

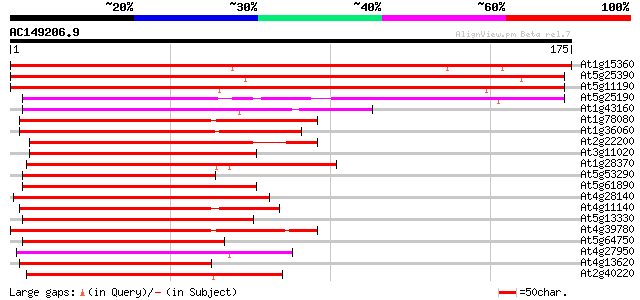
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149206.9 - phase: 0
(175 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g15360 ethylene responsive element like protein 237 3e-63
At5g25390 AP2 domain containing protein 207 3e-54
At5g11190 putative protein 203 3e-53
At5g25190 ethylene-responsive element - like protein 127 4e-30
At1g43160 RAP2.6 (At1g43160) 92 1e-19
At1g78080 putative AP2 domain containing protein (At1g78080) 92 2e-19
At1g36060 putative AP2 domain containing protein RAP2.4 gi|2281633 91 4e-19
At2g22200 pseudogene 89 2e-18
At3g11020 DREB2B transcription factor 88 2e-18
At1g28370 putative ethylene responsive element binding factor 4 ... 88 2e-18
At5g53290 unknown protein 88 3e-18
At5g61890 putative protein 87 3e-18
At4g28140 putative DNA-binding protein 87 3e-18
At4g11140 unknown protein with Ap2 domain 87 3e-18
At5g13330 putative protein 87 6e-18
At4g39780 putative protein 87 6e-18
At5g64750 unknown protein 86 8e-18
At4g27950 unknown protein 86 8e-18
At4g13620 putative protein 86 1e-17
At2g40220 AP2 domain transcription factor (ABI4:abscisic acid-in... 85 2e-17
>At1g15360 ethylene responsive element like protein
Length = 199
Score = 237 bits (604), Expect = 3e-63
Identities = 123/199 (61%), Positives = 144/199 (71%), Gaps = 24/199 (12%)
Query: 1 MVHSKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKT 60
MV +KKFRGVRQRHWGSWV+EIRHPLLKRR+WLGTFETAEEAA+AYDEAA+LMSGRNAKT
Sbjct: 1 MVQTKKFRGVRQRHWGSWVAEIRHPLLKRRIWLGTFETAEEAARAYDEAAVLMSGRNAKT 60
Query: 61 NFPINVEN-----------QTNSISSSSTSSKAFSAVLSAKLRKCCKFPSPSLTCLRLDA 109
NFP+N N +S SSSTSS + S++LSAKLRKCCK PSPSLTCLRLD
Sbjct: 61 NFPLNNNNTGETSEGKTDISASSTMSSSTSSSSLSSILSAKLRKCCKSPSPSLTCLRLDT 120
Query: 110 ENSHIGVWQKGAGPRSESNWIMMVEL-ERKKSASVPEKAKPEEL------------SKNG 156
+SHIGVWQK AG +S+S+W+M VEL S KA + + +
Sbjct: 121 ASSHIGVWQKRAGSKSDSSWVMTVELGPASSSQETTSKASQDAILAPTTEVEIGGSREEV 180
Query: 157 LDDEQKIALQMIEELLNRN 175
LD+E+K+ALQMIEELLN N
Sbjct: 181 LDEEEKVALQMIEELLNTN 199
>At5g25390 AP2 domain containing protein
Length = 189
Score = 207 bits (526), Expect = 3e-54
Identities = 105/177 (59%), Positives = 135/177 (75%), Gaps = 4/177 (2%)
Query: 1 MVHSKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKT 60
MVHSKKFRGVRQR WGSWVSEIRHPLLKRRVWLGTF+TAE AA+AYD+AA+LM+G++AKT
Sbjct: 1 MVHSKKFRGVRQRQWGSWVSEIRHPLLKRRVWLGTFDTAETAARAYDQAAVLMNGQSAKT 60
Query: 61 NFPINVENQTNS--ISSSSTSSKAFSAVLSAKLRKCCKFPSPSLTCLRLDAENSHIGVWQ 118
NFP+ N +NS I+S+ S K+ S +L+AKLRK CK +P LTCLRLD ++SHIGVWQ
Sbjct: 61 NFPVIKSNGSNSLEINSALRSPKSLSELLNAKLRKNCKDQTPYLTCLRLDNDSSHIGVWQ 120
Query: 119 KGAGPRSESNWIMMVELERKKSASVPEKAKPEELSKNGLD--DEQKIALQMIEELLN 173
K AG ++ NW+ +VEL K +A + ++ D ++ ++A+QMIEELLN
Sbjct: 121 KRAGSKTSPNWVKLVELGDKVNARPGGDIETNKMKVRNEDVQEDDQMAMQMIEELLN 177
>At5g11190 putative protein
Length = 189
Score = 203 bits (517), Expect = 3e-53
Identities = 104/187 (55%), Positives = 132/187 (69%), Gaps = 14/187 (7%)
Query: 1 MVHSKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKT 60
MVHS+KFRGVRQR WGSWVSEIRHPLLKRRVWLGTFETAE AA+AYD+AA+LM+G+NAKT
Sbjct: 1 MVHSRKFRGVRQRQWGSWVSEIRHPLLKRRVWLGTFETAEAAARAYDQAALLMNGQNAKT 60
Query: 61 NFPI----NVENQTNSISSSSTSSKAFSAVLSAKLRKCCKFPSPSLTCLRLDAENSHIGV 116
NFP+ + ++S S K+ S +L+AKLRK CK +PSLTCLRLD ++SHIGV
Sbjct: 61 NFPVVKSEEGSDHVKDVNSPLMSPKSLSELLNAKLRKSCKDLTPSLTCLRLDTDSSHIGV 120
Query: 117 WQKGAGPRSESNWIMMVELERKKSASVPEKA----------KPEELSKNGLDDEQKIALQ 166
WQK AG ++ W+M +EL + S + K EE + + DE ++A++
Sbjct: 121 WQKRAGSKTSPTWVMRLELGNVVNESAVDLGLTTMNKQNVEKEEEEEEAIISDEDQLAME 180
Query: 167 MIEELLN 173
MIEELLN
Sbjct: 181 MIEELLN 187
>At5g25190 ethylene-responsive element - like protein
Length = 181
Score = 127 bits (318), Expect = 4e-30
Identities = 78/173 (45%), Positives = 103/173 (59%), Gaps = 16/173 (9%)
Query: 5 KKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPI 64
++FRGVRQRHWGSWVSEIRHPLLK R+WLGTFETAE+AA+AYDEAA LM G A+TNFP
Sbjct: 6 QRFRGVRQRHWGSWVSEIRHPLLKTRIWLGTFETAEDAARAYDEAARLMCGPRARTNFPY 65
Query: 65 NVENQTNSISSSSTSSKAFSAVLSAKLRKCCKFPSPSLTCLRLDAENSHIGVWQKGAGPR 124
N N+I +S SSK SA L+AKL KC + L++ + Q
Sbjct: 66 N----PNAIPTS--SSKLLSATLTAKLHKC------YMASLQMTKQTQTQTQTQTARSQS 113
Query: 125 SESNWIMMVELERKKSASVPEKAKPEE----LSKNGLDDEQKIALQMIEELLN 173
++S+ + E + + + K E+ + +N E+ QMIEELL+
Sbjct: 114 ADSDGVTANESHLNRGVTETTEIKWEDGNANMQQNFRPLEEDHIEQMIEELLH 166
>At1g43160 RAP2.6 (At1g43160)
Length = 192
Score = 92.4 bits (228), Expect = 1e-19
Identities = 54/118 (45%), Positives = 65/118 (54%), Gaps = 11/118 (9%)
Query: 5 KKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPI 64
KK+RGVRQR WG W +EIR P RVWLGTFETAE AA+AYD AA+ G AK NFP
Sbjct: 59 KKYRGVRQRPWGKWAAEIRDPHKATRVWLGTFETAEAAARAYDAAALRFRGSKAKLNFPE 118
Query: 65 NVENQT---------NSISSSSTSSKAFSAVLSAKLRKCCKFPSPSLTCLRLDAENSH 113
NV QT NS+ S T +LS +C + P + L+ E +H
Sbjct: 119 NVGTQTIQRNSHFLQNSMQPSLTYIDQCPTLLS--YSRCMEQQQPLVGMLQPTEEENH 174
>At1g78080 putative AP2 domain containing protein (At1g78080)
Length = 334
Score = 91.7 bits (226), Expect = 2e-19
Identities = 46/93 (49%), Positives = 60/93 (64%), Gaps = 1/93 (1%)
Query: 4 SKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFP 63
+K +RGVRQRHWG WV+EIR P + R+WLGTF+TAEEAA AYD+AA + G A+ NFP
Sbjct: 149 TKLYRGVRQRHWGKWVAEIRLPRNRTRLWLGTFDTAEEAALAYDKAAYKLRGDFARLNFP 208
Query: 64 INVENQTNSISSSSTSSKAFSAVLSAKLRKCCK 96
N+ + + I K + + AKL CK
Sbjct: 209 -NLRHNGSHIGGDFGEYKPLHSSVDAKLEAICK 240
>At1g36060 putative AP2 domain containing protein RAP2.4 gi|2281633
Length = 314
Score = 90.5 bits (223), Expect = 4e-19
Identities = 46/88 (52%), Positives = 58/88 (65%), Gaps = 1/88 (1%)
Query: 4 SKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFP 63
+K +RGVRQRHWG WV+EIR P + R+WLGTF+TAEEAA AYD AA + G +A+ NFP
Sbjct: 140 AKLYRGVRQRHWGKWVAEIRLPRNRTRLWLGTFDTAEEAALAYDRAAFKLRGDSARLNFP 199
Query: 64 INVENQTNSISSSSTSSKAFSAVLSAKL 91
+ QT S S + A + AKL
Sbjct: 200 A-LRYQTGSSPSDTGEYGPIQAAVDAKL 226
>At2g22200 pseudogene
Length = 261
Score = 88.6 bits (218), Expect = 2e-18
Identities = 45/90 (50%), Positives = 56/90 (62%), Gaps = 10/90 (11%)
Query: 7 FRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPINV 66
+RGVRQRHWG WV+EIR P + R+WLGTFETAE+AA AYD+AA + G AK NFP +
Sbjct: 71 YRGVRQRHWGKWVAEIRLPKNRTRLWLGTFETAEKAALAYDQAAFQLRGDIAKLNFPNLI 130
Query: 67 ENQTNSISSSSTSSKAFSAVLSAKLRKCCK 96
N + SS + KL+ CK
Sbjct: 131 HEDMNPLPSS----------VDTKLQAICK 150
>At3g11020 DREB2B transcription factor
Length = 330
Score = 88.2 bits (217), Expect = 2e-18
Identities = 43/71 (60%), Positives = 51/71 (71%)
Query: 7 FRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPINV 66
FRGVRQR WG WV+EIR P + R+WLGTF TAE+AA AYDEAA M G A+ NFP +V
Sbjct: 78 FRGVRQRIWGKWVAEIREPKIGTRLWLGTFPTAEKAASAYDEAATAMYGSLARLNFPQSV 137
Query: 67 ENQTNSISSSS 77
++ S SS S
Sbjct: 138 GSEFTSTSSQS 148
>At1g28370 putative ethylene responsive element binding factor 4
protein
Length = 166
Score = 88.2 bits (217), Expect = 2e-18
Identities = 48/102 (47%), Positives = 62/102 (60%), Gaps = 5/102 (4%)
Query: 6 KFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFP-- 63
++RGVR+R WG + +EIR P K RVWLGTF+T EEAA+AYD+ AI G AKTNFP
Sbjct: 19 RYRGVRKRPWGRYAAEIRDPFKKSRVWLGTFDTPEEAARAYDKRAIEFRGAKAKTNFPCY 78
Query: 64 -INVE--NQTNSISSSSTSSKAFSAVLSAKLRKCCKFPSPSL 102
IN + T S+S SST +F + +FP P +
Sbjct: 79 NINAHCLSLTQSLSQSSTVESSFPNLNLGSDSVSSRFPFPKI 120
>At5g53290 unknown protein
Length = 354
Score = 87.8 bits (216), Expect = 3e-18
Identities = 41/60 (68%), Positives = 46/60 (76%)
Query: 5 KKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPI 64
KKFRGVRQR WG W +EIR P +RR+WLGTFETAEEAA YD AAI + G +A TNF I
Sbjct: 123 KKFRGVRQRPWGKWAAEIRDPEQRRRIWLGTFETAEEAAVVYDNAAIRLRGPDALTNFSI 182
>At5g61890 putative protein
Length = 248
Score = 87.4 bits (215), Expect = 3e-18
Identities = 43/73 (58%), Positives = 49/73 (66%)
Query: 5 KKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPI 64
+ +RGVRQR WG W +EIR P RVWLGTFETAE AA AYDEAA+ G AK NFP
Sbjct: 88 RHYRGVRQRPWGKWAAEIRDPKKAARVWLGTFETAESAALAYDEAALKFKGSKAKLNFPE 147
Query: 65 NVENQTNSISSSS 77
V+ +NS SS
Sbjct: 148 RVQLGSNSTYYSS 160
>At4g28140 putative DNA-binding protein
Length = 292
Score = 87.4 bits (215), Expect = 3e-18
Identities = 39/80 (48%), Positives = 55/80 (68%)
Query: 2 VHSKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTN 61
+ +K +RGVRQR WG WV+EIR P + R+WLGTF+TAEEAA AYD A + G +A N
Sbjct: 138 IATKLYRGVRQRQWGKWVAEIRKPRSRARLWLGTFDTAEEAAMAYDRQAFKLRGHSATLN 197
Query: 62 FPINVENQTNSISSSSTSSK 81
FP + N+ + + S++S +
Sbjct: 198 FPEHFVNKESELHDSNSSDQ 217
>At4g11140 unknown protein with Ap2 domain
Length = 287
Score = 87.4 bits (215), Expect = 3e-18
Identities = 44/81 (54%), Positives = 54/81 (66%), Gaps = 2/81 (2%)
Query: 4 SKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFP 63
++KFRGVRQR WG W +EIR P + RVWLGTF+TAEEAA YD AAI + G NA+ NFP
Sbjct: 84 TRKFRGVRQRPWGKWAAEIRDPSRRVRVWLGTFDTAEEAAIVYDNAAIQLRGPNAELNFP 143
Query: 64 INVENQTNSISSSSTSSKAFS 84
T ++ +ST K S
Sbjct: 144 --PPPVTENVEEASTEVKGVS 162
>At5g13330 putative protein
Length = 212
Score = 86.7 bits (213), Expect = 6e-18
Identities = 41/72 (56%), Positives = 48/72 (65%)
Query: 5 KKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPI 64
+ +RGVRQR WG W +EIR P RVWLGTFETAEEAA AYD AA+ G AK NFP
Sbjct: 37 RHYRGVRQRPWGKWAAEIRDPKKAARVWLGTFETAEEAALAYDRAALKFKGTKAKLNFPE 96
Query: 65 NVENQTNSISSS 76
V+ T + + S
Sbjct: 97 RVQGPTTTTTIS 108
>At4g39780 putative protein
Length = 272
Score = 86.7 bits (213), Expect = 6e-18
Identities = 46/96 (47%), Positives = 60/96 (61%), Gaps = 2/96 (2%)
Query: 1 MVHSKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKT 60
M K +RGVRQRHWG WV+EIR P + R+WLGTF+TAEEAA AYD AA + G A+
Sbjct: 87 MTAQKLYRGVRQRHWGKWVAEIRLPKNRTRLWLGTFDTAEEAAMAYDLAAYKLRGEFARL 146
Query: 61 NFPINVENQTNSISSSSTSSKAFSAVLSAKLRKCCK 96
NFP ++ S + S+V AKL++ C+
Sbjct: 147 NFP-QFRHEDGYYGGGSCFNPLHSSV-DAKLQEICQ 180
>At5g64750 unknown protein
Length = 391
Score = 86.3 bits (212), Expect = 8e-18
Identities = 38/63 (60%), Positives = 46/63 (72%)
Query: 5 KKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFPI 64
+++RGVRQR WG W +EIR P RVWLGTF+ AE AA+AYDEAA+ G AK NFP
Sbjct: 183 RRYRGVRQRPWGKWAAEIRDPFKAARVWLGTFDNAESAARAYDEAALRFRGNKAKLNFPE 242
Query: 65 NVE 67
NV+
Sbjct: 243 NVK 245
>At4g27950 unknown protein
Length = 335
Score = 86.3 bits (212), Expect = 8e-18
Identities = 47/99 (47%), Positives = 58/99 (58%), Gaps = 13/99 (13%)
Query: 3 HSKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNF 62
+ KK+RGVRQR WG W +EIR P +RR+WLGTF TAEEAA YD AAI + G +A TNF
Sbjct: 114 NQKKYRGVRQRPWGKWAAEIRDPEQRRRIWLGTFATAEEAAIVYDNAAIKLRGPDALTNF 173
Query: 63 PINVE-------------NQTNSISSSSTSSKAFSAVLS 88
+ E N + SIS S S+ S+ S
Sbjct: 174 TVQPEPEPVQEQEQEPESNMSVSISESMDDSQHLSSPTS 212
>At4g13620 putative protein
Length = 388
Score = 85.9 bits (211), Expect = 1e-17
Identities = 39/60 (65%), Positives = 46/60 (76%)
Query: 4 SKKFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNFP 63
+K +RGVRQRHWG WV+EIR P + RVWLGTFETAE+AA AYD AA ++ G A NFP
Sbjct: 229 TKLYRGVRQRHWGKWVAEIRLPRNRTRVWLGTFETAEQAAMAYDTAAYILRGEFAHLNFP 288
>At2g40220 AP2 domain transcription factor (ABI4:abscisic
acid-insensitive 4 )
Length = 328
Score = 85.1 bits (209), Expect = 2e-17
Identities = 40/84 (47%), Positives = 59/84 (69%), Gaps = 4/84 (4%)
Query: 6 KFRGVRQRHWGSWVSEIRHPLLKRRVWLGTFETAEEAAKAYDEAAILMSGRNAKTNF--- 62
++RGVRQR WG WV+EIR P + R WLGTF TAE+AA+AYD AA+ + G A+ N
Sbjct: 54 RYRGVRQRSWGKWVAEIREPRKRTRKWLGTFATAEDAARAYDRAAVYLYGSRAQLNLTPS 113
Query: 63 -PINVENQTNSISSSSTSSKAFSA 85
P +V + ++S+S++S+ S + S+
Sbjct: 114 SPSSVSSSSSSVSAASSPSTSSSS 137
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.127 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,901,143
Number of Sequences: 26719
Number of extensions: 148114
Number of successful extensions: 638
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 152
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 436
Number of HSP's gapped (non-prelim): 187
length of query: 175
length of database: 11,318,596
effective HSP length: 93
effective length of query: 82
effective length of database: 8,833,729
effective search space: 724365778
effective search space used: 724365778
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149206.9


