

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149211.1 + phase: 0
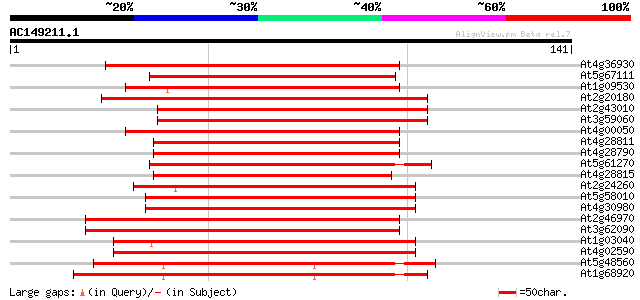
(141 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATU... 123 3e-29
At5g67111 putative bHLH transcription factor (bHLH073/ALCATRAZ) 104 1e-23
At1g09530 putative transcription factor BHLH8 100 3e-22
At2g20180 putative bHLH transcription factor (bHLH015) 97 2e-21
At2g43010 putative transcription factor BHLH9 95 1e-20
At3g59060 putative bHLH transcription factor (bHLH065) 93 5e-20
At4g00050 bHLH like transcription factor (bHLH016) 91 2e-19
At4g28811 AtbHLH119 86 7e-18
At4g28790 bHLH transcription factor like protein (bHLH023) 84 3e-17
At5g61270 bHLH transcription factor like protein 82 1e-16
At4g28815 AtbHLH127 82 1e-16
At2g24260 putative bHLH transcription factor (bHLH066) 80 3e-16
At5g58010 putative bHLH transcription factor (bHLH082) 80 5e-16
At4g30980 putative bHLH transcription factor (bHLH069) 78 1e-15
At2g46970 PIF3 like basic Helix Loop Helix protein (PIL1) 78 1e-15
At3g62090 PIF3 like basic Helix Loop Helix protein 2 (PIL2) 74 3e-14
At1g03040 putative transcription factor (BHLH7) 73 4e-14
At4g02590 putative bHLH transcription factor (bHLH059) 72 1e-13
At5g48560 putative bHLH transcription factor (bHLH078) 70 3e-13
At1g68920 putative bHLH transcription factor (bHLH049) 67 2e-12
>At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATULA
(SPT)
Length = 373
Score = 123 bits (309), Expect = 3e-29
Identities = 63/74 (85%), Positives = 68/74 (91%)
Query: 25 ELPSSKAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDE 84
E PSSK+ P RSSSKR RAAE HNLSEKRRRS+INEK+KALQ+LIPNSNKTDKASMLDE
Sbjct: 179 EAPSSKSGPSSRSSSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSNKTDKASMLDE 238
Query: 85 AIEYLKQLQLQVQV 98
AIEYLKQLQLQVQ+
Sbjct: 239 AIEYLKQLQLQVQM 252
>At5g67111 putative bHLH transcription factor (bHLH073/ALCATRAZ)
Length = 210
Score = 104 bits (260), Expect = 1e-23
Identities = 53/62 (85%), Positives = 58/62 (93%)
Query: 36 RSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQ 95
R+S KR+ A+FHNLSEK+RRSKINEK+KALQ LIPNSNKTDKASMLDEAIEYLKQLQLQ
Sbjct: 86 RNSLKRNIDAQFHNLSEKKRRSKINEKMKALQKLIPNSNKTDKASMLDEAIEYLKQLQLQ 145
Query: 96 VQ 97
VQ
Sbjct: 146 VQ 147
>At1g09530 putative transcription factor BHLH8
Length = 524
Score = 100 bits (249), Expect = 3e-22
Identities = 52/71 (73%), Positives = 59/71 (82%), Gaps = 2/71 (2%)
Query: 30 KAAPPPRSS--SKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIE 87
K A P R+ SKRSR+AE HNLSE+RRR +INEK++ALQ LIPN NK DKASMLDEAIE
Sbjct: 328 KEAGPSRTGLGSKRSRSAEVHNLSERRRRDRINEKMRALQELIPNCNKVDKASMLDEAIE 387
Query: 88 YLKQLQLQVQV 98
YLK LQLQVQ+
Sbjct: 388 YLKSLQLQVQI 398
>At2g20180 putative bHLH transcription factor (bHLH015)
Length = 478
Score = 97.4 bits (241), Expect = 2e-21
Identities = 49/82 (59%), Positives = 62/82 (74%)
Query: 24 SELPSSKAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLD 83
S +K A +S+KRSRAAE HNLSE++RR +INE++KALQ LIP NK+DKASMLD
Sbjct: 265 SRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKASMLD 324
Query: 84 EAIEYLKQLQLQVQVSFLSLKM 105
EAIEY+K LQLQ+Q+ + M
Sbjct: 325 EAIEYMKSLQLQIQMMSMGCGM 346
>At2g43010 putative transcription factor BHLH9
Length = 430
Score = 94.7 bits (234), Expect = 1e-20
Identities = 46/68 (67%), Positives = 59/68 (86%)
Query: 38 SSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQ 97
S++RSRAAE HNLSE+RRR +INE++KALQ LIP+ +KTDKAS+LDEAI+YLK LQLQ+Q
Sbjct: 252 SNRRSRAAEVHNLSERRRRDRINERMKALQELIPHCSKTDKASILDEAIDYLKSLQLQLQ 311
Query: 98 VSFLSLKM 105
V ++ M
Sbjct: 312 VMWMGSGM 319
>At3g59060 putative bHLH transcription factor (bHLH065)
Length = 442
Score = 92.8 bits (229), Expect = 5e-20
Identities = 44/68 (64%), Positives = 59/68 (86%)
Query: 38 SSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQ 97
S++RSRAAE HNLSE+RRR +INE++KALQ LIP+ ++TDKAS+LDEAI+YLK LQ+Q+Q
Sbjct: 251 STRRSRAAEVHNLSERRRRDRINERMKALQELIPHCSRTDKASILDEAIDYLKSLQMQLQ 310
Query: 98 VSFLSLKM 105
V ++ M
Sbjct: 311 VMWMGSGM 318
>At4g00050 bHLH like transcription factor (bHLH016)
Length = 399
Score = 91.3 bits (225), Expect = 2e-19
Identities = 45/69 (65%), Positives = 54/69 (78%)
Query: 30 KAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYL 89
KA S+KRSRAA HN SE++RR KIN+++K LQ L+PNS+KTDKASMLDE IEYL
Sbjct: 200 KAGGKSSVSTKRSRAAAIHNQSERKRRDKINQRMKTLQKLVPNSSKTDKASMLDEVIEYL 259
Query: 90 KQLQLQVQV 98
KQLQ QV +
Sbjct: 260 KQLQAQVSM 268
>At4g28811 AtbHLH119
Length = 470
Score = 85.9 bits (211), Expect = 7e-18
Identities = 39/62 (62%), Positives = 51/62 (81%)
Query: 37 SSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQV 96
+S KRSRAA+ HNLSE+RRR +INE++K LQ L+P KTDK SML++ IEY+K LQLQ+
Sbjct: 274 TSRKRSRAADMHNLSERRRRERINERMKTLQELLPRCRKTDKVSMLEDVIEYVKSLQLQI 333
Query: 97 QV 98
Q+
Sbjct: 334 QM 335
>At4g28790 bHLH transcription factor like protein (bHLH023)
Length = 413
Score = 83.6 bits (205), Expect = 3e-17
Identities = 40/62 (64%), Positives = 50/62 (80%)
Query: 37 SSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQV 96
+SSKRSRAA H LSE+RRR KINE +KALQ L+P KTD++SMLD+ IEY+K LQ Q+
Sbjct: 271 TSSKRSRAAIMHKLSERRRRQKINEMMKALQELLPRCTKTDRSSMLDDVIEYVKSLQSQI 330
Query: 97 QV 98
Q+
Sbjct: 331 QM 332
>At5g61270 bHLH transcription factor like protein
Length = 366
Score = 81.6 bits (200), Expect = 1e-16
Identities = 39/71 (54%), Positives = 55/71 (76%), Gaps = 2/71 (2%)
Query: 36 RSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQ 95
RS+ +R RAA HN SE+RRR +IN++++ LQ L+P ++K DK S+LD+ IE+LKQLQ Q
Sbjct: 159 RSNGRRGRAAAIHNESERRRRDRINQRMRTLQKLLPTASKADKVSILDDVIEHLKQLQAQ 218
Query: 96 VQVSFLSLKMN 106
VQ F+SL+ N
Sbjct: 219 VQ--FMSLRAN 227
>At4g28815 AtbHLH127
Length = 307
Score = 81.6 bits (200), Expect = 1e-16
Identities = 37/60 (61%), Positives = 49/60 (81%)
Query: 37 SSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQV 96
+S KRSRAAE HNL+E+RRR KINE++K LQ LIP NK+ K SML++ IEY+K L++Q+
Sbjct: 144 TSRKRSRAAEMHNLAERRRREKINERMKTLQQLIPRCNKSTKVSMLEDVIEYVKSLEMQI 203
>At2g24260 putative bHLH transcription factor (bHLH066)
Length = 350
Score = 80.5 bits (197), Expect = 3e-16
Identities = 41/75 (54%), Positives = 57/75 (75%), Gaps = 4/75 (5%)
Query: 32 APPPRSSSK----RSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIE 87
A PP+S +K R +A + H+++E+ RR +I E++KALQ L+PN NKTDKASMLDE I+
Sbjct: 129 AAPPQSRTKIRARRGQATDPHSIAERLRRERIAERMKALQELVPNGNKTDKASMLDEIID 188
Query: 88 YLKQLQLQVQVSFLS 102
Y+K LQLQV+V +S
Sbjct: 189 YVKFLQLQVKVLSMS 203
>At5g58010 putative bHLH transcription factor (bHLH082)
Length = 274
Score = 79.7 bits (195), Expect = 5e-16
Identities = 37/68 (54%), Positives = 55/68 (80%)
Query: 35 PRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQL 94
PR ++R +A + H+++E+ RR +I E++K+LQ L+PN+NKTDKASMLDE IEY++ LQL
Sbjct: 74 PRVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNTNKTDKASMLDEIIEYVRFLQL 133
Query: 95 QVQVSFLS 102
QV+V +S
Sbjct: 134 QVKVLSMS 141
>At4g30980 putative bHLH transcription factor (bHLH069)
Length = 310
Score = 78.2 bits (191), Expect = 1e-15
Identities = 36/68 (52%), Positives = 54/68 (78%)
Query: 35 PRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQL 94
P+ ++R +A + H+++E+ RR +I E++K+LQ L+PN NKTDKASMLDE I+Y+K LQL
Sbjct: 128 PKVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNGNKTDKASMLDEIIDYVKFLQL 187
Query: 95 QVQVSFLS 102
QV+V +S
Sbjct: 188 QVKVLSMS 195
>At2g46970 PIF3 like basic Helix Loop Helix protein (PIL1)
Length = 416
Score = 78.2 bits (191), Expect = 1e-15
Identities = 37/79 (46%), Positives = 56/79 (70%)
Query: 20 NDEGSELPSSKAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKA 79
+DE + + A + +KR R+ E H L E++RR + N+K++ALQ+L+PN K DKA
Sbjct: 206 DDESDDAKTQVHARTRKPVTKRKRSTEVHKLYERKRRDEFNKKMRALQDLLPNCYKDDKA 265
Query: 80 SMLDEAIEYLKQLQLQVQV 98
S+LDEAI+Y++ LQLQVQ+
Sbjct: 266 SLLDEAIKYMRTLQLQVQM 284
>At3g62090 PIF3 like basic Helix Loop Helix protein 2 (PIL2)
Length = 363
Score = 73.6 bits (179), Expect = 3e-14
Identities = 37/79 (46%), Positives = 52/79 (64%)
Query: 20 NDEGSELPSSKAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKA 79
+DE + A ++ KR R AE +N E+ +R+ IN+K++ LQNL+PNS+K D
Sbjct: 165 DDESDDARPQVPARTRKALVKRKRNAEAYNSPERNQRNDINKKMRTLQNLLPNSHKDDNE 224
Query: 80 SMLDEAIEYLKQLQLQVQV 98
SMLDEAI Y+ LQLQVQ+
Sbjct: 225 SMLDEAINYMTNLQLQVQM 243
>At1g03040 putative transcription factor (BHLH7)
Length = 302
Score = 73.2 bits (178), Expect = 4e-14
Identities = 35/85 (41%), Positives = 59/85 (69%), Gaps = 9/85 (10%)
Query: 27 PSSKAAPP---------PRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTD 77
P S+ APP PR ++R +A + H+++E+ RR +I E++++LQ L+P NKTD
Sbjct: 125 PMSQPAPPMPHQQSTIRPRVRARRGQATDPHSIAERLRRERIAERIRSLQELVPTVNKTD 184
Query: 78 KASMLDEAIEYLKQLQLQVQVSFLS 102
+A+M+DE ++Y+K L+LQV+V +S
Sbjct: 185 RAAMIDEIVDYVKFLRLQVKVLSMS 209
>At4g02590 putative bHLH transcription factor (bHLH059)
Length = 310
Score = 71.6 bits (174), Expect = 1e-13
Identities = 32/76 (42%), Positives = 55/76 (72%)
Query: 27 PSSKAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAI 86
P + PR ++R +A + H+++E+ RR +I E+++ALQ L+P NKTD+A+M+DE +
Sbjct: 136 PHQPTSIRPRVRARRGQATDPHSIAERLRRERIAERIRALQELVPTVNKTDRAAMIDEIV 195
Query: 87 EYLKQLQLQVQVSFLS 102
+Y+K L+LQV+V +S
Sbjct: 196 DYVKFLRLQVKVLSMS 211
>At5g48560 putative bHLH transcription factor (bHLH078)
Length = 498
Score = 70.5 bits (171), Expect = 3e-13
Identities = 39/91 (42%), Positives = 62/91 (67%), Gaps = 7/91 (7%)
Query: 22 EGSELPSSKAAPPPRS----SSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNK-T 76
EG++ ++K PP+ ++R +A + H+L+E+ RR KI E++K LQ+L+P NK T
Sbjct: 282 EGNKSNNTKPPEPPKDYIHVRARRGQATDSHSLAERVRREKIGERMKLLQDLVPGCNKVT 341
Query: 77 DKASMLDEAIEYLKQLQLQVQVSFLSLKMNN 107
KA MLDE I Y++ LQ QV+ FLS+K+++
Sbjct: 342 GKALMLDEIINYVQSLQRQVE--FLSMKLSS 370
>At1g68920 putative bHLH transcription factor (bHLH049)
Length = 486
Score = 67.4 bits (163), Expect = 2e-12
Identities = 39/94 (41%), Positives = 59/94 (62%), Gaps = 7/94 (7%)
Query: 17 GLKNDEGSELPSSKAAPPPRS----SSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPN 72
G K++ G + + PP ++R +A H+L+E+ RR KI+E++K LQ+L+P
Sbjct: 279 GKKSNSGKQQGKQSSDPPKDGYIHVRARRGQATNSHSLAERVRREKISERMKFLQDLVPG 338
Query: 73 SNK-TDKASMLDEAIEYLKQLQLQVQVSFLSLKM 105
NK T KA MLDE I Y++ LQ QV+ FLS+K+
Sbjct: 339 CNKVTGKAVMLDEIINYVQSLQRQVE--FLSMKL 370
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,096,748
Number of Sequences: 26719
Number of extensions: 111880
Number of successful extensions: 797
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 138
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 618
Number of HSP's gapped (non-prelim): 187
length of query: 141
length of database: 11,318,596
effective HSP length: 89
effective length of query: 52
effective length of database: 8,940,605
effective search space: 464911460
effective search space used: 464911460
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149211.1


