

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149212.6 - phase: 0 /pseudo
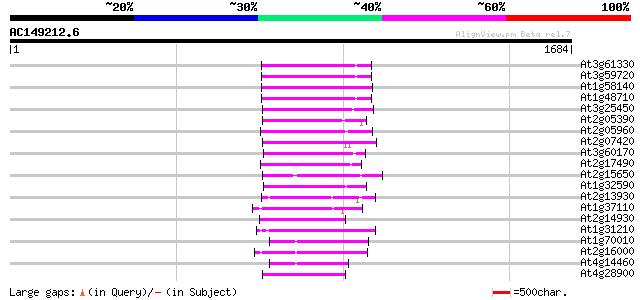
(1684 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g61330 copia-type polyprotein 207 4e-53
At3g59720 copia-type reverse transcriptase-like protein 203 6e-52
At1g58140 hypothetical protein 200 5e-51
At1g48710 hypothetical protein 199 9e-51
At3g25450 hypothetical protein 194 4e-49
At2g05390 putative retroelement pol polyprotein 188 2e-47
At2g05960 putative retroelement pol polyprotein 187 3e-47
At2g07420 putative retroelement pol polyprotein 182 1e-45
At3g60170 putative protein 179 2e-44
At2g17490 putative retroelement pol polyprotein 177 6e-44
At2g15650 putative retroelement pol polyprotein 177 6e-44
At1g32590 hypothetical protein, 5' partial 171 4e-42
At2g13930 putative retroelement pol polyprotein 165 2e-40
At1g37110 162 2e-39
At2g14930 pseudogene 161 3e-39
At1g31210 putative reverse transcriptase 161 3e-39
At1g70010 hypothetical protein 160 4e-39
At2g16000 putative retroelement pol polyprotein 160 8e-39
At4g14460 retrovirus-related like polyprotein 159 1e-38
At4g28900 putative protein 158 2e-38
>At3g61330 copia-type polyprotein
Length = 1352
Score = 207 bits (527), Expect = 4e-53
Identities = 113/330 (34%), Positives = 170/330 (51%), Gaps = 3/330 (0%)
Query: 755 NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSR 814
N C GK K SF + +PLEL+H D+ G + SL S Y L+ +DD+SR
Sbjct: 499 NQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSR 558
Query: 815 WTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHE 874
TWV F+K K E+F F ++ E L I +RSD GGEF ++ F +CE +GI +
Sbjct: 559 KTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQ 618
Query: 875 FSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEK 934
+ PR+PQQNGVVERKNRT+ EMAR+M+ L K WAEA+ + Y+ NR + + K
Sbjct: 619 LTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGK 678
Query: 935 TAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSE 994
T E + G +P +S+ FG + + K D K+++ IF+GY SK Y++YN +
Sbjct: 679 TPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPD 738
Query: 995 THCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEI 1054
T S ++ FD+ + + N ED E + E SE ++P
Sbjct: 739 TKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDEPEPTR---EEPPSEEPTTPPTSP 795
Query: 1055 TPEAESNSEAEPSPEIQNENASEDIQDNSQ 1084
T S +E +P ++ ++ +N +
Sbjct: 796 TSSQIEESSSERTPRFRSIQELYEVTENQE 825
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 203 bits (517), Expect = 6e-52
Identities = 111/330 (33%), Positives = 168/330 (50%), Gaps = 3/330 (0%)
Query: 755 NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSR 814
N C G K SF + +PLEL+H D+ G + SL S Y L+ +DD+SR
Sbjct: 499 NQVCEGCLLGNQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSR 558
Query: 815 WTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHE 874
TWV F+K K E+F F ++ E L I +RSD GGEF ++ F +CE +GI +
Sbjct: 559 KTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDSGGEFTSKEFLKYCEDNGIRRQ 618
Query: 875 FSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEK 934
+ PR+PQQNGV ERKNRT+ EMAR+M+ L K WAEA+ + Y+ NR + + K
Sbjct: 619 LTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGK 678
Query: 935 TAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSE 994
T E + G +P +S+ FG + + K D K+++ IF+GY SK Y++YN +
Sbjct: 679 TPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRNKLDDKSEKYIFIGYDNNSKGYKLYNPD 738
Query: 995 THCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEI 1054
T S ++ FD+ + + N ED E + E SE ++P
Sbjct: 739 TKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTR---EEPPSEEPTTPPTSP 795
Query: 1055 TPEAESNSEAEPSPEIQNENASEDIQDNSQ 1084
T S +E +P ++ ++ +N +
Sbjct: 796 TSSQIEESSSERTPRFRSIQELYEVTENQE 825
>At1g58140 hypothetical protein
Length = 1320
Score = 200 bits (509), Expect = 5e-51
Identities = 111/334 (33%), Positives = 167/334 (49%), Gaps = 2/334 (0%)
Query: 755 NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSR 814
N C GK K SF + +PLEL+H D+ G + SL S Y L+ +DD+SR
Sbjct: 499 NQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSR 558
Query: 815 WTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHE 874
TWV F+K K E+F F ++ E L I +RSD GGEF ++ F +CE +GI +
Sbjct: 559 KTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQ 618
Query: 875 FSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEK 934
+ PR+PQQNGV ERKNRT+ EMAR+M+ L K WAEA+ + Y+ NR + + K
Sbjct: 619 LTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGK 678
Query: 935 TAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSE 994
T E + G +P +S+ FG + + K D K+++ IF+GY SK Y++YN +
Sbjct: 679 TPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPD 738
Query: 995 THCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEI 1054
T S ++ FD+ + + N ED E + + E + P +
Sbjct: 739 TKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTR--EEPPSEEPTTPPTSPT 796
Query: 1055 TPEAESNSEAEPSPEIQNENASEDIQDNSQQAIQ 1088
+ + E E E + + D ++IQ
Sbjct: 797 SSQIEEKCEPMDFQEAIEKKTWRNAMDEEIKSIQ 830
>At1g48710 hypothetical protein
Length = 1352
Score = 199 bits (507), Expect = 9e-51
Identities = 110/330 (33%), Positives = 167/330 (50%), Gaps = 3/330 (0%)
Query: 755 NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSR 814
N C GK K SF + + LEL+H D+ G + SL S Y L+ +DD+SR
Sbjct: 499 NQVCEGCLLGKQFKMSFPKESSSRAQKSLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSR 558
Query: 815 WTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHE 874
TWV F+K K E+F F ++ E L I +RSD GGEF ++ F +CE +GI +
Sbjct: 559 KTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQ 618
Query: 875 FSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEK 934
+ PR+PQQNGV ERKNRT+ EMAR+M+ L K WAEA+ + Y+ NR + + K
Sbjct: 619 LTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGK 678
Query: 935 TAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSE 994
T E + G + +S+ FG + + K D K+++ IF+GY SK Y++YN +
Sbjct: 679 TPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPD 738
Query: 995 THCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEI 1054
T S ++ FD+ + + N ED E + E SE ++P
Sbjct: 739 TKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDEPEPTR---EEPPSEEPTTPPTSP 795
Query: 1055 TPEAESNSEAEPSPEIQNENASEDIQDNSQ 1084
T S +E +P ++ ++ +N +
Sbjct: 796 TSSQIEESSSERTPRFRSIQELYEVTENQE 825
>At3g25450 hypothetical protein
Length = 1343
Score = 194 bits (493), Expect = 4e-49
Identities = 111/340 (32%), Positives = 182/340 (52%), Gaps = 10/340 (2%)
Query: 759 GACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWV 818
G+C GK + SF ++ LEL+H DL G ++ ++ +Y V++DD+SR+ W
Sbjct: 461 GSCLFGKQARHSFPKATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWS 520
Query: 819 KFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSP 878
+K K A F F ++ E I R+D GGEF + F+ FC K GI ++P
Sbjct: 521 ILLKEKSEAFGKFKEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGINRHLTAP 580
Query: 879 RTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYE 938
TPQQNGVVER+NRTL M R+++ N+ + W EA+ S Y+ NR+ R + +T YE
Sbjct: 581 YTPQQNGVVERRNRTLLGMTRSILKHMNMPNYLWGEAVRHSTYLINRVGTRSLSNQTPYE 640
Query: 939 LFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCV 998
+FK +PN+ + FGC Y LKK D +++ ++LG SKAYR+ + +
Sbjct: 641 VFKHKKPNVEHLRVFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRI 700
Query: 999 EESMHVKFDD------REPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEA 1052
S V FD+ +E S+T ++S + T+ ++ +++++ +E E + EA
Sbjct: 701 FVSRDVVFDENRSWMWQESSSETDKESGTFTITLSE---FGNNGVTENDISTEPEETEEA 757
Query: 1053 EITPEAES-NSEAEPSPEIQNENASEDIQDNSQQAIQPKF 1091
EI E E+ EAE Q++ + ++ + +Q I+P +
Sbjct: 758 EINGEDENIIEEAETEEHDQSQEEPQPVRRSQRQVIRPNY 797
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 188 bits (478), Expect = 2e-47
Identities = 106/324 (32%), Positives = 168/324 (51%), Gaps = 16/324 (4%)
Query: 759 GACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWV 818
G+C GK + F S+ LEL+H DL G + ++ +Y LV++DD++R+ W
Sbjct: 429 GSCLLGKQARQPFPKATTYRASQVLELVHGDLCGPITQSTTAKKRYILVLIDDHTRYMWS 488
Query: 819 KFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSP 878
+K K A E F F T+++ E +KI R+D GGEF ++ F+ FC K GI ++P
Sbjct: 489 MLLKEKSEAFEKFRDFKTKVEQESGVKIKTFRTDKGGEFVSQEFQDFCAKEGINRHLTAP 548
Query: 879 RTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYE 938
TPQQNGVVER+NRTL M R+++ + + W EA+ S YI NR+ R + +T YE
Sbjct: 549 YTPQQNGVVERRNRTLLGMTRSILKHMKMPNYLWGEAVRHSTYIINRVGTRSLQNQTPYE 608
Query: 939 LFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCV 998
+FK +PN+ + FGC Y +L+K D +++ ++LG SKAYR+ + +
Sbjct: 609 VFKQRKPNVEHLRVFGCIGYAKIEGPHLRKLDDRSKMLVYLGTEPGSKAYRLLDPTNRKI 668
Query: 999 EESMHVKFDDREPGSKTPEQSES-NAGTIDSEDASESDQLSDSEKHSEVESSPEAE---- 1053
+K+++ + ++ + S G + ESD + + E E+S E E
Sbjct: 669 -----IKWNNSDSETRDISGTFSLTLGEFGNNGIQESDDIETEKNGEESENSHEEEGENE 723
Query: 1054 ------ITPEAESNSEAEPSPEIQ 1071
I E S A P P ++
Sbjct: 724 HNEQEQIDAEETQPSHATPLPTLR 747
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 187 bits (476), Expect = 3e-47
Identities = 110/347 (31%), Positives = 180/347 (51%), Gaps = 13/347 (3%)
Query: 752 PS*NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDD 811
P +C GK + F S+ LEL+H DL G ++ + +Y LV++DD
Sbjct: 418 PKEKEICASCMLGKQARQVFPKATTYRASQILELIHGDLCGPISPPTAAKRRYILVLIDD 477
Query: 812 YSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGI 871
+SR+ W +K K A F +F ++ E KI +R+D GGEF ++ F++FCE+ GI
Sbjct: 478 HSRFMWSFLLKEKSEAFGKFKTFKATVEQETGEKIKTLRTDRGGEFLSQEFQTFCEEEGI 537
Query: 872 LHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPM 931
+ ++P TPQQNGVVER+NRTL M R+++ ++ + W EAI + Y+ NR+ R +
Sbjct: 538 IRHLTAPYTPQQNGVVERRNRTLLGMTRSIMKHMSVPNYLWGEAIRHATYLINRVGTRAL 597
Query: 932 LEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY 991
+ +T YE K +PN+ + FGC Y +L+K D +++ ++LG SKAY +
Sbjct: 598 INQTPYEALKKKKPNVEHLRVFGCVSYAKVEFPHLRKLDERSRILVYLGTETGSKAYTLL 657
Query: 992 NSETHCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPE 1051
+ T + S V FD+ +K+ + + S I E + ++ + EVE+
Sbjct: 658 DPTTRKIIVSRDVVFDE----NKSWKWANSELIEIQKEPGMFTLAQTEFHNNEEVENETS 713
Query: 1052 AEI---TPEAESNSEAE---PSPEIQNENASEDI---QDNSQQAIQP 1089
EI E +N +AE P + N S+D+ + +Q I+P
Sbjct: 714 EEIEENEAEDSTNKDAEDGLDEPSVPEPNHSDDVPVFTRSGRQVIKP 760
>At2g07420 putative retroelement pol polyprotein
Length = 1664
Score = 182 bits (463), Expect = 1e-45
Identities = 107/377 (28%), Positives = 187/377 (49%), Gaps = 39/377 (10%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC GK KS F + + +L+H D++ S KY + +D+ S++TW
Sbjct: 472 ACILGKHCKSVFPKSSTIY-EKCFDLIHSDVWTSP-CLSRENHKYFVTFIDEKSKFTWFT 529
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
+ SKD E F++F T + + + KI +RSD+ GE+ + F+ KHGI+H+ S P
Sbjct: 530 LLPSKDRVLEAFTNFQTYVTNHYDAKIKILRSDNRGEYTSHAFKQHLNKHGIIHQTSCPY 589
Query: 880 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYEL 939
TPQQNGV ERKNR L E+ R M+ N+ KHFW + + ++CY+ N+ + +L+ + +E+
Sbjct: 590 TPQQNGVAERKNRHLMEVRRVMMFHTNVPKHFWIDGVVSACYLINQTPTKILLDSSPFEV 649
Query: 940 FKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVE 999
+P I++ FGC C++L + + K K+ +G+F+GYS K Y+ Y ET V
Sbjct: 650 LNKVKPFINHLRVFGCVCFVLISGEQRNKLQPKSTKGMFIGYSINQKGYKCYVLETRKVL 709
Query: 1000 ESMHVKF------------DDREPGSKTPE------------------QSESNAGTIDSE 1029
S VKF +D + + +P Q+++ T + E
Sbjct: 710 ISRDVKFLESKSYYDKKNWEDIQDLTDSPSDRATNLRIILERLGVSNIQTQTTPRTSNPE 769
Query: 1030 DASESDQLSDSEKHSEVESSPEAE-----ITPEAESNSEAEPSPEIQNEN--ASEDIQDN 1082
++ + + + E+ E E + + E ES+ E + N++ + + +++
Sbjct: 770 TITQPENMEEEEEEEEEEEEKQGKEQELITLEETESSKVQEKDTSLLNDDNGHTNNQEED 829
Query: 1083 SQQAIQPKFKHKSSHLR 1099
S +P+ +S HL+
Sbjct: 830 SNSREEPRIPRRSEHLK 846
>At3g60170 putative protein
Length = 1339
Score = 179 bits (453), Expect = 2e-44
Identities = 106/309 (34%), Positives = 156/309 (50%), Gaps = 11/309 (3%)
Query: 761 CQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKF 820
C GK + S K +S L+L+H D+ G + S G +Y L +DD++R TWV F
Sbjct: 473 CLTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYF 532
Query: 821 IKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRT 880
+ K A F F ++ E + +R+D GGEF + F FC HGI + ++ T
Sbjct: 533 LHEKSEAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGISRQLTAAFT 592
Query: 881 PQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELF 940
PQQNGV ERKNRT+ R+M+ E + K FW+EA S +IQNR + T E +
Sbjct: 593 PQQNGVAERKNRTIMNAVRSMLSERQVPKMFWSEATKWSVHIQNRSPTAAVEGMTPEEAW 652
Query: 941 KGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEE 1000
G +P + YF FGC Y+ K D K+++ +FLG SE SKA+R+Y+ +
Sbjct: 653 SGRKPVVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLYDPVMKKIVI 712
Query: 1001 SMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPE--- 1057
S V FD+ + E+ T++ D D EK+SEV P A +P
Sbjct: 713 SKDVVFDEDKSWDWDQADVEAKEVTLECGD-------EDDEKNSEV-VEPIAVASPNHVG 764
Query: 1058 AESNSEAEP 1066
+++N + P
Sbjct: 765 SDNNVSSSP 773
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 177 bits (448), Expect = 6e-44
Identities = 98/308 (31%), Positives = 161/308 (51%), Gaps = 4/308 (1%)
Query: 752 PS*NPFSGACQKGKIVKSS---FKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVI 808
P N G C+ + K S F + LEL+H D+ G + +S+ GS+Y L
Sbjct: 185 PKFNVEEGKCESCILSKHSRDPFPKESETRAKHKLELIHSDVCGPMQNSSINGSRYILTF 244
Query: 809 VDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEK 868
+DD +R WV F+K+K + F F +++ +I K+R D G E+ ++ F F E
Sbjct: 245 IDDATRMVWVYFLKAKSEVFQTFKKFKNLVENNANCRIKKLRIDRGTEYLSKEFSEFLEG 304
Query: 869 HGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYI 928
+GI + ++ +PQQN V ER+NR+L EMAR MI +L WAEA++ + Y QNR
Sbjct: 305 NGIERQLTAAYSPQQNEVSERRNRSLVEMARAMIKAKDLPLKLWAEAVHVAAYAQNRTPT 364
Query: 929 RPMLEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAY 988
R + KT E + ++P++S+ FG CY+ + +K+D K++R IF+GYS ++K Y
Sbjct: 365 RTLKNKTPLEAWSDSKPSVSHMKVFGSICYVHIPDEKRRKWDDKSKRAIFVGYSSQTKGY 424
Query: 989 RVYNSETHCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVES 1048
RVY + + ++ S V FD+ ++ + + E DQ +D + + E+
Sbjct: 425 RVYLLKENKIDISRDVIFDEDSKWDWEKKEVIKHY-DMSREPEDRGDQQADEQNSRDNEA 483
Query: 1049 SPEAEITP 1056
+ P
Sbjct: 484 RGRRNVLP 491
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 177 bits (448), Expect = 6e-44
Identities = 110/362 (30%), Positives = 175/362 (47%), Gaps = 14/362 (3%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC GK + SF + T LE++H D+ G + S+ GS+Y ++ +DDY+ WV
Sbjct: 492 ACNLGKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVY 551
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
F+K K F F ++ + I +R E FCE GI + + P
Sbjct: 552 FLKQKSETFATFKKFKALVEKQSNCSIKTLRP----------MEVFCEDEGINRQVTLPY 601
Query: 880 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEK-TAYE 938
+PQQNG ERKNR+L EMAR+M+ E +L WAEA+ TS Y+QNR+ + + + T E
Sbjct: 602 SPQQNGAAERKNRSLVEMARSMLVEQDLPLKLWAEAVYTSAYLQNRLPSKAIEDDVTPME 661
Query: 939 LFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCV 998
+ G +PN+S+ FG CY+ +K DAKA+ GI +GYS ++K YRV+ E V
Sbjct: 662 KWCGHKPNVSHLRIFGSICYVHIPDQKRRKLDAKAKCGILIGYSNQTKGYRVFLLEDEKV 721
Query: 999 EESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEA 1058
E S V F + + ++ + D ES ++ H + A E
Sbjct: 722 EVSRDVVFQEDKKWDWDKQEEVKKTFVMSINDIQESRDQQETSSHDLSQIDDHAN-NGEG 780
Query: 1059 ESNSE--AEPSPEIQNENASEDIQDNSQQAIQPKFKHKSSHLRS*SLEAKIVLEEQDHIS 1116
E++S ++ + + + E + + S + I K + + +EA +V E+
Sbjct: 781 ETSSHVLSQVNDQEERETSESPKKYKSMKEILEKAPRMENDEAAQGIEACLVANEEPQTY 840
Query: 1117 DK 1118
D+
Sbjct: 841 DE 842
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 171 bits (432), Expect = 4e-42
Identities = 92/310 (29%), Positives = 159/310 (50%), Gaps = 1/310 (0%)
Query: 761 CQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKF 820
C KGK ++ S + +++ L+L+H D+ G +N S G +Y L +DD+SR W
Sbjct: 435 CLKGKQIRESIPKESAWKSTQVLQLVHTDICGPINPASTSGKRYILNFIDDFSRKCWTYL 494
Query: 821 IKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRT 880
+ K + F F +++ E K++ +RSD GGE+ + F+ +C++ GI + ++ T
Sbjct: 495 LSEKSETFQFFKEFKAEVERESGKKLVCLRSDRGGEYNSREFDEYCKEFGIKRQLTAAYT 554
Query: 881 PQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELF 940
PQQNGV ERKNR++ M R M+ E ++ + FW EA+ + YI NR + + + T E +
Sbjct: 555 PQQNGVAERKNRSVMNMTRCMLMEMSVPRKFWPEAVQYAVYILNRSPSKALNDITPEEKW 614
Query: 941 KGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEE 1000
+P++ + FG Y L K D K+ + + G S+ SKAYR+Y+ T +
Sbjct: 615 SSWKPSVEHLRIFGSLAYALVPYQKRIKLDEKSIKCVMFGVSKESKAYRLYDPATGKILI 674
Query: 1001 SMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAES 1060
S V+FD+ E G + ++S D+ D + + H+ + E E E +
Sbjct: 675 SRDVQFDE-ERGWEWEDKSLEEELVWDNSDHEPAGEEGPEINHNGQQDQEETEEEEETVA 733
Query: 1061 NSEAEPSPEI 1070
+ + P +
Sbjct: 734 ETVHQNLPAV 743
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 165 bits (418), Expect = 2e-40
Identities = 108/362 (29%), Positives = 178/362 (48%), Gaps = 28/362 (7%)
Query: 757 FSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQV-NTTSLYGSKYGLVIVDDYSRW 815
F C GK + SF V T L +H DL+G N SL S+Y + VDDYSR
Sbjct: 448 FCEDCVYGKQHRVSFAPAQHV-TKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRK 506
Query: 816 TWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEF 875
W+ F++ KD A E F + ++++ + K+ K+R+D+G E+ N FE FC++ GI+
Sbjct: 507 VWIYFLRKKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVRHK 566
Query: 876 SSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKT 935
+ TPQQNG+ ER NRT+ + R+M+ + + K FWAEA +T+ Y+ NR +
Sbjct: 567 TCAYTPQQNGIAERLNRTIMDKVRSMLSRSGMEKKFWAEAASTAVYLINRSPSTAINFDL 626
Query: 936 AYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSET 995
E + G P++S +FGC YI + K + ++++GIF Y E K Y+V+ E
Sbjct: 627 PEEKWTGALPDLSSLRKFGCLAYIHADQG---KLNPRSKKGIFTSYPEGVKGYKVWVLED 683
Query: 996 HCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSE-------------- 1041
S +V F ++ S++ D ED + ++D E
Sbjct: 684 KKCVISRNVIFREQVMFKDLKGDSQNTISESDLEDLRVNPDMNDQEFTDQGGATQDNSNP 743
Query: 1042 -----KHSEVESSPEAEITPEAESNSEAEPSPEIQNENASEDIQDNSQQAIQPKFKHKSS 1096
H+ V +SP T + SE E S +++ + + ++D ++ I+ K+ S
Sbjct: 744 SEATTSHNPVLNSP----THSQDEESEEEDSDAVEDLSTYQLVRDRVRRTIKANPKYNES 799
Query: 1097 HL 1098
++
Sbjct: 800 NM 801
>At1g37110
Length = 1356
Score = 162 bits (409), Expect = 2e-39
Identities = 118/358 (32%), Positives = 183/358 (50%), Gaps = 41/358 (11%)
Query: 729 INDLIKIRFNGISSAKRIKFLKNPS*NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHI 788
+N+L + G+ K I L+ F C GK K SF S L +H
Sbjct: 449 MNNLKVLAGKGLIDRKEINELE------FCEHCVMGKSKKVSFNVGKHTSEDA-LSYVHA 501
Query: 789 DLFGQVNTT-SLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKIL 847
DL+G N T S+ G +Y L I+DD +R W+ F+KSKD + F + + ++++ K+
Sbjct: 502 DLWGSPNVTPSISGKQYFLSIIDDKTRKVWLYFLKSKDETFDKFCEWKSLVENQVNKKVK 561
Query: 848 KVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNL 907
+R+D+G EF N F+S+C++HGI + TPQQNGV ER NRT+ E R +++++ +
Sbjct: 562 CLRTDNGLEFCNSRFDSYCKEHGIERHRTCTYTPQQNGVAERMNRTIMEKVRCLLNKSGV 621
Query: 908 AKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLK 967
+ FWAEA T+ Y+ NR + E++ +P + +FG Y+ + LK
Sbjct: 622 EEVFWAEAAATAAYLINRSPASAINHNVPEEMWLNRKPGYKHLRKFGSIAYVHQDQGKLK 681
Query: 968 KFDAKAQRGIFLGYSERSKAYRVY-NSETHCV-------EES-----MHVKFDD------ 1008
+A +G FLGY +K Y+V+ E CV +ES + VK DD
Sbjct: 682 ---PRALKGFFLGYPAGTKGYKVWLLEEEKCVISRNVVFQESVVYRDLKVKEDDTDNLNQ 738
Query: 1009 REPGSKTPEQSE----SNAGTI-----DSEDASESDQLSDSEKHSEVESSPEAEITPE 1057
+E S EQ++ S +G + DSE +E +Q SDSE+ EVE S + + TP+
Sbjct: 739 KETTSSEVEQNKFAEASGSGGVIQLQSDSEPITEGEQSSDSEE--EVEYSEKTQETPK 794
>At2g14930 pseudogene
Length = 1149
Score = 161 bits (408), Expect = 3e-39
Identities = 89/259 (34%), Positives = 141/259 (54%), Gaps = 2/259 (0%)
Query: 751 NPS*NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVD 810
N S + +C+ GK + F + D ++ SRPLE +H DL+G +S+ G +Y ++ +D
Sbjct: 473 NKSTSKMCESCRLGKSSRLPFIASDFIA-SRPLERVHCDLWGPAPVSSIQGFQYYVIFID 531
Query: 811 DYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHG 870
+ SR+ W +K K C +F F + +++ + KI +SD GGEF + F ++ G
Sbjct: 532 NRSRFCWFYPLKHKSDFCSLFMKFQSFVENLLQTKIGTFQSDGGGEFTSNRFLQHLQESG 591
Query: 871 ILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRP 930
I H S P TPQQNG+ ERK+R L E T++ ++ + FW EA T+ ++ N +
Sbjct: 592 IQHYISCPHTPQQNGLAERKHRQLTERGLTLMFQSKAPQRFWVEAFFTANFLSNLLPTSA 651
Query: 931 M-LEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYR 989
+ T Y++ G P+ S FGC C+ KFD ++ + IFLGY+E+ K YR
Sbjct: 652 LDSSTTPYQVLFGKAPDYSALRTFGCACFPTLRAYARNKFDPRSLKCIFLGYTEKYKGYR 711
Query: 990 VYNSETHCVEESMHVKFDD 1008
+ T+ V S HV FD+
Sbjct: 712 CFFPPTNRVYLSRHVLFDE 730
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 161 bits (408), Expect = 3e-39
Identities = 109/366 (29%), Positives = 182/366 (48%), Gaps = 13/366 (3%)
Query: 740 ISSAKRIKFLKNPS*NPFSGACQKGKIVKSSFKSKDIVSTSR---PLELLHIDLFGQVNT 796
+ ++K I+ K+ + +P CQ GK + F ++S SR PL+ +H DL+G
Sbjct: 471 LQNSKAIQINKSRT-SPVCEPCQMGKSSRLPF----LISDSRVLHPLDRIHCDLWGPSPV 525
Query: 797 TSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGE 856
S G KY + VDDYSR++W + +K VF SF ++++ KI +SD GGE
Sbjct: 526 VSNQGLKYYAIFVDDYSRYSWFYPLHNKSEFLSVFISFQKLVENQLNTKIKVFQSDGGGE 585
Query: 857 FENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAI 916
F + ++ +HGI H S P TPQQNG+ ERK+R L E+ +M+ ++ + FW E+
Sbjct: 586 FVSNKLKTHLSEHGIHHRISCPYTPQQNGLAERKHRHLVELGLSMLFHSHTPQKFWVESF 645
Query: 917 NTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRG 976
T+ YI NR+ + + YE G +P+ S FG CY KFD ++ +
Sbjct: 646 FTANYIINRLPSSVLKNLSPYEALFGEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSLQC 705
Query: 977 IFLGYSERSKAYRVYNSETHCVEESMHVKFDDREPGSKTPEQS---ESNAGTIDSEDASE 1033
+FLGY+ + K YR + T V S +V F++ E K QS + + + + ++
Sbjct: 706 VFLGYNSQYKGYRCFYPPTGKVYISRNVIFNESELPFKEKYQSLVPQYSTPLLQAWQHNK 765
Query: 1034 SDQLSDSEKHSEVESSPEAEITPEAESNSE--AEPSPEIQNENASEDIQDNSQQAIQPKF 1091
++S ++ S P T +E +P P NE + E++ +++ +
Sbjct: 766 ISEISVPAAPVQLFSKPIDLNTYAGSQVTEQLTDPEPTSNNEGSDEEVNPVAEEIAANQE 825
Query: 1092 KHKSSH 1097
+ +SH
Sbjct: 826 QVINSH 831
>At1g70010 hypothetical protein
Length = 1315
Score = 160 bits (406), Expect = 4e-39
Identities = 94/300 (31%), Positives = 152/300 (50%), Gaps = 4/300 (1%)
Query: 779 TSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQI 838
+SRP +L+HID +G + + G +Y L IVDDYSR TWV +++K V +F T +
Sbjct: 459 SSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLRNKSDVLTVIPTFVTMV 518
Query: 839 QSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMA 898
+++ E I VRSD+ E F F GI+ S P TPQQN VVERK++ + +A
Sbjct: 519 ENQFETTIKGVRSDNAPELN---FTQFYHSKGIVPYHSCPETPQQNSVVERKHQHILNVA 575
Query: 899 RTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCY 958
R++ ++++ +W + I T+ Y+ NR+ + +K +E+ T P + FGC CY
Sbjct: 576 RSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCY 635
Query: 959 ILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESMHVKFDDREPGSKTPEQ 1018
+ KF +A+ F+GY K Y++ + ETH + S HV F + +
Sbjct: 636 ASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVFHEELFPFLGSDL 695
Query: 1019 SESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAE-SNSEAEPSPEIQNENASE 1077
S+ + + Q S+ + +SS EI P A +N+ EPS + + A +
Sbjct: 696 SQEEQNFFPDLNPTPPMQRQSSDHVNPSDSSSSVEILPSANPTNNVPEPSVQTSHRKAKK 755
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 160 bits (404), Expect = 8e-39
Identities = 102/338 (30%), Positives = 172/338 (50%), Gaps = 7/338 (2%)
Query: 736 RFNGISSAKRIKFLKNPS*NPFSGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVN 795
R + IS + KN + F C K K SF + + V +LLHID++G +
Sbjct: 569 RLDAISDSLGTTRHKNKG-SDFCHVCHLAKQRKLSFPTSNKVC-KEIFDLLHIDVWGPFS 626
Query: 796 TTSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGG 855
++ G KY L IVDD+SR TW+ +K+K VF +F Q++++ ++K+ VRSD+
Sbjct: 627 VETVEGYKYFLTIVDDHSRATWMYLLKTKSEVLTVFPAFIQQVENQYKVKVKAVRSDNAP 686
Query: 856 EFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEA 915
E + F SF + GI+ S P TP+QN VVERK++ + +AR ++ ++ + W +
Sbjct: 687 ELK---FTSFYAEKGIVSFHSCPETPEQNSVVERKHQHILNVARALMFQSQVPLSLWGDC 743
Query: 916 INTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQR 975
+ T+ ++ NR + ++ KT YE+ GT P FGC CY + KF +++
Sbjct: 744 VLTAVFLINRTPSQLLMNKTPYEILTGTAPVYEQLRTFGCLCYSSTSPKQRHKFQPRSRA 803
Query: 976 GIFLGYSERSKAYRVYNSETHCVEESMHVKFDDRE-PGSKTPEQSESNAGTIDSEDASES 1034
+FLGY K Y++ + E++ V S +V+F + P +K P SES+ S
Sbjct: 804 CLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFPLAKNP-GSESSLKLFTPMVPVSS 862
Query: 1035 DQLSDSEKHSEVESSPEAEITPEAESNSEAEPSPEIQN 1072
+SD+ S +++ P+ S +P + +
Sbjct: 863 GIISDTTHSPSSLPSQISDLPPQISSQRVRKPPAHLND 900
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 159 bits (402), Expect = 1e-38
Identities = 80/238 (33%), Positives = 132/238 (54%), Gaps = 3/238 (1%)
Query: 780 SRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQ 839
S P +L+H+D++G + S+ G +Y L +VDD +R TWV +++K VF F +
Sbjct: 630 SNPFDLVHLDIWGPFSIESIEGFRYFLTVVDDCTRTTWVYMLRNKKDVSSVFPEFIKLVS 689
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ KI +RSD+ E F ++HG+LH FS TPQQN VVERK++ + +AR
Sbjct: 690 TQFNAKIKAIRSDNAPELG---FTEIVKEHGMLHHFSCAYTPQQNSVVERKHQHILNVAR 746
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
++ ++N+ +W++ + T+ ++ NR+ + K+ YEL +P+ S FGC C++
Sbjct: 747 ALLFQSNIPMQYWSDCVTTAVFLINRLPSPLLNNKSPYELILNKQPDYSLLKNFGCLCFV 806
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESMHVKFDDREPGSKTPE 1017
KF +A+ +FLGY K Y+V + E+H V S +V F + KT E
Sbjct: 807 STNAHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSVTVSRNVVFKEHVFPFKTSE 864
>At4g28900 putative protein
Length = 1415
Score = 158 bits (400), Expect = 2e-38
Identities = 91/251 (36%), Positives = 138/251 (54%), Gaps = 4/251 (1%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
ACQ GKI K F S D VS SR LE +H DL+G S G +Y ++ +D+YSR+TW
Sbjct: 460 ACQMGKICKLPFASSDFVS-SRLLERVHCDLWGPAPVVSSQGFRYYVIFIDNYSRFTWFY 518
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
++ K VF +F ++++ + KI + D GGEF + F S + GI S P
Sbjct: 519 PLRLKSDFFSVFLTFQKMVENQCQQKIASFQCDGGGEFISNQFVSHLAECGIRQLISCPY 578
Query: 880 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPML-EKTAYE 938
TPQQNG+ ERK+R + E+ +M+ + + + W EA TS ++ N + + +K+ YE
Sbjct: 579 TPQQNGIAERKHRHITELGSSMMFQGKVPQFLWVEAFYTSNFLCNLLPSSVLKDQKSPYE 638
Query: 939 LFKGTRPNISYFHQFGCTCYILNTKDYL-KKFDAKAQRGIFLGYSERSKAYRVYNSETHC 997
+ G P + FGC CY N + Y KFD K+ +F GY+E+ K Y+ ++ T
Sbjct: 639 VLMGKAPVYTSLRVFGCACY-PNLRPYASNKFDPKSLLCVFTGYNEKYKGYKCFHPPTGK 697
Query: 998 VEESMHVKFDD 1008
+ + HV FD+
Sbjct: 698 IYINRHVLFDE 708
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.359 0.159 0.563
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,034,979
Number of Sequences: 26719
Number of extensions: 1120648
Number of successful extensions: 8085
Number of sequences better than 10.0: 193
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 7376
Number of HSP's gapped (non-prelim): 541
length of query: 1684
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1571
effective length of database: 8,299,349
effective search space: 13038277279
effective search space used: 13038277279
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC149212.6


