

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149210.5 + phase: 0
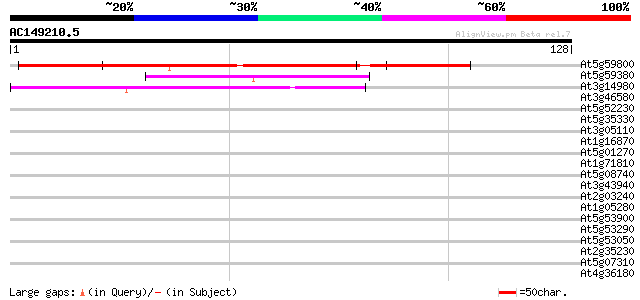
(128 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59800 unknown protein 64 2e-11
At5g59380 unknown protein 45 8e-06
At3g14980 PHD-finger protein, putative 39 7e-04
At3g46580 unknown protein 37 0.004
At5g52230 unknown protein 35 0.014
At5g35330 unknown protein 33 0.040
At3g05110 hypothetical protein 28 1.7
At1g16870 Unknown protein (F6I1.13) 27 2.2
At5g01270 hypothetical protein 27 2.8
At1g71810 unknown protein 27 2.8
At5g08740 NADH dehydrogenase-like protein 27 3.7
At3g43940 putative protein 27 3.7
At2g03240 unknown protein 27 3.7
At1g05280 hypothetical protein 27 3.7
At5g53900 unknown protein 26 4.9
At5g53290 unknown protein 26 6.3
At5g53050 unknown protein 26 6.3
At2g35230 unknown protein 26 6.3
At5g07310 putative transcription factor 25 8.3
At4g36180 receptor protein kinase like protein 25 8.3
>At5g59800 unknown protein
Length = 306
Score = 63.9 bits (154), Expect = 2e-11
Identities = 34/77 (44%), Positives = 49/77 (63%), Gaps = 1/77 (1%)
Query: 3 RSMDVEMEHLNLNRNVMNLREKELQIVVQSPSKVEVPDGWLVEQRPRPSNPNHIDRYYIE 62
RS+ +L +RN + + + LQ +PDGW+VE++PR S+ +HIDR YIE
Sbjct: 147 RSLVSVERYLRESRNSIEQQLRVLQNRRGHSKDFRLPDGWIVEEKPRRSS-SHIDRSYIE 205
Query: 63 PHTGQKFRSLVSVQRYL 79
P TG KFRS+ +V+RYL
Sbjct: 206 PGTGNKFRSMAAVERYL 222
Score = 58.5 bits (140), Expect = 9e-10
Identities = 35/85 (41%), Positives = 52/85 (61%), Gaps = 4/85 (4%)
Query: 22 REKELQIVVQSPSK-VEVPDGWLVEQRPRPSNPNHIDRYYIEPHTGQKFRSLVSVQRYLN 80
R K+ V+ SK +P GW VE+ PR N ++ID+YY+E TG++FRSLVSV+RYL
Sbjct: 99 RTKQDHCGVEYASKGFRLPRGWSVEEVPR-KNSHYIDKYYVERKTGKRFRSLVSVERYLR 157
Query: 81 GGETGDYLPTQRIISENNNNVSSNF 105
E+ + + Q + +N S +F
Sbjct: 158 --ESRNSIEQQLRVLQNRRGHSKDF 180
Score = 53.9 bits (128), Expect = 2e-08
Identities = 28/65 (43%), Positives = 39/65 (59%), Gaps = 1/65 (1%)
Query: 22 REKELQIVVQSPSKVEVPDGWLVEQRPRPSNPNHIDRYYIEPHTGQKFRSLVSVQRYLNG 81
RE +LQI + ++ GW V RPR SN +D Y+IEP TG++F SL ++ R+L
Sbjct: 15 RETQLQIADPTSFCGKIMPGWTVVNRPRSSNNGVVDTYFIEPGTGRQFSSLEAIHRHL-A 73
Query: 82 GETGD 86
GE D
Sbjct: 74 GEVND 78
>At5g59380 unknown protein
Length = 225
Score = 45.4 bits (106), Expect = 8e-06
Identities = 23/53 (43%), Positives = 30/53 (56%), Gaps = 2/53 (3%)
Query: 32 SPSKVEVPDGWLVEQRPRPSNPN--HIDRYYIEPHTGQKFRSLVSVQRYLNGG 82
+P +P GW VE + R S +D+YY EP+TG+KFRS V YL G
Sbjct: 75 APGDNWLPPGWRVEDKIRTSGATAGSVDKYYYEPNTGRKFRSRTEVLYYLEHG 127
>At3g14980 PHD-finger protein, putative
Length = 1189
Score = 38.9 bits (89), Expect = 7e-04
Identities = 23/83 (27%), Positives = 39/83 (46%), Gaps = 3/83 (3%)
Query: 1 MNRSMDVEMEHLNLNRNVMNLREKE--LQIVVQSPSKVEVPDGWLVEQRPRPSNPNHIDR 58
+N V L +++ + + KE ++ + KV W +E+R R + H+D
Sbjct: 213 LNNGSKVSPNELIMSKTCLKIDPKEDPRPLLYKYVCKVLTAARWKIEKRERSAGRKHVDT 272
Query: 59 YYIEPHTGQKFRSLVSVQRYLNG 81
+YI P G+KFR S + L G
Sbjct: 273 FYISPE-GRKFREFGSAWKALGG 294
>At3g46580 unknown protein
Length = 182
Score = 36.6 bits (83), Expect = 0.004
Identities = 20/47 (42%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Query: 38 VPDGWLVEQRPRPSNPNH--IDRYYIEPHTGQKFRSLVSVQRYLNGG 82
+P W E R R S +D++Y EP TG+KFRS V YL G
Sbjct: 35 LPPDWRTEIRVRTSGTKAGTVDKFYYEPITGRKFRSKNEVLYYLEHG 81
>At5g52230 unknown protein
Length = 746
Score = 34.7 bits (78), Expect = 0.014
Identities = 16/63 (25%), Positives = 31/63 (48%), Gaps = 2/63 (3%)
Query: 27 QIVVQSPSKVEVPDGWL--VEQRPRPSNPNHIDRYYIEPHTGQKFRSLVSVQRYLNGGET 84
+++V+ + +P+GW+ +E R D ++I+P + F+S RY+ G
Sbjct: 28 KVIVEKSAAQGLPEGWIKKLEITNRSGRKTRRDPFFIDPKSEYIFQSFKDASRYVETGNI 87
Query: 85 GDY 87
G Y
Sbjct: 88 GHY 90
>At5g35330 unknown protein
Length = 272
Score = 33.1 bits (74), Expect = 0.040
Identities = 20/52 (38%), Positives = 26/52 (49%), Gaps = 2/52 (3%)
Query: 30 VQSPSKVEVPDGWLVEQRPRPSNPNHI-DRYYIEPHTGQKFRSLVSVQRYLN 80
+ P+ P GW R R D YY+ P +G+K RS V VQ+YLN
Sbjct: 120 IDKPNISRPPAGWQRLLRIRGEGGTRFADVYYVAP-SGKKLRSTVEVQKYLN 170
>At3g05110 hypothetical protein
Length = 372
Score = 27.7 bits (60), Expect = 1.7
Identities = 13/55 (23%), Positives = 26/55 (46%)
Query: 3 RSMDVEMEHLNLNRNVMNLREKELQIVVQSPSKVEVPDGWLVEQRPRPSNPNHID 57
R ++ + + +L + +EKEL++ + + + DG V+ P NH D
Sbjct: 77 REVEAKRKFFDLKEKELEEKEKELELKQRQVQERSIQDGPSVDAEPLTQQRNHND 131
>At1g16870 Unknown protein (F6I1.13)
Length = 480
Score = 27.3 bits (59), Expect = 2.2
Identities = 11/27 (40%), Positives = 15/27 (54%)
Query: 40 DGWLVEQRPRPSNPNHIDRYYIEPHTG 66
+GWLV P+ + H +Y PHTG
Sbjct: 233 EGWLVLYAPKGRDWTHGGYFYKNPHTG 259
>At5g01270 hypothetical protein
Length = 771
Score = 26.9 bits (58), Expect = 2.8
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Query: 27 QIVVQSPSK-VEVPDGWLVEQRPRPSNPNHIDRYY 60
+I +Q PS + P GWLV+ RPS P Y
Sbjct: 549 KIPMQPPSSSMHSPGGWLVDDENRPSFPGRPSGLY 583
>At1g71810 unknown protein
Length = 692
Score = 26.9 bits (58), Expect = 2.8
Identities = 12/34 (35%), Positives = 17/34 (49%)
Query: 59 YYIEPHTGQKFRSLVSVQRYLNGGETGDYLPTQR 92
Y+ +PH G R+ YL+ G GD+ P R
Sbjct: 340 YHADPHPGNFLRTYDGQLAYLDFGMMGDFRPELR 373
>At5g08740 NADH dehydrogenase-like protein
Length = 519
Score = 26.6 bits (57), Expect = 3.7
Identities = 14/57 (24%), Positives = 32/57 (55%), Gaps = 3/57 (5%)
Query: 9 MEHLNLNRNVMNLR---EKELQIVVQSPSKVEVPDGWLVEQRPRPSNPNHIDRYYIE 62
++ +N+++N++ +E + V + KV++ G+LV+ R SN + Y++E
Sbjct: 274 VQSINVSKNILTSAPDGNREAAMKVLTSRKVQLLLGYLVQSIKRASNLEEDEGYFLE 330
>At3g43940 putative protein
Length = 202
Score = 26.6 bits (57), Expect = 3.7
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 6/43 (13%)
Query: 13 NLNRNVMNLREKELQIVVQSPSKV------EVPDGWLVEQRPR 49
N+ R ++RE E+ V ++ S+ + PDGW +E+R R
Sbjct: 62 NVERFRRHIRESEICQVCKAASETIIHLLRDCPDGWYLEERVR 104
>At2g03240 unknown protein
Length = 776
Score = 26.6 bits (57), Expect = 3.7
Identities = 11/25 (44%), Positives = 16/25 (64%)
Query: 23 EKELQIVVQSPSKVEVPDGWLVEQR 47
EK++ ++ KVE PDGW E+R
Sbjct: 166 EKQMDALIAFRVKVEHPDGWPWEER 190
>At1g05280 hypothetical protein
Length = 461
Score = 26.6 bits (57), Expect = 3.7
Identities = 10/23 (43%), Positives = 14/23 (60%)
Query: 41 GWLVEQRPRPSNPNHIDRYYIEP 63
G + +RP PS+ NH D Y + P
Sbjct: 102 GCVFVERPLPSSQNHTDSYLLPP 124
>At5g53900 unknown protein
Length = 377
Score = 26.2 bits (56), Expect = 4.9
Identities = 16/63 (25%), Positives = 27/63 (42%), Gaps = 1/63 (1%)
Query: 43 LVEQRPRPSNPNHIDRYY-IEPHTGQKFRSLVSVQRYLNGGETGDYLPTQRIISENNNNV 101
L Q P N + + + +E H KFR ++ ++ + L Q+ + NNN
Sbjct: 302 LFNQEQNPMNVENQNEFLNLEGHHPNKFRRSYTLPARMDSSSSSTSLDQQQPLEFRNNNS 361
Query: 102 SSN 104
SN
Sbjct: 362 GSN 364
>At5g53290 unknown protein
Length = 354
Score = 25.8 bits (55), Expect = 6.3
Identities = 13/39 (33%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query: 2 NRSMDVEMEHLNLNRNVMNLREKELQIVVQSPSKVEVPD 40
+ S + +++HL+ +V+N R +E+Q VQ P K P+
Sbjct: 214 SESTEEDLQHLSSPTSVLNHRSEEIQ-QVQQPFKSAKPE 251
>At5g53050 unknown protein
Length = 396
Score = 25.8 bits (55), Expect = 6.3
Identities = 14/42 (33%), Positives = 19/42 (44%), Gaps = 1/42 (2%)
Query: 45 EQRPRPSNPNHIDRYYIEPHTGQKFRSLVSVQRYLNG-GETG 85
EQR H + Y+E G R + Q+Y+ G ETG
Sbjct: 182 EQRAAVDLDTHYSKDYLEESVGTNTRRAILYQQYVKGISETG 223
>At2g35230 unknown protein
Length = 402
Score = 25.8 bits (55), Expect = 6.3
Identities = 13/43 (30%), Positives = 19/43 (43%)
Query: 49 RPSNPNHIDRYYIEPHTGQKFRSLVSVQRYLNGGETGDYLPTQ 91
RP P H R Y+ + Q + + Y+ G E Y+P Q
Sbjct: 179 RPYIPGHEQRPYVPGNEQQPYMPGNEQRPYIPGHEQRSYMPAQ 221
>At5g07310 putative transcription factor
Length = 263
Score = 25.4 bits (54), Expect = 8.3
Identities = 14/53 (26%), Positives = 25/53 (46%)
Query: 50 PSNPNHIDRYYIEPHTGQKFRSLVSVQRYLNGGETGDYLPTQRIISENNNNVS 102
P + ++YY + Q S ++ L GGETG + + +S N++ S
Sbjct: 182 PQTIPYFNQYYYNQYLHQGGNSNDALSYSLAGGETGGSMYNHQTLSTTNSSSS 234
>At4g36180 receptor protein kinase like protein
Length = 1136
Score = 25.4 bits (54), Expect = 8.3
Identities = 10/24 (41%), Positives = 14/24 (57%)
Query: 88 LPTQRIISENNNNVSSNFTFTSFC 111
LP ++S +NNN S F+ FC
Sbjct: 257 LPKLEVLSLSNNNFSGTVPFSLFC 280
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,234,888
Number of Sequences: 26719
Number of extensions: 138495
Number of successful extensions: 336
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 316
Number of HSP's gapped (non-prelim): 25
length of query: 128
length of database: 11,318,596
effective HSP length: 87
effective length of query: 41
effective length of database: 8,994,043
effective search space: 368755763
effective search space used: 368755763
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149210.5


