

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149210.3 + phase: 0
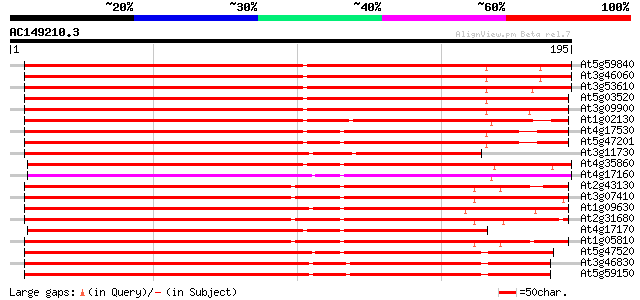
(195 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59840 GTP-binding protein ara-3 289 6e-79
At3g46060 GTP-binding protein ara-3 288 1e-78
At3g53610 GTPase AtRAB8 285 1e-77
At5g03520 GTP-binding protein - like 281 1e-76
At3g09900 putative Ras-like GTP-binding protein 276 4e-75
At1g02130 GTP-binding protein, ara-5 196 7e-51
At4g17530 ras-related small GTP-binding protein RAB1c 181 2e-46
At5g47201 ras-related small GTP-binding protein-like 180 5e-46
At3g11730 putative GTP-binding protein (ATFP8) 179 6e-46
At4g35860 GTP-binding protein GB2 156 6e-39
At4g17160 GTP-binding RAB2A like protein 156 6e-39
At2g43130 Ras-related GTP-binding protein (ARA-4) 152 1e-37
At3g07410 GTP-binding protein like 151 2e-37
At1g09630 putative RAS-related protein, RAB11C 150 5e-37
At2g31680 putative RAS superfamily GTP-binding protein 149 7e-37
At4g17170 GTP-binding RAB2A like protein 148 2e-36
At1g05810 RAS-related protein ARA-1 148 2e-36
At5g47520 GTP-binding protein-like 147 5e-36
At3g46830 GTP-binding protein Rab11 145 1e-35
At5g59150 GTP-binding protein rab11 - like 143 5e-35
>At5g59840 GTP-binding protein ara-3
Length = 216
Score = 289 bits (740), Expect = 6e-79
Identities = 149/193 (77%), Positives = 169/193 (87%), Gaps = 4/193 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRFSD FTTS+ITT+GID+K R IELDGK+I LQ+WDTAGQERFRTITTA
Sbjct: 24 SGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTA 83
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGAMGILLVYDVTDESSFNNI NWIR+IEQH+SDN VNKILVGNK DMDESKRAVP S
Sbjct: 84 YYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN-VNKILVGNKADMDESKRAVPKS 142
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL--PDSKKTPSRIRINKHNNAT 183
KGQALA++YGIKFFETSAKTNLNV+E FFSIA+DIKQRL DS+ P+ I+I++ + A
Sbjct: 143 KGQALADEYGIKFFETSAKTNLNVEEVFFSIAKDIKQRLADTDSRAEPATIKISQTDQAA 202
Query: 184 -AARGSQKSECCG 195
A + +QKS CCG
Sbjct: 203 GAGQATQKSACCG 215
>At3g46060 GTP-binding protein ara-3
Length = 216
Score = 288 bits (737), Expect = 1e-78
Identities = 148/193 (76%), Positives = 168/193 (86%), Gaps = 4/193 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRFSD FTTS+ITT+GID+K R IELDGK+I LQ+WDTAGQERFRTITTA
Sbjct: 24 SGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTA 83
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGAMGILLVYDVTDESSFNNI NWIR+IEQH+SDN VNKILVGNK DMDESKRAVP +
Sbjct: 84 YYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN-VNKILVGNKADMDESKRAVPTA 142
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL--PDSKKTPSRIRINKHNNAT 183
KGQALA++YGIKFFETSAKTNLNV+E FFSI RDIKQRL DS+ P+ I+I++ + A
Sbjct: 143 KGQALADEYGIKFFETSAKTNLNVEEVFFSIGRDIKQRLSDTDSRAEPATIKISQTDQAA 202
Query: 184 -AARGSQKSECCG 195
A + +QKS CCG
Sbjct: 203 GAGQATQKSACCG 215
>At3g53610 GTPase AtRAB8
Length = 216
Score = 285 bits (729), Expect = 1e-77
Identities = 146/193 (75%), Positives = 168/193 (86%), Gaps = 4/193 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRFSD FTTS+ITT+GID+K R IELDGK+I LQ+WDTAGQERFRTITTA
Sbjct: 24 SGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTA 83
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGAMGILLVYDVTDESSFNNI NWIR+IEQH+SD+ VNKILVGNK DMDESKRAVP S
Sbjct: 84 YYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDS-VNKILVGNKADMDESKRAVPKS 142
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL--PDSKKTPSRIRINKHN-NA 182
KGQALA++YG+KFFETSAKTNLNV+E FFSIA+DIKQRL D++ P I+IN+ + A
Sbjct: 143 KGQALADEYGMKFFETSAKTNLNVEEVFFSIAKDIKQRLADTDARAEPQTIKINQSDQGA 202
Query: 183 TAARGSQKSECCG 195
++ +QKS CCG
Sbjct: 203 GTSQATQKSACCG 215
>At5g03520 GTP-binding protein - like
Length = 216
Score = 281 bits (720), Expect = 1e-76
Identities = 141/191 (73%), Positives = 164/191 (85%), Gaps = 3/191 (1%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRFSD FTTS+ITT+GID+K R +ELDGK+I LQ+WDTAGQERFRTITTA
Sbjct: 24 SGVGKSCLLLRFSDDTFTTSFITTIGIDFKIRTVELDGKRIKLQIWDTAGQERFRTITTA 83
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGAMGILLVYDVTDESSFNNI NW+++IEQH+SDN VNKILVGNK DMDESKRAVP +
Sbjct: 84 YYRGAMGILLVYDVTDESSFNNIRNWMKNIEQHASDN-VNKILVGNKADMDESKRAVPTA 142
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL--PDSKKTPSRIRINKHNNAT 183
KGQALA++YGIKFFETSAKTNLNV+ F SIA+DIKQRL D+K P I+I K + A
Sbjct: 143 KGQALADEYGIKFFETSAKTNLNVENVFMSIAKDIKQRLTETDTKAEPQGIKITKQDTAA 202
Query: 184 AARGSQKSECC 194
++ ++KS CC
Sbjct: 203 SSSTAEKSACC 213
>At3g09900 putative Ras-like GTP-binding protein
Length = 218
Score = 276 bits (707), Expect = 4e-75
Identities = 141/193 (73%), Positives = 165/193 (85%), Gaps = 5/193 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRFSD FTTS+ITT+GID+K R +ELDGK+I LQ+WDTAGQERFRTITTA
Sbjct: 24 SGVGKSCLLLRFSDDTFTTSFITTIGIDFKIRTVELDGKRIKLQIWDTAGQERFRTITTA 83
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGAMGILLVYDVTDESSFNNI NW+++IEQH+SD+ VNKILVGNK DMDESKRAVP S
Sbjct: 84 YYRGAMGILLVYDVTDESSFNNIRNWMKNIEQHASDS-VNKILVGNKADMDESKRAVPTS 142
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL--PDSKKTPSRIRINKH--NN 181
KGQALA++YGIKFFETSAKTN NV++ F SIA+DIKQRL D+K P I+I K N
Sbjct: 143 KGQALADEYGIKFFETSAKTNQNVEQVFLSIAKDIKQRLTESDTKAEPQGIKITKQDANK 202
Query: 182 ATAARGSQKSECC 194
A+++ ++KS CC
Sbjct: 203 ASSSSTNEKSACC 215
>At1g02130 GTP-binding protein, ara-5
Length = 203
Score = 196 bits (498), Expect = 7e-51
Identities = 100/193 (51%), Positives = 137/193 (70%), Gaps = 12/193 (6%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRFSD + SYI+T+G+D+K R +E DGK I LQ+WDTAGQERFRTIT++
Sbjct: 17 SGVGKSCLLLRFSDDSYVESYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQERFRTITSS 76
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA GI++VYDVTDE SFNN+ W+ I++++SDN VNK+LVGNK D+ E+ RA+P+
Sbjct: 77 YYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDN-VNKLLVGNKSDLTEN-RAIPYE 134
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPD----SKKTPSRIRINKHNN 181
+A A++ GI F ETSAK NV++AF +++ IK+R+ + P ++I
Sbjct: 135 TAKAFADEIGIPFMETSAKDATNVEQAFMAMSASIKERMASQPAGNNARPPTVQIRGQPV 194
Query: 182 ATAARGSQKSECC 194
A QK+ CC
Sbjct: 195 A------QKNGCC 201
>At4g17530 ras-related small GTP-binding protein RAB1c
Length = 202
Score = 181 bits (459), Expect = 2e-46
Identities = 95/192 (49%), Positives = 130/192 (67%), Gaps = 11/192 (5%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRF+D + SYI+T+G+D+K R +E DGK I LQ+WDTAGQERFRTIT++
Sbjct: 17 SGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQERFRTITSS 76
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA GI++ YDVTD SFNN+ W+ I++++S+N VNK+LVGNK D+ S++ V
Sbjct: 77 YYRGAHGIIVTYDVTDLESFNNVKQWLNEIDRYASEN-VNKLLVGNKCDL-TSQKVVSTE 134
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL---PDSKKTPSRIRINKHNNA 182
+A A++ GI F ETSAK NV+EAF ++ IK R+ P P ++I
Sbjct: 135 TAKAFADELGIPFLETSAKNATNVEEAFMAMTAAIKTRMASQPAGGSKPPTVQIR----- 189
Query: 183 TAARGSQKSECC 194
+Q+S CC
Sbjct: 190 -GQPVNQQSGCC 200
>At5g47201 ras-related small GTP-binding protein-like
Length = 202
Score = 180 bits (456), Expect = 5e-46
Identities = 95/192 (49%), Positives = 130/192 (67%), Gaps = 11/192 (5%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS LLLRF+D + SYI+T+G+D+K R +E DGK I LQ+WDTAGQERFRTIT++
Sbjct: 17 SGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQERFRTITSS 76
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA GI++ YDVTD SFNN+ W+ I++++S+N VNK+LVGNK D+ S++ V
Sbjct: 77 YYRGAHGIIVTYDVTDLESFNNVKQWLNEIDRYASEN-VNKLLVGNKNDL-TSQKVVSTE 134
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL---PDSKKTPSRIRINKHNNA 182
+A A++ GI F ETSAK NV+EAF ++ IK R+ P P ++I
Sbjct: 135 TAKAFADELGIPFLETSAKNATNVEEAFMAMTAAIKTRMASQPAGGAKPPTVQIR----- 189
Query: 183 TAARGSQKSECC 194
+Q+S CC
Sbjct: 190 -GQPVNQQSGCC 200
>At3g11730 putative GTP-binding protein (ATFP8)
Length = 205
Score = 179 bits (455), Expect = 6e-46
Identities = 87/159 (54%), Positives = 124/159 (77%), Gaps = 2/159 (1%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+ VGKS LLLRF+D + SYI+T+G+D+K R IE DGK I LQ+WDTAGQERFRTIT++
Sbjct: 17 SSVGKSCLLLRFADDAYIDSYISTIGVDFKIRTIEQDGKTIKLQIWDTAGQERFRTITSS 76
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA GI++VYD T+ SFNN+ W+ I++++++++ K+L+GNK DM ESK V
Sbjct: 77 YYRGAHGIIIVYDCTEMESFNNVKQWLSEIDRYANESVC-KLLIGNKNDMVESK-VVSTE 134
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRL 164
G+ALA++ GI F ETSAK ++NV++AF +IA +IK+++
Sbjct: 135 TGRALADELGIPFLETSAKDSINVEQAFLTIAGEIKKKM 173
>At4g35860 GTP-binding protein GB2
Length = 211
Score = 156 bits (395), Expect = 6e-39
Identities = 86/198 (43%), Positives = 120/198 (60%), Gaps = 11/198 (5%)
Query: 7 GVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTAY 66
GVGKS LLL+F+D F + T+G+++ R + +DG+ I LQ+WDTAGQE FR+IT +Y
Sbjct: 16 GVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMVTVDGRPIKLQIWDTAGQESFRSITRSY 75
Query: 67 YRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFSK 126
YRGA G LLVYD+T +FN++ +W+ QH++ N ++ +L+GNK D+ KRAV +
Sbjct: 76 YRGAAGALLVYDITRRETFNHLASWLEDARQHANPN-MSIMLIGNKCDL-AHKRAVSKEE 133
Query: 127 GQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPDS----KKTPSRIRINKHNNA 182
GQ A ++G+ F E SA+T NV+EAF A I Q + D S I+I
Sbjct: 134 GQQFAKEHGLLFLEASARTAQNVEEAFIETAAKILQNIQDGVFDVSNESSGIKIGYGRTQ 193
Query: 183 TAARG-----SQKSECCG 195
AA G SQ CCG
Sbjct: 194 GAAGGRDGTISQGGGCCG 211
>At4g17160 GTP-binding RAB2A like protein
Length = 205
Score = 156 bits (395), Expect = 6e-39
Identities = 80/192 (41%), Positives = 116/192 (59%), Gaps = 5/192 (2%)
Query: 7 GVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTAY 66
GVGKS LLL+F+D F + T+G+++ + I +D K I LQ+WDTAGQE FR++T +Y
Sbjct: 16 GVGKSCLLLKFTDKRFQAVHDLTIGVEFGAKTITIDNKPIKLQIWDTAGQESFRSVTRSY 75
Query: 67 YRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFSK 126
YRG G LLVYD+T +FN++ +W+ QH+S+N+ +L+GNK D+ E KR V +
Sbjct: 76 YRGRAGTLLVYDITRRETFNHLASWLEEARQHASENMTT-MLIGNKCDL-EDKRTVSTEE 133
Query: 127 GQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPD---SKKTPSRIRINKHNNAT 183
G+ A ++G+ F E SAKT NV+EAF A I +R+ D + I
Sbjct: 134 GEQFAREHGLIFMEASAKTAHNVEEAFVETAATIYKRIQDGVVDEANEPGITPGPFGGKD 193
Query: 184 AARGSQKSECCG 195
A+ Q+ CCG
Sbjct: 194 ASSSQQRRGCCG 205
>At2g43130 Ras-related GTP-binding protein (ARA-4)
Length = 214
Score = 152 bits (383), Expect = 1e-37
Identities = 83/198 (41%), Positives = 126/198 (62%), Gaps = 15/198 (7%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+ VGKS+LL R++ + F + T+G++++ +++ +DGK++ Q+WDTAGQERFR +T+A
Sbjct: 21 SAVGKSNLLTRYARNEFNPNSKATIGVEFQTQSMLIDGKEVKAQIWDTAGQERFRAVTSA 80
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G L+VYD+T S+F N+ W+ + H SD V K+L+GNK D+ ES RAV
Sbjct: 81 YYRGAVGALVVYDITRSSTFENVGRWLDELNTH-SDTTVAKMLIGNKCDL-ESIRAVSVE 138
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDI------KQRLPDSKK---TPSRIRI 176
+G++LA G+ F ETSA + NV AF + R+I KQ DS K T +R+ +
Sbjct: 139 EGKSLAESEGLFFMETSALDSTNVKTAFEMVIREIYSNISRKQLNSDSYKEELTVNRVSL 198
Query: 177 NKHNNATAARGSQKSECC 194
K+ N G++ CC
Sbjct: 199 VKNEN----EGTKTFSCC 212
>At3g07410 GTP-binding protein like
Length = 217
Score = 151 bits (381), Expect = 2e-37
Identities = 80/197 (40%), Positives = 124/197 (62%), Gaps = 10/197 (5%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+ VGKS+LL RFS F T+ T+G++++ + +E++GK++ Q+WDTAGQERFR +T+A
Sbjct: 21 SAVGKSNLLSRFSRDEFDTNSKATIGVEFQTQLVEIEGKEVKAQIWDTAGQERFRAVTSA 80
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA G L+VYD+T +F ++ W++ + H D V ++LVGNK D+ E RAV
Sbjct: 81 YYRGAFGALIVYDITRGDTFESVKRWLQELNTH-CDTAVAQMLVGNKCDL-EDIRAVSVE 138
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDI----KQRLPDSKKTPSRIRINKHNN 181
+G+ALA + G+ F ETSA NVD+AF + R+I ++L +S + + +N+ +
Sbjct: 139 EGKALAEEEGLFFMETSALDATNVDKAFEIVIREIFNNVSRKLLNSDAYKAELSVNRVSL 198
Query: 182 ATAARGSQKS----ECC 194
GS+ S CC
Sbjct: 199 VNNQDGSESSWRNPSCC 215
>At1g09630 putative RAS-related protein, RAB11C
Length = 217
Score = 150 bits (378), Expect = 5e-37
Identities = 79/196 (40%), Positives = 122/196 (61%), Gaps = 9/196 (4%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS+LL RF+ + F +T+G+++ R ++++G+ + Q+WDTAGQER+R IT+A
Sbjct: 21 SGVGKSNLLSRFTRNEFCLESKSTIGVEFATRTLQVEGRTVKAQIWDTAGQERYRAITSA 80
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G LLVYDVT ++F N++ W++ + H+ NIV +L+GNK D+ + RAV
Sbjct: 81 YYRGALGALLVYDVTKPTTFENVSRWLKELRDHADSNIV-IMLIGNKTDL-KHLRAVATE 138
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIA----RDIKQRLPDSKKTPSRIRINKHNN 181
Q+ A K G+ F ETSA LNV++AF +I R I ++ S +T + I +
Sbjct: 139 DAQSYAEKEGLSFIETSALEALNVEKAFQTILSEVYRIISKKSISSDQTTANANIKEGQT 198
Query: 182 ---ATAARGSQKSECC 194
A + + K CC
Sbjct: 199 IDVAATSESNAKKPCC 214
>At2g31680 putative RAS superfamily GTP-binding protein
Length = 219
Score = 149 bits (377), Expect = 7e-37
Identities = 83/199 (41%), Positives = 128/199 (63%), Gaps = 13/199 (6%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+ VGKS+LL R++ + F T+G++++ + +E++GK++ Q+WDTAGQERFR +T+A
Sbjct: 21 SAVGKSNLLSRYARNEFNAHSKATIGVEFQTQNMEIEGKEVKAQIWDTAGQERFRAVTSA 80
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G L+VYD++ S+F ++ W+ ++ H SD V ++LVGNK D+ ES RAV
Sbjct: 81 YYRGAVGALVVYDISRRSTFESVGRWLDELKTH-SDTTVARMLVGNKCDL-ESIRAVSVE 138
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDI------KQRLPDSKKT----PSRIR 175
+G+ALA G+ F ETSA + NV AF + RDI KQ D+ KT +R+
Sbjct: 139 EGKALAETEGLFFMETSALDSTNVKTAFEMVIRDIYTNISRKQLNSDTYKTELSMKNRVS 198
Query: 176 INKHNNATAARGSQKSECC 194
+ K +N ++ +G S CC
Sbjct: 199 LVKDDNKSSTQGFGFS-CC 216
>At4g17170 GTP-binding RAB2A like protein
Length = 211
Score = 148 bits (374), Expect = 2e-36
Identities = 74/160 (46%), Positives = 107/160 (66%), Gaps = 2/160 (1%)
Query: 7 GVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTAY 66
GVGKS LLL+F+D F + T+G+++ R I +D K I LQ+WDTAGQE FR+IT +Y
Sbjct: 16 GVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDTAGQESFRSITRSY 75
Query: 67 YRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFSK 126
YRGA G LLVYD+T +FN++ +W+ QH++ N + +L+GNK D+ +RAV +
Sbjct: 76 YRGAAGALLVYDITRRETFNHLASWLEDARQHANAN-MTIMLIGNKCDL-AHRRAVSTEE 133
Query: 127 GQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPD 166
G+ A ++G+ F E SAKT NV+EAF A I +++ D
Sbjct: 134 GEQFAKEHGLIFMEASAKTAQNVEEAFIKTAATIYKKIQD 173
>At1g05810 RAS-related protein ARA-1
Length = 261
Score = 148 bits (374), Expect = 2e-36
Identities = 81/198 (40%), Positives = 131/198 (65%), Gaps = 12/198 (6%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+ VGKS+LL R++ + F+ + T+G++++ +++E++GK++ Q+WDTAGQERFR +T+A
Sbjct: 64 SAVGKSNLLSRYARNEFSANSKATIGVEFQTQSMEIEGKEVKAQIWDTAGQERFRAVTSA 123
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G L+VYD+T ++F ++ W+ ++ H SD V ++LVGNK D+ E+ RAV
Sbjct: 124 YYRGAVGALVVYDITRRTTFESVGRWLDELKIH-SDTTVARMLVGNKCDL-ENIRAVSVE 181
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDI------KQRLPDSKK---TPSRIRI 176
+G+ALA + G+ F ETSA + NV AF + DI KQ D+ K T +R+ +
Sbjct: 182 EGKALAEEEGLFFVETSALDSTNVKTAFEMVILDIYNNVSRKQLNSDTYKDELTVNRVSL 241
Query: 177 NKHNNATAARGSQKSECC 194
K +N +A++ S CC
Sbjct: 242 VKDDN-SASKQSSGFSCC 258
>At5g47520 GTP-binding protein-like
Length = 221
Score = 147 bits (370), Expect = 5e-36
Identities = 77/184 (41%), Positives = 122/184 (65%), Gaps = 4/184 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+ VGKS+LL RF+ F + +T+G++++ + ++++GK+I Q+WDTAGQERFR +T+A
Sbjct: 23 SAVGKSNLLARFARDEFYPNSKSTIGVEFQTQKMDINGKEIKAQIWDTAGQERFRAVTSA 82
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G LLVYD++ +F++I W+ + HS N+V ILVGNK D+ + R V +
Sbjct: 83 YYRGAVGALLVYDISRRQTFHSIGRWLNELHTHSDMNVVT-ILVGNKSDL-KDLREVSTA 140
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPDSKKTPSRIRINKHNNATAA 185
+G+ALA G+ F ETSA + NV AF ++ ++I L S+K S +NK + A+ +
Sbjct: 141 EGKALAEAQGLFFMETSALDSSNVAAAFETVVKEIYNIL--SRKVMSSQELNKQDPASLS 198
Query: 186 RGSQ 189
G +
Sbjct: 199 NGKK 202
>At3g46830 GTP-binding protein Rab11
Length = 217
Score = 145 bits (367), Expect = 1e-35
Identities = 77/183 (42%), Positives = 117/183 (63%), Gaps = 4/183 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGKS++L RF+ + F +T+G+++ R +++GK I Q+WDTAGQER+R IT+A
Sbjct: 21 SGVGKSNILSRFTRNEFCLESKSTIGVEFATRTTQVEGKTIKAQIWDTAGQERYRAITSA 80
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G LLVYD+T +F+N+ W+R + H+ NIV ++ GNK D++ R+V
Sbjct: 81 YYRGAVGALLVYDITKRQTFDNVLRWLRELRDHADSNIV-IMMAGNKSDLNH-LRSVAEE 138
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPDSKKTPSRIRINKHNNATAA 185
GQ+LA K G+ F ETSA NV++AF +I +I + SKK + N+A
Sbjct: 139 DGQSLAEKEGLSFLETSALEATNVEKAFQTILGEIYHII--SKKALAAQEAAAANSAIPG 196
Query: 186 RGS 188
+G+
Sbjct: 197 QGT 199
>At5g59150 GTP-binding protein rab11 - like
Length = 217
Score = 143 bits (361), Expect = 5e-35
Identities = 73/183 (39%), Positives = 116/183 (62%), Gaps = 4/183 (2%)
Query: 6 AGVGKSSLLLRFSDSYFTTSYITTLGIDYKNRAIELDGKKIMLQVWDTAGQERFRTITTA 65
+GVGK+++L RF+ + F +T+G+++ R ++++GK + Q+WDTAGQER+R IT+A
Sbjct: 21 SGVGKTNILSRFTRNEFCLESKSTIGVEFATRTLQVEGKTVKAQIWDTAGQERYRAITSA 80
Query: 66 YYRGAMGILLVYDVTDESSFNNITNWIRSIEQHSSDNIVNKILVGNKVDMDESKRAVPFS 125
YYRGA+G LLVYD+T +F+N+ W+R + H+ NIV ++ GNK D++ R+V
Sbjct: 81 YYRGAVGALLVYDITKRQTFDNVLRWLRELRDHADSNIV-IMMAGNKADLNH-LRSVAEE 138
Query: 126 KGQALANKYGIKFFETSAKTNLNVDEAFFSIARDIKQRLPDSKKTPSRIRINKHNNATAA 185
GQ LA G+ F ETSA NV++AF ++ +I + SKK + N+A
Sbjct: 139 DGQTLAETEGLSFLETSALEATNVEKAFQTVLAEIYHII--SKKALAAQEAAAANSAIPG 196
Query: 186 RGS 188
+G+
Sbjct: 197 QGT 199
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,889,728
Number of Sequences: 26719
Number of extensions: 144944
Number of successful extensions: 592
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 389
Number of HSP's gapped (non-prelim): 109
length of query: 195
length of database: 11,318,596
effective HSP length: 94
effective length of query: 101
effective length of database: 8,807,010
effective search space: 889508010
effective search space used: 889508010
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149210.3


