%), Gaps = 13/180 (7%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKC-VCEAYRNLRESQEGFFEKENF 103
++R R+ + + +++K ++ RK K E ++ + E R RE +E +E
Sbjct: 425 EERKRREEEEIERRRKEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKREEER 484
Query: 104 FDGEE--KNDEEKGENVLEEVIEANLIIHDIGNEE----RKIDEEEGVEEENEEKGNCSS 157
EE K EE+ + EE +A + EE ++ +E + E E E+
Sbjct: 485 KREEEEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRKEREEVERKRREE 544
Query: 158 MVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKE---KNEEEEENDKKNKVMK 214
++RRE EEAR E + + K E+ K+ +E K EE+E ++ ++ K
Sbjct: 545 QERKRRE---EEARKREEERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAK 601
Score = 38.1 bits (87), Expect = 0.010
Identities = 41/179 (22%), Positives = 76/179 (41%), Gaps = 19/179 (10%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
++R ++ + +K+ ++ ++ + K E + K E R RE QE K
Sbjct: 496 EERKKREEEAEQARKREEEREKEEEMAKKREEERQRKEREEVERKRREEQE---RKRREE 552
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENE---------EKGNC 155
+ ++ +E K E + + E + ERKI EE+ + E E +K
Sbjct: 553 EARKREEERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKER 612
Query: 156 SSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
M +++RE EEAR E K ++ ER +++ E+ EEE ++ + K
Sbjct: 613 EEMERKKRE---EEARKREEEMAK----IREEERQRKEREDVERKRREEEAMRREEERK 664
Score = 36.6 bits (83), Expect = 0.028
Identities = 40/166 (24%), Positives = 75/166 (45%), Gaps = 21/166 (12%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
+++ R+R+ A+K++ ++ R+ + K E + K E R +RE QE E+E
Sbjct: 543 EEQERKRREEEARKREEERK-REEEMAKRREQERQRKEREEVERKIREEQERKREEEMAK 601
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEG-VEEENEEKGNCSSMVKRRR 163
E++ +++ E + + E E RK +EE + EE ++ + ++RR
Sbjct: 602 RREQERQKKEREEMERKKRE---------EEARKREEEMAKIREEERQRKEREDVERKRR 652
Query: 164 EKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKK 209
E EEA E + K E E+ + +EEEE ++
Sbjct: 653 E---EEAMRREEERKREEEAAKRAE-------EERRKKEEEEEKRR 688
Score = 32.3 bits (72), Expect = 0.53
Identities = 31/138 (22%), Positives = 58/138 (41%), Gaps = 5/138 (3%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
++R R+ + ++++ + R+ V K E + + A R +E Q+ E+E
Sbjct: 559 EERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKK--EREEME 616
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGN---CSSMVKR 161
+ + + K E + ++ E + + ERK EEE + E E K +
Sbjct: 617 RKKREEEARKREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKREEEAAKRAEEE 676
Query: 162 RREKVMEEARNSVPENGK 179
RR+K EE + P K
Sbjct: 677 RRKKEEEEEKRRWPPQPK 694
Score = 32.0 bits (71), Expect = 0.69
Identities = 25/91 (27%), Positives = 43/91 (46%), Gaps = 7/91 (7%)
Query: 124 EANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHL 183
E + ++ +I +R+ +EE + EE+ KRR E EEA+ E +
Sbjct: 416 ELSKLMREIEERKRREEEEIERRRKEEEEARKREEAKRREE---EEAKRREEEETE---- 468
Query: 184 VKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
K E + K+E+E+ EEEE ++ + K
Sbjct: 469 RKKREEEEARKREEERKREEEEAKRREEERK 499
>At2g02200 putative protein on transposon FARE2.10 (cds2)
Length = 586
Score = 41.6 bits (96), Expect = 9e-04
Identities = 42/153 (27%), Positives = 71/153 (45%), Gaps = 24/153 (15%)
Query: 91 RESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKI-----DEEEGV 145
RE Q+ EK+ EEK +EE E LE++ D G E+++I +E EG
Sbjct: 203 REEQKEEDEKKE--QEEEKQEEEGKEEKLEKIEYRG----DEGTEKQEIPKQGDEEMEGE 256
Query: 146 EEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEE 205
EE+ EE+G +E+ E+ E + + K + + ++EK+K E +EE
Sbjct: 257 EEKQEEEG---------KEEEEEKVEYRGDEGTEKQEIPKQGDEEMEGEEEKQKEEGKEE 307
Query: 206 NDKK----NKVMKWALPGLQFQQPVKDGDEQSE 234
++K + + + Q+ K GDE+ E
Sbjct: 308 EEEKVEYRDHHSTCNVEETEKQENPKQGDEEME 340
>At4g37820 unknown protein
Length = 532
Score = 40.8 bits (94), Expect = 0.001
Identities = 37/180 (20%), Positives = 73/180 (40%), Gaps = 17/180 (9%)
Query: 57 KKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGE 116
K++K D S ++ + EN + + ++E++ E+ + +G E + EK
Sbjct: 355 KREKEDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESSSQEGNENKETEKKS 414
Query: 117 NVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPE 176
+ + + N E+KI++ E + N +KG+ + +RE + + +
Sbjct: 415 SESQR--------KENTNSEKKIEQVESTDSSNTQKGDEQKTDESKRESGNDTSNKETED 466
Query: 177 NGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQPVKDGDEQSEVV 236
+ K K+E +N E EE + + K AL + Q VKD E +
Sbjct: 467 DSSKTESEK--------KEENNRNGETEETQNEQEQTKSALE-ISHTQDVKDARTDLETL 517
Score = 32.0 bits (71), Expect = 0.69
Identities = 45/182 (24%), Positives = 72/182 (38%), Gaps = 27/182 (14%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENE- 150
E +EG +N G+ D+E G+ ++E E + + E EEG E+ N
Sbjct: 84 EDEEG---SKNEGGGDVSTDKENGDEIVEREEEEKAVEENNEKEAEGTGNEEGNEDSNNG 140
Query: 151 EKGNCSSMVKRRREKVMEEAR----NSVPENGKVMH-LVKAFERLLSIKKEKEKNE---- 201
E + E EEAR + +VMH + + ++ E + N
Sbjct: 141 ESEKVVDESEGGNEISNEEAREINYKGDDASSEVMHGTEEKSNEKVEVEGESKSNSTENV 200
Query: 202 --EEEENDKKNKVMKWAL---PGLQFQQPVKDGDEQSEVVSSCCDDDDFDF*TTWIGSKG 256
E+E+ KN+V++ ++ L + D EQ E S + D T G KG
Sbjct: 201 SVHEDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKS------ELDSKT---GEKG 251
Query: 257 FS 258
FS
Sbjct: 252 FS 253
Score = 31.2 bits (69), Expect = 1.2
Identities = 29/120 (24%), Positives = 47/120 (39%), Gaps = 29/120 (24%)
Query: 107 EEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEK----------GNCS 156
+ KN+E++ E V E+ + ++ ++E EE+ E K G
Sbjct: 292 QTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQGEGKEE 351
Query: 157 SMVKRRRE--KVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
KR +E EE++ PEN KEKE + +EEN+ K +K
Sbjct: 352 EPEKREKEDSSSQEESKEEEPEN-----------------KEKEASSSQEENEIKETEIK 394
>At5g44280 putative protein
Length = 486
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 5/93 (5%)
Query: 85 EAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEG 144
EA ++L+E E EKE D E K+DE + + +EV++ N D ++ + DEEE
Sbjct: 27 EATQDLQEKDETKEEKEG--DEEVKHDEAEED---QEVVKPNDAEEDDDGDDAEEDEEEE 81
Query: 145 VEEENEEKGNCSSMVKRRREKVMEEARNSVPEN 177
VE E +E+ + E+ E+++ P +
Sbjct: 82 VEAEEDEEAEEEEEEEEEEEEEEEDSKERSPSS 114
Score = 34.3 bits (77), Expect = 0.14
Identities = 22/64 (34%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRRE 164
D EE +D + E EE +EA D EE + +EEE EEE + K S + +
Sbjct: 64 DAEEDDDGDDAEEDEEEEVEAE---EDEEAEEEEEEEEEEEEEEEDSKERSPSSISGDQS 120
Query: 165 KVME 168
+ ME
Sbjct: 121 EFME 124
>At3g43590 putative protein
Length = 551
Score = 40.4 bits (93), Expect = 0.002
Identities = 40/150 (26%), Positives = 66/150 (43%), Gaps = 16/150 (10%)
Query: 91 RESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNE-ERKIDEEEGVEEEN 149
RE E +N GEE++D+E E++ +++E L D+GN+ + + + GV
Sbjct: 20 REDPVAITEVDN---GEEEDDDEANEDLSLKILEKALSRRDVGNKLDSDLSSDSGVVSTV 76
Query: 150 EEKGNCSSMVKRRREKVME----EARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEE 205
G S + K K M+ EA + +P K E + +K E E +E E
Sbjct: 77 MVNGVKSKVKKSESSKKMKRNKLEADHEIPIVWNDQDEEKVVEEI--VKGEGEDDEVERS 134
Query: 206 NDKK------NKVMKWALPGLQFQQPVKDG 229
++ K N V+K L G ++ P G
Sbjct: 135 DEPKTEETASNLVLKKLLRGARYFDPPDAG 164
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.339 0.149 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,501,731
Number of Sequences: 26719
Number of extensions: 508689
Number of successful extensions: 7246
Number of sequences better than 10.0: 314
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 273
Number of HSP's that attempted gapping in prelim test: 5145
Number of HSP's gapped (non-prelim): 978
length of query: 439
length of database: 11,318,596
effective HSP length: 102
effective length of query: 337
effective length of database: 8,593,258
effective search space: 2895927946
effective search space used: 2895927946
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149305.8


