

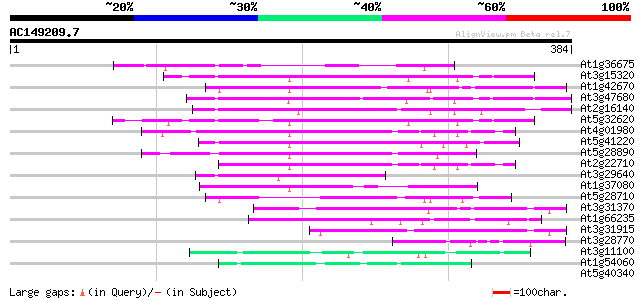
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149209.7 - phase: 0
(384 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g36675 putative protein 108 5e-24
At3g15320 hypothetical protein 107 1e-23
At1g42670 unknown protein 102 5e-22
At3g47680 unknown protein 101 8e-22
At2g16140 pseudogene; similar to MURA transposase of maize Muta... 91 8e-19
At5g32620 putative protein 91 1e-18
At4g01980 hypothetical protein 89 5e-18
At5g41220 glutathione transferase-like 88 9e-18
At5g28890 putative protein 84 1e-16
At2g22710 hypothetical protein 84 2e-16
At3g29640 hypothetical protein 76 3e-14
At1g37080 hypothetical protein 61 1e-09
At5g28710 putative protein 53 2e-07
At3g31370 hypothetical protein 50 2e-06
At1g66235 putative protein 49 4e-06
At3g31915 hypothetical protein 46 4e-05
At3g28770 hypothetical protein 45 5e-05
At3g11100 unknown protein 44 1e-04
At1g54060 unknown protein 43 3e-04
At5g40340 unknown protein 40 0.002
>At1g36675 putative protein
Length = 268
Score = 108 bits (270), Expect = 5e-24
Identities = 85/241 (35%), Positives = 108/241 (44%), Gaps = 81/241 (33%)
Query: 72 NFNPYFRSMMGYSSQTPPFNGYMPMVNENFQSVG--EYPDFSSQINRGGMTRDNEVAPTP 129
N PY S+M SSQ P ++ PM NE +VG E+P+FS+Q+ G M+ +E P
Sbjct: 7 NHRPYHGSIMSPSSQAPSYSS-TPMGNETNTNVGATEFPEFSTQMALGVMSGVHETIPNE 65
Query: 130 EDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFD 189
ED T R+ + W T+QNLVL+S WIKY +V +T +
Sbjct: 66 EDLTCNHSRSSK--WTTDQNLVLLSRWIKYE--QIVSLVETKK----------------- 104
Query: 190 SPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTL 249
D+ KRMQ SGWS+D V KA ELY
Sbjct: 105 --------------------------DNVKRMQQSGWSKDGVLAKAHELY---------- 128
Query: 250 MEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYE------DSVGSSARPMDRDAAKKKGKK 303
GN SGSSGSKR++E +SVGSSARP+ D AKKK KK
Sbjct: 129 ---------------SNAGNTGSGSSGSKRTHESDVRDANSVGSSARPIGSDIAKKKTKK 173
Query: 304 K 304
K
Sbjct: 174 K 174
>At3g15320 hypothetical protein
Length = 287
Score = 107 bits (266), Expect = 1e-23
Identities = 76/279 (27%), Positives = 131/279 (46%), Gaps = 35/279 (12%)
Query: 106 EYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVV 165
E P FSSQ++ G +++ + ++ PK +++ W ++++VL+S W+ V+
Sbjct: 10 EVPAFSSQLSEEG----SDLEGSEDEVKPKQSISRK-KWTAKEDIVLVSAWLNTSKDPVI 64
Query: 166 GRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQG 223
G +Q +S+W +IA Y S D R+ C++R+ +NK + K+VG Y + +
Sbjct: 65 GNDQQGQSFWKRIAAYVAASPSLDGLPKREHAKCKHRWGKVNKSVTKFVGCYKTTTTHKT 124
Query: 224 SGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGSQVGGNVD---------SGS 274
SG SEDDV K A E+Y FTL WR LR ++ D G+
Sbjct: 125 SGQSEDDVMKLAYEIYFNDTKKNFTLDHAWRELRYDQKWCEATSRKGDKNAKRKKCGDGN 184
Query: 275 SGSKRSYEDSVGSSARPMDRDAAKKKGKKKS--------------KGETLEKVEKEWVQF 320
+ S+ + + +RP AAK K +K + G+++E + W
Sbjct: 185 ASSQPIHVEDDSVMSRPPGVKAAKAKARKSATIKEGKKPATVKDDSGQSVEHFQNLW--- 241
Query: 321 KELKEQEIEQLKELTLVKQQKNKLLQEKTQAKKMKMYLK 359
ELKE++ ++ KE QQ +LL + + ++LK
Sbjct: 242 -ELKEKDWDR-KEKQSKHQQLERLLSKTEPLSDIDLFLK 278
>At1g42670 unknown protein
Length = 579
Score = 102 bits (253), Expect = 5e-22
Identities = 81/257 (31%), Positives = 127/257 (48%), Gaps = 18/257 (7%)
Query: 135 KSKRNQQP--SWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS-- 190
K K+ ++P +W++ +++VLISGW+ VVG Q ++W +IA Y N
Sbjct: 329 KGKKAKKPRRNWSSTEDVVLISGWLNTSKDPVVGNEQKGAAFWERIAVYYNSSPKLKGVE 388
Query: 191 PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLM 250
R + C+ R++ +N +NK+VG+Y +A + Q SG ++DDV A +++ +FT
Sbjct: 389 KRGHICCKQRWSKVNDAVNKFVGSYLAASKQQTSGQNDDDVVSLAHQIFSKDYGCKFTCE 448
Query: 251 EEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDS--VG---SSARPMDRDAAKKKGKKKS 305
WR + RY + G + ++ DS VG ARP+ AAK K K
Sbjct: 449 HAWR----ERRYDQKWIAQSTHGKAKRRKCEADSEPVGVEDKEARPIGVKAAKAAAKAKG 504
Query: 306 KGETLEKVEKEWVQFKELKE-QEIEQLKELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEE 364
K + L VE E + LKE Q I ++KE ++K ++++K K L L E
Sbjct: 505 KAK-LSPVEGE--ETNALKEIQSIWEIKEKDHAAKEKLIIIKDKKNRTKFFERL-LGKTE 560
Query: 365 HLDDRKNELLGKLEREL 381
L D + EL KL EL
Sbjct: 561 PLSDIEIELKNKLINEL 577
>At3g47680 unknown protein
Length = 302
Score = 101 bits (251), Expect = 8e-22
Identities = 76/282 (26%), Positives = 139/282 (48%), Gaps = 28/282 (9%)
Query: 122 DNEVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEY 181
D++V+P TPK KR ++ W+ ++LVL+S W+ +V+G Q ++W +IA Y
Sbjct: 29 DDDVSPNQAAHTPKVKRERR-KWSAGEDLVLVSAWLNTSKDAVIGNEQKGYAFWSRIAAY 87
Query: 182 CNEHCSFD--SPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELY 239
+ R+ + R+ +N+ + K+VG+Y++A + + SG ++DDV A E+Y
Sbjct: 88 YGASPKLNGVEKRETGHIKQRWTKINEGVGKFVGSYEAATKQKSSGQNDDDVVALAHEIY 147
Query: 240 GCGKNVQFTLMEEWRALRDQPRYGSQVGGNV---------DSGSSGSKRSYEDSVGSSAR 290
++ +FTL WR LR + ++ S D S+ +++E S R
Sbjct: 148 N-SEHGKFTLEHAWRVLRFEQKWLSAPSTKATVMSKRRKSDKASTSQPQTHEAEEAMS-R 205
Query: 291 PMDRDAAKKKGKK--------KSKGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKN 342
P+ AAK K KK + KG + +++ W E+K+++ E L++ +++++
Sbjct: 206 PIGVKAAKAKAKKAVSKTTTVEDKGNVMLEIQSIW----EIKQKDWE-LRQKDREQEKED 260
Query: 343 KLLQEKTQAKKMKMYLKLRDEEHLDDRKNELLGKLERELFEN 384
QE+ K+ L +E L D + L KL E+ N
Sbjct: 261 FEKQERLSRTKLLESL-FTKKEPLTDIEVALKNKLINEMLSN 301
>At2g16140 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 311
Score = 91.3 bits (225), Expect = 8e-19
Identities = 77/282 (27%), Positives = 136/282 (47%), Gaps = 36/282 (12%)
Query: 126 APTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEH 185
+P+ E+ PK KR + W+ ++++VL+S W+ +V+G Q + ++W +IA Y +
Sbjct: 43 SPSLEEHVPKVKRERM-KWSAKEDMVLVSSWLNTSKDAVIGNEQKANTFWSRIAAYYDAS 101
Query: 186 CSFDSPRDIVA--CRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGK 243
+ R + + R+ +N + K+VG+Y++A R + SG +++DV A E++
Sbjct: 102 PQLNGLRKRMQGNIKQRWAKINDGVCKFVGSYEAASREKSSGQNDNDVISLAHEIFNNDY 161
Query: 244 NVQFTLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDSVG---------SSARPMDR 294
+F L WR LR ++ SQ +V S ++ + S + +RP+
Sbjct: 162 GYKFPLEHAWRVLRHDQKWCSQ--ASVMSKRRKCDKAAQPSTSQPPSHGVEEAMSRPIGV 219
Query: 295 DAAKKKGKK--------KSKGETLEKVEKEW-VQFK--ELKEQEIEQLKELTLVKQQKNK 343
AAK K KK + KG + +++ W ++ K EL++++ EQ KE K + +K
Sbjct: 220 KAAKAKAKKTVTKTTTVEDKGNAMLEIQSIWEIKQKDWELRQKDREQEKEDFEKKDRLSK 279
Query: 344 -LLQEKTQAKKMKMYLKLRDEEHLDDRKNELLGKLERELFEN 384
L E AKK E L D + L KL L E+
Sbjct: 280 TTLLESLIAKK----------EPLTDNEVTLKNKLIDFLLEH 311
>At5g32620 putative protein
Length = 301
Score = 90.5 bits (223), Expect = 1e-18
Identities = 77/321 (23%), Positives = 140/321 (42%), Gaps = 62/321 (19%)
Query: 71 NNFNPYFRSMMGYSSQTPPFNGYMPMVNENFQSVGEY-------PDFSSQINRGGMTRDN 123
++ NP+++S +G+M ++ + + ++ P FSSQ++ G +
Sbjct: 2 DDMNPFYQS-----------SGFMDLLQSQQEDMSQFASGSTEVPAFSSQLSEEG----S 46
Query: 124 EVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCN 183
E+ + ++ PK +++ W ++++VL+S W+ V+G +Q + Y
Sbjct: 47 ELEGSEDEVKPKQSISRK-KWTAKEDIVLVSAWLNPSKDPVIGNDQQA---------YVA 96
Query: 184 EHCSFDS--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGC 241
S D R+ C+ R+ +NK + K+V Y + + SG SEDDV K A E+Y
Sbjct: 97 ASPSLDGLPKREHAKCKQRWGKVNKSVTKFVACYKTTTTHKTSGQSEDDVMKLAYEIYFN 156
Query: 242 GKNVQFTLMEEWRALRDQPRYGSQVGGNVD---------SGSSGSKRSYEDSVGSSARPM 292
FTL WR LR ++ D G++ S + + +RP
Sbjct: 157 DTKKNFTLDHAWRELRYDQKWCEATSRKGDENAKRRKCGDGNASSHPIHVEDDSIMSRPP 216
Query: 293 DRDAAKKKGKKKS--------------KGETLEKVEKEWVQFKELKEQEIEQLKELTLVK 338
AAK KG+K + G+++E + W ELKE++ ++ KE
Sbjct: 217 GVKAAKAKGRKSAIVKEGKKPATVKDDSGQSVEHFQNLW----ELKEKDWDR-KEKQSKH 271
Query: 339 QQKNKLLQEKTQAKKMKMYLK 359
QQ +LL + + ++LK
Sbjct: 272 QQLERLLSKTEPLSDIDLFLK 292
>At4g01980 hypothetical protein
Length = 302
Score = 88.6 bits (218), Expect = 5e-18
Identities = 78/279 (27%), Positives = 126/279 (44%), Gaps = 36/279 (12%)
Query: 91 NGYMPMVNENFQS----VGEYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNT 146
+G+M +++ +S +Y SS+I G R E + E+ + W
Sbjct: 11 SGFMDLLHSQQESHNNETNQYEVGSSEIPVFGTQRCQE---SHEERYASRVAIARKKWAA 67
Query: 147 EQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRFNYM 204
++++VLIS W+ VVG Q + ++W +IA Y + D R C+ R+ M
Sbjct: 68 KEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAANPDLDGVPKRASAQCKQRWAKM 127
Query: 205 NKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGS 264
N+L+ K+VG Y + + SG +E+DV A EL+ +F+L WR LR ++
Sbjct: 128 NELVMKFVGCYATTTNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLLRHDQKW-- 185
Query: 265 QVGGNVDSGSSGSKRSYEDSVGSSA---------------RPMDRDAAKKKGKKKS--KG 307
+ N G +KR + VG+ A RP AAK K KK KG
Sbjct: 186 -LISNAPKGKGIAKRR-KVRVGTQAASSQPIDLEDDDVMRRPPGVKAAKAKAKKTPTVKG 243
Query: 308 ETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQ 346
E + VE +LKE++ ++ KQ KN++L+
Sbjct: 244 EEVNSVE-HLTSLWDLKERDWDRKD-----KQSKNQMLE 276
>At5g41220 glutathione transferase-like
Length = 590
Score = 87.8 bits (216), Expect = 9e-18
Identities = 66/251 (26%), Positives = 116/251 (45%), Gaps = 35/251 (13%)
Query: 130 EDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFD 189
++TT + K ++ W+ ++ +LIS W+ +V + ++W +I Y N S
Sbjct: 261 QNTTDRRKHRRK--WSRAEDAILISAWLNTSKDPIVDNEHKACAFWKRIGAYFNNSASLA 318
Query: 190 S--PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQF 247
+ R+ C+ R++ +N + K+VG YD A + SG SEDDVF+ A ++Y F
Sbjct: 319 NLPKREPSHCKQRWSKLNDKVCKFVGCYDQALNQRSSGQSEDDVFQVAYQVYTNNYKSNF 378
Query: 248 TLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRS--------YEDSVGSSARPMDRD---- 295
TL WR LR ++ S G SKR+ Y S + P+ D
Sbjct: 379 TLEHAWRELRHSKKWCSLYPFENSKGGGSSKRTKLNNGDRVYSSSSNPESVPIALDEEEQ 438
Query: 296 ---------AAKKKGKKKSKGETLE--------KVEKEWVQFKELKEQEIEQLKELTLVK 338
++K+K KK + T+E ++E WV +E EQ +++ + ++
Sbjct: 439 VMDLPLGVKSSKQKEKKVATIITIEEREADSGSRLENLWVLDEE--EQVMDRPLGVKSLE 496
Query: 339 QQKNKLLQEKT 349
Q++NK+ + T
Sbjct: 497 QKENKVAPKPT 507
Score = 28.9 bits (63), Expect = 5.0
Identities = 20/82 (24%), Positives = 42/82 (50%), Gaps = 4/82 (4%)
Query: 278 KRSYEDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKELTLV 337
K + E+ + +R + K K++ + ++ ++E W LKE++IE+ K+LT +
Sbjct: 505 KPTIEEREAADSRSRLENLWALKEKEEREADSRSRLENLWA----LKEKDIEEQKKLTRM 560
Query: 338 KQQKNKLLQEKTQAKKMKMYLK 359
+ K+ L + Q + + LK
Sbjct: 561 EVLKSLLGRTTDQLSEKEDILK 582
>At5g28890 putative protein
Length = 232
Score = 84.0 bits (206), Expect = 1e-16
Identities = 63/239 (26%), Positives = 109/239 (45%), Gaps = 25/239 (10%)
Query: 91 NGYMPMVNENFQSVGEYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNTEQNL 150
+G+M ++ +S ++++ N D+E TP+ PK K W+ ++++
Sbjct: 6 SGFMDLLQSQQES------YNNENNPQLSDEDHEDVNTPKVAIPKKK------WSAKEDV 53
Query: 151 VLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRFNYMNKLI 208
+LIS W+ VVG Q + ++W +IA Y R+ C+ R+ MN+L+
Sbjct: 54 ILISAWLNTSKDPVVGNEQKAPAFWKRIATYVAASPDLVGFPKRESAQCKQRWAKMNELV 113
Query: 209 NKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGSQVGG 268
K+VG Y + + SG +E+DV A EL+ +F+L WR +R + ++ +
Sbjct: 114 MKFVGCYATETNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLVRHEQKW---IIS 170
Query: 269 NVDSGSSGSKRSYEDSVGSSARP--------MDRDAAKKKGKKKSKGETLEKVEKEWVQ 319
N SKR S S++P M R K K K+K K +K+ +Q
Sbjct: 171 NTPKEKRMSKRRKVGSQAKSSQPINLEDYDVMARPLRVKVPKAKTKKIPNVKKKKKLIQ 229
>At2g22710 hypothetical protein
Length = 300
Score = 83.6 bits (205), Expect = 2e-16
Identities = 66/222 (29%), Positives = 102/222 (45%), Gaps = 29/222 (13%)
Query: 144 WNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS--PRDIVACRNRF 201
W ++++VLIS W+ VVG Q + ++W +IA Y D R C+ R+
Sbjct: 63 WAAKEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAASPDLDGVPKRASAQCKQRW 122
Query: 202 NYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPR 261
MN+L+ K+VG Y + + SG +E+DV A EL+ +F+L WR LR +
Sbjct: 123 AKMNELVMKFVGCYATTTNQKASGQTENDVMLFANELFKNDMKKKFSLDHAWRLLRHDQK 182
Query: 262 YGSQVGGNVDSGSSGSKRSYEDSVGSSA---------------RPMDRDAAKKKGKKKS- 305
+ + N G +KR + VG+ A RP AAK K K
Sbjct: 183 W---LISNAPKGKGIAKRR-KVRVGTQAASSQPIDLEDDDVMRRPPGVKAAKAKAKMTPT 238
Query: 306 -KGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQ 346
KGE + VE +LKE++ ++ KQ KN++L+
Sbjct: 239 VKGEEVNSVE-HLTSLWDLKEKDWDRKD-----KQSKNQMLE 274
>At3g29640 hypothetical protein
Length = 171
Score = 76.3 bits (186), Expect = 3e-14
Identities = 42/132 (31%), Positives = 70/132 (52%), Gaps = 3/132 (2%)
Query: 128 TPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYC--NEH 185
T ++ PK +Q+ W ++++VL+ W+ V+G +Q +S+ +IA Y +
Sbjct: 16 TQDEVKPKVGLSQK-KWFPKEDIVLVRAWLNTNKDHVIGNDQQCQSFLKQIASYVAISPQ 74
Query: 186 CSFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNV 245
+ R+ C+ R++ +NK + K+VG Y +A + SG SEDDV K A E+Y
Sbjct: 75 LDYLPKREHAKCKQRWSKVNKSVTKFVGCYKTATTHKTSGQSEDDVMKLAYEIYYNDTKK 134
Query: 246 QFTLMEEWRALR 257
+F L WR LR
Sbjct: 135 KFNLEHTWRKLR 146
>At1g37080 hypothetical protein
Length = 224
Score = 60.8 bits (146), Expect = 1e-09
Identities = 50/194 (25%), Positives = 85/194 (43%), Gaps = 31/194 (15%)
Query: 131 DTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDS 190
+TT K + W +++++VLIS W+ SVVG Q ++++W ++ Y + S
Sbjct: 58 NTTTKRTTTTRNKWTSKEDIVLISTWLNTSKDSVVGNEQRADAFWKRVVVYFASTLNVSS 117
Query: 191 --PRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFT 248
R+ + R+ +NK++ + G++++A SG +ED + K K +
Sbjct: 118 QIKREPSHSKQRWGKINKIVCMFGGSHEAANTQMASGMNEDVLMNKK-------KKAEAK 170
Query: 249 LMEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDSVG-SSARPMDRDAAKKKGKKKSK- 306
EE S S + +D G + ARP AAK KGK K+
Sbjct: 171 KAEE--------------------NESPSVVNLDDHHGETEARPPSVKAAKSKGKMKANT 210
Query: 307 GETLEKVEKEWVQF 320
T+E EK +F
Sbjct: 211 SATMEGDEKPTERF 224
>At5g28710 putative protein
Length = 264
Score = 53.1 bits (126), Expect = 2e-07
Identities = 50/220 (22%), Positives = 92/220 (41%), Gaps = 61/220 (27%)
Query: 135 KSKRNQQP--SWNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPR 192
K K+ ++P +W++ +++VLI GW+ +VG Q
Sbjct: 64 KGKKAKKPRRNWSSIEDIVLICGWLNTSKDPMVGNEQKG--------------------- 102
Query: 193 DIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEE 252
AY +A + Q SG ++D V A +++ +FT
Sbjct: 103 --------------------AAYLTASKQQTSGQNDDFVVSLAHQIFSKDYGCKFTCEHA 142
Query: 253 WRALRDQPRYGSQVGGNVDSGSSGSKRSYE---DSVG---SSARPMDRDAAKKKGKKKSK 306
WR LR ++ +Q + +R E DSVG ARP+ AAK + +
Sbjct: 143 WRELRYDQKWIAQ-----STHGKAKRRKCEADSDSVGVEDKEARPISVKAAKAAKQSPVE 197
Query: 307 GE---TLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNK 343
GE L++++ W E+K+++ ++L ++K++KN+
Sbjct: 198 GEETNALKEIQSIW----EIKDKDHAAKEKLIIIKEKKNR 233
>At3g31370 hypothetical protein
Length = 272
Score = 50.1 bits (118), Expect = 2e-06
Identities = 58/221 (26%), Positives = 92/221 (41%), Gaps = 25/221 (11%)
Query: 168 NQTSESYWGKIAEYCNEHCSFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWS 227
N+ S WG + + ++ +D V C K+VG+Y + + + SG +
Sbjct: 60 NKKSRRKWGLTEDVVLSNAWLNTSKDHVGCC-----------KFVGSYVATTKEKTSGEN 108
Query: 228 EDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDSV-- 285
++DV A + + TL WR + ++ S NV + S D V
Sbjct: 109 DNDVMNFAHHSFHTDYKTKLTLEHVWREVMFDQKWCSSNALNVPAKQRKQNESVGDHVTV 168
Query: 286 ---GSSARPMDRDAAKKKGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKN 342
G RP+D AAK K K K+K T VE + Q I +KE L+ ++K
Sbjct: 169 DLEGYQTRPIDVKAAKAKAKGKAK--TRLTVEGD-ANNTMTDMQYIWGIKEKNLIFKEKP 225
Query: 343 KLLQEKTQAKKMKMYLKLRDEEHLDD--RKNELLGKLEREL 381
+EK ++K KL +E L+ KNE + +E L
Sbjct: 226 LNGKEKFLSEKN----KLSNERLLESLLTKNESVSDIELAL 262
>At1g66235 putative protein
Length = 265
Score = 49.3 bits (116), Expect = 4e-06
Identities = 56/232 (24%), Positives = 99/232 (42%), Gaps = 37/232 (15%)
Query: 164 VVGRNQTSESYWGKIAE-YCNEHCSFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQ 222
++G Q+S+ +W ++AE + N S R + + R + K K + +
Sbjct: 8 IIGVYQSSDHFWDRVAESFENRKNPTWSKRSKKSLQCRLQTIEKASKKLHACIKLCENRR 67
Query: 223 GSGWSEDDVFKKAQELYGCGKNVQ--------FTLMEEWRALRDQPRYGSQV-------G 267
SG S DD+F +A+E+ K+ + + ++ + +D +V
Sbjct: 68 SSGASSDDIFNQAKEMLMQDKHFKSGWKFDHVWNIIRNFEKFKDGATQAKKVLNLCGLEN 127
Query: 268 GNVDSGSSGSKRSY---------EDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEKEWV 318
+DS S S S+ +D +G S P R KK K K K + + V K
Sbjct: 128 PTLDSVSQASSGSFSFSLNLDDEDDIIGRS--PYQRPIRVKKSKLKRKNDQILDVIK--- 182
Query: 319 QFKELKEQEIEQLKELTLVKQQ------KNKLLQEKTQAKKMKMYLKLRDEE 364
F+E +Q +EQLK+ + +QQ K+ L+E+ + K+ +Y L E
Sbjct: 183 TFEEGNKQLMEQLKKTSAQRQQYLEMQSKSLALREQKEENKV-LYRNLNSIE 233
>At3g31915 hypothetical protein
Length = 239
Score = 45.8 bits (107), Expect = 4e-05
Identities = 49/184 (26%), Positives = 85/184 (45%), Gaps = 16/184 (8%)
Query: 206 KLINKW----VGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPR 261
+L +W +G+Y +A + + SG +++DV + + + +FTL WR +R +
Sbjct: 54 RLRGRWHQIDMGSYVAATKEKTSGQNDNDVLNFSHQSFYTDYKPKFTLEHAWREVRVDQK 113
Query: 262 YGSQVGGNVDSGSSGSKRSYEDSVGSSAR-PMDRDAAKKKGKKKSKGETLEKVEKEWVQF 320
+ S NV + +R +SVG ++ D + G K +KG+ K+ E
Sbjct: 114 WCSSNALNVPA----KRRKQNESVGDHVTIDLEGDQTRPIGVKATKGKAKAKLTLEGDAN 169
Query: 321 KELKE-QEIEQLKELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDD--RKNELLGKL 377
+ E Q I +KE L ++K +EK ++K KLR E L+ KNE + +
Sbjct: 170 NTMTEMQYIWGIKEKDLAFKEKLLNGKEKFLSEKK----KLRKERLLESLLTKNESVFDI 225
Query: 378 EREL 381
E L
Sbjct: 226 ELAL 229
>At3g28770 hypothetical protein
Length = 2081
Score = 45.4 bits (106), Expect = 5e-05
Identities = 37/125 (29%), Positives = 66/125 (52%), Gaps = 13/125 (10%)
Query: 263 GSQVGGNVDSGSSGSKRSYEDSVGSSARPMDRDAAKKKGKKKSKGETLEKVE---KEWVQ 319
G V D G+K +D++ +S++ +D KK KK+SK ++K E KE+V
Sbjct: 911 GESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKK--KKESKNSNMKKKEEDKKEYVN 968
Query: 320 FKELKEQEIEQLKELTLVKQQKNKLLQEKTQAKKMK----MYLKLRDEEHLDDRKNELLG 375
ELK+QE + KE T K + +KL +E K+ K K R+++ +++K++
Sbjct: 969 -NELKKQE-DNKKETT--KSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKE 1024
Query: 376 KLERE 380
+ ++E
Sbjct: 1025 EAKKE 1029
Score = 37.4 bits (85), Expect = 0.014
Identities = 32/113 (28%), Positives = 57/113 (50%), Gaps = 9/113 (7%)
Query: 279 RSYEDSVGSSARPMDRDAAKKKGKK--KSKGETLEKVEKEWVQFKELKEQEIEQLKELTL 336
R E+ + D+++ KKK K K K + ++ V+KE + ++ + +E + KE+
Sbjct: 1111 RKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIES 1170
Query: 337 VKQQKNKL-----LQEKTQAKKMKMYLKLRDEEHLDDRKNELLGKLERELFEN 384
K QKN++ K Q KK + +K +E+ L +KNE K + + EN
Sbjct: 1171 SKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKL--KKNEEDRKKQTSVEEN 1221
Score = 35.0 bits (79), Expect = 0.069
Identities = 27/98 (27%), Positives = 48/98 (48%), Gaps = 2/98 (2%)
Query: 274 SSGSKRSYEDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKE 333
S KR +DS ++ ++ K KKK + ET EK E E + K+ KE + E
Sbjct: 1033 SQDKKREEKDSEERKSKKEKEESRDLKAKKKEE-ETKEKKESENHKSKK-KEDKKEHEDN 1090
Query: 334 LTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDDRKN 371
++ K++ K ++ ++K K +D E L+D+ +
Sbjct: 1091 KSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNS 1128
Score = 33.1 bits (74), Expect = 0.26
Identities = 25/98 (25%), Positives = 46/98 (46%), Gaps = 7/98 (7%)
Query: 277 SKRSYEDSVGSSARPMDRDAAKKKGK--KKSKGETLEKVEKEWVQFKELKEQEIEQLKEL 334
S +S ++ V + +D KKK K K+S+ + L+K E++ K+ +E+ K+
Sbjct: 1170 SSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEED-----RKKQTSVEENKKQ 1224
Query: 335 TLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDDRKNE 372
K++KNK +K K K E + +N+
Sbjct: 1225 KETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQ 1262
Score = 32.7 bits (73), Expect = 0.34
Identities = 27/104 (25%), Positives = 47/104 (44%), Gaps = 9/104 (8%)
Query: 274 SSGSKRSYEDSVGSSARPMDRDAAKKKGKK-----KSKGETLEKVEKEWVQFKELKEQEI 328
S SK E+ + + A+K + KK KSK + K EK+ Q K+ +E++
Sbjct: 984 SENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDS 1043
Query: 329 EQLKELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDDRKNE 372
E+ K K++K + K + K+ + K E H +K +
Sbjct: 1044 EERKS----KKEKEESRDLKAKKKEEETKEKKESENHKSKKKED 1083
Score = 32.3 bits (72), Expect = 0.45
Identities = 23/94 (24%), Positives = 51/94 (53%), Gaps = 3/94 (3%)
Query: 278 KRSYEDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKELT-- 335
K+ EDS + + + K K K+++K E + +K+ + K+ +E++ ++ KE +
Sbjct: 999 KKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREE-KDSEERKSKKEKEESRD 1057
Query: 336 LVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDDR 369
L ++K + +EK +++ K K +EH D++
Sbjct: 1058 LKAKKKEEETKEKKESENHKSKKKEDKKEHEDNK 1091
Score = 30.4 bits (67), Expect = 1.7
Identities = 35/130 (26%), Positives = 58/130 (43%), Gaps = 17/130 (13%)
Query: 271 DSGSSGSKRSYEDSVGSSARPMDRDAAKKK------GKKK--------SKGETLEKVEKE 316
DS SK+ E+S A+ + + +KK KKK +K E+ +KE
Sbjct: 1042 DSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKE 1101
Query: 317 WVQFKELKEQEIEQLKE--LTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDDRKNELL 374
+ +E K ++ E+ K+ L Q NK ++K + KK ++KL +E K E
Sbjct: 1102 KKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNE-KKKSQHVKLVKKESDKKEKKENE 1160
Query: 375 GKLERELFEN 384
K E + E+
Sbjct: 1161 EKSETKEIES 1170
Score = 30.0 bits (66), Expect = 2.2
Identities = 26/101 (25%), Positives = 45/101 (43%), Gaps = 15/101 (14%)
Query: 275 SGSKRSYEDSVGSSARPMDRDAAKK----KGKKKSKGETLEKVEKEWVQFKELKEQEIEQ 330
S K E+ S + ++ ++K K +KKS + +K EKE KE E ++
Sbjct: 1151 SDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEM------KESEEKK 1204
Query: 331 LKELTLVKQQKNKLLQEKTQAKKMKMYLKLRDEEHLDDRKN 371
LK K ++++ Q + K + K + DD+KN
Sbjct: 1205 LK-----KNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKN 1240
>At3g11100 unknown protein
Length = 249
Score = 43.9 bits (102), Expect = 1e-04
Identities = 61/261 (23%), Positives = 98/261 (37%), Gaps = 52/261 (19%)
Query: 124 EVAPTPEDTTPKSKRNQQPSWNTEQNLVLISGWIKYGTCSV-VGRNQTSESYWGKIAEYC 182
E P + T + ++ W+ + LI W G V + R ++ W ++A+
Sbjct: 2 ETTPETQSKTHRLPAGREDWWSEDATATLIEAW---GDRYVNLNRGNLRQNDWKEVADAV 58
Query: 183 NEHCSFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQGSGWSEDD--------VFKK 234
N P+ V C+NR + + K Y + K S W D V KK
Sbjct: 59 NSSHGNGRPKTDVQCKNRIDTLKK-------KYKTEKAKPLSNWCFFDRLDFLIGPVMKK 111
Query: 235 AQELYGCGKNVQFTLMEEWRALRDQPRYGSQVGGNVDSGSSGSK---------------R 279
+ G V+ LM GS + + D + R
Sbjct: 112 SS-----GAVVKSALMNPNLNPTGSKSTGSSLDDDDDDDDDDEEDDDDAGDWGFVVRKHR 166
Query: 280 SYED---SVGSSARPMDRDAAKKKGKKKSKGETLEKVE-KEWVQFKELKEQEIEQLKELT 335
ED S GS+ R + R K GE E++E K+ EL++Q +E KEL
Sbjct: 167 KVEDVDSSEGSAFRELARSILK-------LGEAFERIEGKKQQMMIELEKQRMEVAKELE 219
Query: 336 LVKQQKNKLLQEKTQAKKMKM 356
L Q+ N L++ + + +K K+
Sbjct: 220 L--QRMNMLMEMQLELEKSKL 238
>At1g54060 unknown protein
Length = 383
Score = 42.7 bits (99), Expect = 3e-04
Identities = 41/173 (23%), Positives = 67/173 (38%), Gaps = 13/173 (7%)
Query: 144 WNTEQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPRDIVACRNRFNY 203
W+ E VLI W S G+ + +W ++AE N+ P+ + C+NR +
Sbjct: 94 WSEEATKVLIEAW--GDRFSEPGKGTLKQQHWKEVAEIVNKSRQCKYPKTDIQCKNRIDT 151
Query: 204 MNKLINKWVGAYDSAKRMQGSGWSEDDVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYG 263
+ K + + AK G G S+ FKK + L G T + +A P G
Sbjct: 152 VKKKYKQ-----EKAKIASGDGPSKWVFFKKLESLIG----GTTTFIASSKASEKAPMGG 202
Query: 264 SQVGGNVDSGSSGSKRSYEDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEKE 316
+ GN S + V D+ + +K+S ET + + E
Sbjct: 203 AL--GNSRSSMFKRQTKGNQIVQQQQEKRGSDSMRWHFRKRSASETESESDPE 253
>At5g40340 unknown protein
Length = 1008
Score = 40.4 bits (93), Expect = 0.002
Identities = 44/213 (20%), Positives = 85/213 (39%), Gaps = 15/213 (7%)
Query: 180 EYCNEHCSFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQ----------GSGWSED 229
+Y + S D R+ + F Y + +K++G+YDS+ + + G +
Sbjct: 640 QYPKDSSSRDMVREFMTIYRSFTYHDGANHKFLGSYDSSDKEKEELSEMGKPVTKGKEKK 699
Query: 230 DVFKKAQELYGCGKNVQFTLMEEWRALRDQPRYGSQVGGNVDSGSSGSKRSYEDSVGSSA 289
D KA++ + ++ T EE + + +S G + + +
Sbjct: 700 DKKGKAKQK---AEEIEVTGKEENETDKHGKMKKERKRKKSESKKEGGEGEETQKEANES 756
Query: 290 RPMDRDAAKKKGKKKSKGETLEKVEKEWVQFKELKEQEIEQLKELTLVKQQKNKLLQEKT 349
+R K + KK+S GE + E KE K + E K+ V++++ + ++
Sbjct: 757 TKKERKRKKSESKKQSDGEEETQKEPSESTKKERKRKNPESKKKAEAVEEEETRKESVES 816
Query: 350 QAKKMKMYLKLRDEEHL--DDRKNELLGKLERE 380
K+ K DEE + + K E K +RE
Sbjct: 817 TKKERKRKKPKHDEEEVPNETEKPEKKKKKKRE 849
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,026,856
Number of Sequences: 26719
Number of extensions: 492144
Number of successful extensions: 2820
Number of sequences better than 10.0: 277
Number of HSP's better than 10.0 without gapping: 82
Number of HSP's successfully gapped in prelim test: 195
Number of HSP's that attempted gapping in prelim test: 2320
Number of HSP's gapped (non-prelim): 519
length of query: 384
length of database: 11,318,596
effective HSP length: 101
effective length of query: 283
effective length of database: 8,619,977
effective search space: 2439453491
effective search space used: 2439453491
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149209.7


