

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.6 - phase: 0 /pseudo/partial
(543 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
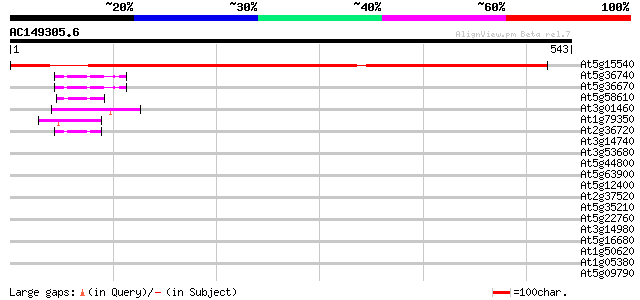
At5g15540 putative protein 593 e-170
At5g36740 putative protein 47 3e-05
At5g36670 putative protein 47 3e-05
At5g58610 putative protein 44 3e-04
At3g01460 unknown protein 44 3e-04
At1g79350 hypothetical protein 43 5e-04
At2g36720 putative PHD-type zinc finger protein 42 9e-04
At3g14740 unknown protein 41 0.002
At3g53680 putative protein 40 0.003
At5g44800 helicase-like protein 40 0.003
At5g63900 putative protein 40 0.004
At5g12400 putative protein 40 0.004
At2g37520 unknown protein 40 0.004
At5g35210 putative protein 39 0.007
At5g22760 putative protein 39 0.007
At3g14980 PHD-finger protein, putative 39 0.010
At5g16680 putative protein 38 0.013
At1g50620 hypothetical protein 38 0.016
At1g05380 unknown protein 37 0.021
At5g09790 unknown protein 37 0.037
>At5g15540 putative protein
Length = 1755
Score = 593 bits (1530), Expect = e-170
Identities = 308/521 (59%), Positives = 376/521 (72%), Gaps = 45/521 (8%)
Query: 1 MAIDILGTIAARLKRDAVICSQEKFWVLQDLLSEDAAPQHYPKDTCCVCLGGRVENLFKC 60
MAI++LGTIAARLKRDAV+CS+++FW L + SE + Q
Sbjct: 596 MAIELLGTIAARLKRDAVLCSKDRFWTLLESDSEISVDQ--------------------- 634
Query: 61 SGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCNSQRKDDAKKNRKVS-KD 119
E ++ +RNW+C +C+C +QLLVLQSYC + K K + S ++
Sbjct: 635 ---------------ELDISSRNWHCPLCVCKRQLLVLQSYCKTDTKGTGKLESEESIEN 679
Query: 120 DSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPIYYFARMKSR 179
S + E+VQQ+LLNY QDV SADD+H FICWFYLC WYK+ PK Q K YY AR+K++
Sbjct: 680 PSMITKTEVVQQMLLNYLQDVGSADDVHTFICWFYLCLWYKDVPKSQNKFKYYIARLKAK 739
Query: 180 TIVRDSGSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRA 239
+I+R+SG+ +S LTRD+IK+ITLALG NSSF RGFDKI + LL SL+EN+P IRAKALRA
Sbjct: 740 SIIRNSGATTSFLTRDAIKQITLALGMNSSFSRGFDKILNMLLASLRENAPNIRAKALRA 799
Query: 240 VSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITE 299
VSIIVEADPEVL DK VQ +VEGRFCD+AISVREAALELVGRHIASHPDVG KYFEK+ E
Sbjct: 800 VSIIVEADPEVLCDKRVQLAVEGRFCDSAISVREAALELVGRHIASHPDVGIKYFEKVAE 859
Query: 300 RIKDTGVSVRKRAIKIIRDMCCSDGNFLRVYKSLHRDNFTCY***IEYPAYSASDLQDLV 359
RIKDTGVSVRKRAIKIIRDMC S+ NF + + + S +QDLV
Sbjct: 860 RIKDTGVSVRKRAIKIIRDMCTSNPNFSEFTSACAEI--------LSRISDDESSVQDLV 911
Query: 360 CKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTL 419
CKTFYEFWFEEP TQ D S++PLE+ KKT+Q+V +L R PN QLLVT+IKR L L
Sbjct: 912 CKTFYEFWFEEPPGHHTQFASDASSIPLELEKKTKQMVGLLSRTPNQQLLVTIIKRALAL 971
Query: 420 DFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAF 479
DF PQ+AKA+G+NPV+L +VR+RCELMCKCLLEKILQV+EM+ E E LPYVLVLHAF
Sbjct: 972 DFFPQAAKAAGINPVALASVRRRCELMCKCLLEKILQVEEMSREEGEVQVLPYVLVLHAF 1031
Query: 480 CLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
CLVDP LC PAS+P++FV+TLQPYLK+Q + +L S+
Sbjct: 1032 CLVDPGLCTPASDPTKFVITLQPYLKSQADSRTGAQLLESI 1072
>At5g36740 putative protein
Length = 960
Score = 47.0 bits (110), Expect = 3e-05
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 15/70 (21%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCN 103
DTC +C G +L C GC FH CLD+K + P+ WYC C C +C
Sbjct: 651 DTCGICGDGG--DLICCDGCPSTFHQSCLDIK--KFPSGAWYCYNCSC--------KFC- 697
Query: 104 SQRKDDAKKN 113
KD+A K+
Sbjct: 698 --EKDEAAKH 705
>At5g36670 putative protein
Length = 1030
Score = 47.0 bits (110), Expect = 3e-05
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 15/70 (21%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCN 103
DTC +C G +L C GC FH CLD+K + P+ WYC C C +C
Sbjct: 498 DTCGICGDGG--DLICCDGCPSTFHQSCLDIK--KFPSGAWYCYNCSC--------KFC- 544
Query: 104 SQRKDDAKKN 113
KD+A K+
Sbjct: 545 --EKDEAAKH 552
>At5g58610 putative protein
Length = 1065
Score = 43.5 bits (101), Expect = 3e-04
Identities = 20/47 (42%), Positives = 28/47 (59%), Gaps = 6/47 (12%)
Query: 46 CCVC-LGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCIC 91
C VC GG+ L C GC FHA+CL ++ +VP+ +W+C C C
Sbjct: 697 CSVCHYGGK---LILCDGCPSAFHANCLGLE--DVPDGDWFCQSCCC 738
>At3g01460 unknown protein
Length = 2176
Score = 43.5 bits (101), Expect = 3e-04
Identities = 22/89 (24%), Positives = 44/89 (48%), Gaps = 3/89 (3%)
Query: 41 YPKDTCCVC-LGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLL--V 97
+ + C VC + +++ C CD +H CL+ +P+ NWYC C+ +K++
Sbjct: 1285 WDEGVCKVCGVDKDDDSVLLCDTCDAEYHTYCLNPPLIRIPDGNWYCPSCVIAKRMAQEA 1344
Query: 98 LQSYCNSQRKDDAKKNRKVSKDDSTFSNH 126
L+SY +R+ K ++++ + H
Sbjct: 1345 LESYKLVRRRKGRKYQGELTRASMELTAH 1373
Score = 33.5 bits (75), Expect = 0.31
Identities = 17/58 (29%), Positives = 25/58 (42%), Gaps = 5/58 (8%)
Query: 37 APQHYP---KDTCCVCLGG--RVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
AP P +D C G +E + C C+R FH C++ P+ +W C C
Sbjct: 73 APAEVPEPDRDASCGACGRPESIELVVVCDACERGFHMSCVNDGVEAAPSADWMCSDC 130
>At1g79350 hypothetical protein
Length = 1318
Score = 42.7 bits (99), Expect = 5e-04
Identities = 21/66 (31%), Positives = 32/66 (47%), Gaps = 5/66 (7%)
Query: 29 QDLLSEDAAPQHYPKDT----CCVCLG-GRVENLFKCSGCDRLFHADCLDVKENEVPNRN 83
+++ S D+A D C +C G + L CS CD+LFH DC+ ++P+
Sbjct: 665 ENMYSADSADDSNDSDDEFQICQICSGEDERKKLLHCSECDKLFHPDCVVPPVIDLPSEA 724
Query: 84 WYCLMC 89
W C C
Sbjct: 725 WICFSC 730
>At2g36720 putative PHD-type zinc finger protein
Length = 958
Score = 42.0 bits (97), Expect = 9e-04
Identities = 17/46 (36%), Positives = 23/46 (49%), Gaps = 4/46 (8%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
D C +C G NL C C R FH +C+ + +P NW+C C
Sbjct: 578 DLCVICADGG--NLLLCDSCPRAFHIECVSLP--SIPRGNWHCKYC 619
Score = 30.4 bits (67), Expect = 2.6
Identities = 12/33 (36%), Positives = 17/33 (51%), Gaps = 4/33 (12%)
Query: 60 CSGCDRLFHADCLD----VKENEVPNRNWYCLM 88
C C++ +H CL V E+P NW+C M
Sbjct: 678 CDQCEKEYHIGCLSSQNIVDLKELPKGNWFCSM 710
>At3g14740 unknown protein
Length = 341
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/71 (30%), Positives = 31/71 (42%), Gaps = 4/71 (5%)
Query: 46 CCVCL---GGRVENLFKCSGCDRLFHADCL-DVKENEVPNRNWYCLMCICSKQLLVLQSY 101
C VC G + + C GCD + HA C + +P +W+C C+ SK + S
Sbjct: 153 CAVCQSTDGDPLNPIVFCDGCDLMVHASCYGNPLVKAIPEGDWFCRQCLSSKNREKIFSC 212
Query: 102 CNSQRKDDAKK 112
C K A K
Sbjct: 213 CLCTTKGGAMK 223
>At3g53680 putative protein
Length = 839
Score = 40.4 bits (93), Expect = 0.003
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 4/46 (8%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
D C +C G +L C+GC + FH CL K +P WYC C
Sbjct: 487 DMCSIC--GNGGDLLLCAGCPQAFHTACL--KFQSMPEGTWYCSSC 528
Score = 30.0 bits (66), Expect = 3.4
Identities = 17/47 (36%), Positives = 24/47 (50%), Gaps = 9/47 (19%)
Query: 60 CSGCDRLFHADCLDVKENE------VPNRNWYCLMCICSKQLLVLQS 100
C C++ +H CL +ENE +P W+C CS+ VLQS
Sbjct: 585 CDQCEKEYHVGCL--RENELCDLKGIPQDKWFC-CSDCSRIHRVLQS 628
>At5g44800 helicase-like protein
Length = 2228
Score = 40.0 bits (92), Expect = 0.003
Identities = 23/75 (30%), Positives = 32/75 (42%), Gaps = 8/75 (10%)
Query: 46 CCVC-LGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLV----LQS 100
C +C LGG +L C C R +H CL+ +PN W C C + + L L +
Sbjct: 64 CVICDLGG---DLLCCDSCPRTYHTACLNPPLKRIPNGKWICPKCSPNSEALKPVNRLDA 120
Query: 101 YCNSQRKDDAKKNRK 115
R K+N K
Sbjct: 121 IAKRARTKTKKRNSK 135
>At5g63900 putative protein
Length = 557
Score = 39.7 bits (91), Expect = 0.004
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 5/49 (10%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRN-WYCLMCIC 91
D CCVC G +L C GC FH CL + + +P + W+C C C
Sbjct: 257 DVCCVCHWGG--DLLLCDGCPSAFHHACLGL--SSLPEEDLWFCPCCCC 301
Score = 35.0 bits (79), Expect = 0.11
Identities = 19/65 (29%), Positives = 29/65 (44%), Gaps = 6/65 (9%)
Query: 46 CCVCLGGRVEN-----LFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQS 100
CC + G +E+ L C C R FH CL V +R W+C C++ L++
Sbjct: 299 CCCDICGSMESPANSKLMACEQCQRRFHLTCLKEDSCIVSSRGWFC-SSQCNRVFSALEN 357
Query: 101 YCNSQ 105
S+
Sbjct: 358 LLGSK 362
>At5g12400 putative protein
Length = 1595
Score = 39.7 bits (91), Expect = 0.004
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query: 44 DTCCVC-LGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
D CC C + G +L C GC +H+ C+ + + +P +WYC C
Sbjct: 609 DDCCFCKMDG---SLLCCDGCPAAYHSKCVGLASHLLPEGDWYCPEC 652
>At2g37520 unknown protein
Length = 825
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 4/46 (8%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
D C +C G +L C+GC + FH CL K +P WYC C
Sbjct: 465 DMCSICGDGG--DLLLCAGCPQAFHTACL--KFQSMPEGTWYCSSC 506
>At5g35210 putative protein
Length = 1606
Score = 38.9 bits (89), Expect = 0.007
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQ 94
D C +C G L C GC +H+ C+ V + +P+ W+C C +K+
Sbjct: 412 DECRIC--GMDGTLLCCDGCPLAYHSRCIGVVKMYIPDGPWFCPECTINKK 460
>At5g22760 putative protein
Length = 1516
Score = 38.9 bits (89), Expect = 0.007
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSK 93
D C +C G L C GC +H+ C+ V + +P+ WYC C K
Sbjct: 414 DECRLC--GMDGTLLCCDGCPLAYHSRCIGVVKMYIPDGPWYCPECTIKK 461
>At3g14980 PHD-finger protein, putative
Length = 1189
Score = 38.5 bits (88), Expect = 0.010
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 4/58 (6%)
Query: 34 EDAAPQHYPKDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCIC 91
E A+ D+C VC G L C C FH CL ++ +P +WYC C C
Sbjct: 717 EKASDDDPNDDSCGVCGDGG--ELICCDNCPSTFHQACLSMQV--LPEGSWYCSSCTC 770
>At5g16680 putative protein
Length = 1280
Score = 38.1 bits (87), Expect = 0.013
Identities = 26/95 (27%), Positives = 43/95 (44%), Gaps = 12/95 (12%)
Query: 33 SEDAAPQHYPKDTCCVCL-GGRVENLFKCSGC-DRLFHADCLDVKENEVPNRNWYCLMCI 90
S+D+ + C +C GR + L CSGC D H C+ +EVP +W C C
Sbjct: 263 SDDSEMVEHDVKVCDICGDAGREDLLAICSGCSDGAEHTYCMREMLDEVPEGDWLCEECA 322
Query: 91 CSKQLLVLQSYCNSQRKDDAKKNRKVSKDDSTFSN 125
+ ++K +AK+ R+ +T+S+
Sbjct: 323 EEAE----------KQKQEAKRKRETEVTFNTYSS 347
>At1g50620 hypothetical protein
Length = 629
Score = 37.7 bits (86), Expect = 0.016
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 4/66 (6%)
Query: 29 QDLLSEDAAPQHYPKD-TCCVCLG--GRVENLFKCSGCDRLFHADCLDVKE-NEVPNRNW 84
Q+LL + ++ K TC +C G +E + C C++ +H CL VP W
Sbjct: 306 QNLLWNPPSREYMSKAMTCQICQGTINEIETVLICDACEKGYHLKCLHAHNIKGVPKSEW 365
Query: 85 YCLMCI 90
+C C+
Sbjct: 366 HCSRCV 371
>At1g05380 unknown protein
Length = 1138
Score = 37.4 bits (85), Expect = 0.021
Identities = 16/48 (33%), Positives = 26/48 (53%), Gaps = 4/48 (8%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCIC 91
D C +C G +L C GC +H +CL ++ +P+ +W+C C C
Sbjct: 626 DACGICGDGG--DLICCDGCPSTYHQNCLGMQ--VLPSGDWHCPNCTC 669
>At5g09790 unknown protein
Length = 352
Score = 36.6 bits (83), Expect = 0.037
Identities = 25/85 (29%), Positives = 33/85 (38%), Gaps = 11/85 (12%)
Query: 34 EDAAPQHYPKDTCCVCLGGRVEN-LFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICS 92
E+ Y TC C G ++ L C CDR FH CL VP W C+ C
Sbjct: 55 EEEDEDSYSNVTCEKCGSGEGDDELLLCDKCDRGFHMKCLRPIVVRVPIGTWLCVDCSDQ 114
Query: 93 KQLLVLQSYCNSQRKDDAKKNRKVS 117
+ + RK+ K+ R S
Sbjct: 115 RPV----------RKETRKRRRSCS 129
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,024,679
Number of Sequences: 26719
Number of extensions: 503712
Number of successful extensions: 2018
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 1928
Number of HSP's gapped (non-prelim): 129
length of query: 543
length of database: 11,318,596
effective HSP length: 104
effective length of query: 439
effective length of database: 8,539,820
effective search space: 3748980980
effective search space used: 3748980980
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149305.6


