

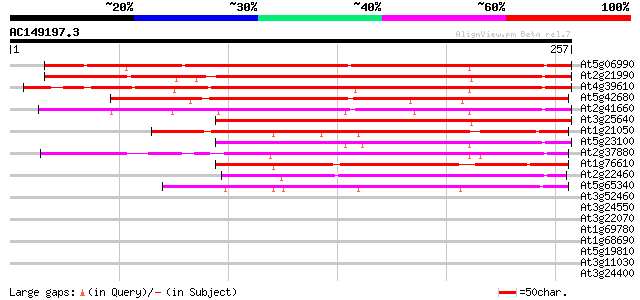
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.3 - phase: 0
(257 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g06990 putative protein 280 7e-76
At2g21990 unknown protein 231 3e-61
At4g39610 unknown protein 226 9e-60
At5g42680 putative protein 161 4e-40
At2g41660 unknown protein 159 2e-39
At3g25640 unknown protein (At3g25640) 144 4e-35
At1g21050 unknown protein 142 2e-34
At5g23100 putative protein 137 4e-33
At2g37880 unknown protein 132 2e-31
At1g76610 hypothetical protein 130 5e-31
At2g22460 unknown protein 124 5e-29
At5g65340 putative protein 113 1e-25
At3g52460 putative protein 36 0.023
At3g24550 protein kinase, putative 35 0.040
At3g22070 unknown protein 35 0.040
At1g69780 homeobox gene 13 protein 35 0.040
At1g68690 protein kinase, putative 35 0.052
At5g19810 proline-rich protein 34 0.067
At3g11030 unknown protein 34 0.067
At3g24400 protein kinase, putative 34 0.088
>At5g06990 putative protein
Length = 261
Score = 280 bits (715), Expect = 7e-76
Identities = 151/254 (59%), Positives = 182/254 (71%), Gaps = 17/254 (6%)
Query: 17 SPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQ--NSTNKLFGKFRSMFRSFPIIVP 74
+PTT P SP S + TVR I L+ + ++K + + KLF + RS+FRS PI+ P
Sbjct: 12 TPTTMSPLGSPKSKKSTA-TVRPEITLEQPSGRNKTTGSKSTKLFRRVRSVFRSLPIMSP 70
Query: 75 SCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGK 134
CK P + G R E +HGG R+TGTLFGYRK RVNLA QE+ + P LLLELAIPTGK
Sbjct: 71 MCKFP-VGGGGRLHENHVHGGTRVTGTLFGYRKTRVNLAVQENPRSLPILLLELAIPTGK 129
Query: 135 LLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQM 194
LLQD+G+GL RIALECEK S +KTKI+DEPIW L+CNGKK GYGVKR PT++DL V+QM
Sbjct: 130 LLQDLGVGLVRIALECEKKPS-EKTKIIDEPIWALYCNGKKSGYGVKRQPTEEDLVVMQM 188
Query: 195 LHSVSVAVGELPSDM-----------SDPQDGELSYMRAHFERVIGSKDSETYYMMMPDG 243
LH+VS+ G LP Q+G+L+YMRAHFERVIGS+DSETYYMM PDG
Sbjct: 189 LHAVSMGAGVLPVSSGAITEQSGGGGGGQQEGDLTYMRAHFERVIGSRDSETYYMMNPDG 248
Query: 244 NSNGPELSVFFVRV 257
NS GPELS+FFVRV
Sbjct: 249 NS-GPELSIFFVRV 261
>At2g21990 unknown protein
Length = 252
Score = 231 bits (589), Expect = 3e-61
Identities = 124/248 (50%), Positives = 169/248 (68%), Gaps = 13/248 (5%)
Query: 17 SPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMFRSFPIIVPS- 75
S T PSP++ + P R + L + + K+ S K+F FRS+FRSFPII P+
Sbjct: 11 SSTDSYSTPSPSASPSPSPAPRQHVTLLEPSHQHKKKS-KKVFRVFRSVFRSFPIITPAA 69
Query: 76 CKMPTMNGN-----HRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAI 130
CK+P + G HR+ + G R+TGTLFGYRK RV+L+ QE +C P L++ELA+
Sbjct: 70 CKIPVLPGGSLPDQHRSGSS----GSRVTGTLFGYRKGRVSLSIQESPRCLPSLVVELAM 125
Query: 131 PTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLY 190
T L +++ G+ RIALE EK +K KI+DEP+WT+F NGKK GYGVKRD T++DL
Sbjct: 126 QTMVLQKELSGGMVRIALETEKRGDKEKIKIMDEPLWTMFSNGKKTGYGVKRDATEEDLN 185
Query: 191 VIQMLHSVSVAVGELPSDMS-DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPE 249
V+++L VS+ G LP + + D E++YMRA+FERV+GSKDSET+YM+ P+GN NGPE
Sbjct: 186 VMELLRPVSMGAGVLPGNTEFEGPDSEMAYMRAYFERVVGSKDSETFYMLSPEGN-NGPE 244
Query: 250 LSVFFVRV 257
LS+FFVRV
Sbjct: 245 LSIFFVRV 252
>At4g39610 unknown protein
Length = 264
Score = 226 bits (576), Expect = 9e-60
Identities = 130/263 (49%), Positives = 174/263 (65%), Gaps = 23/263 (8%)
Query: 7 SQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMF 66
S +++ PP SPT PSP S T P R LQP +SK K+N TN +F R++F
Sbjct: 13 SSSSSSIPPSSPT-----PSPTSTTTPP---RQHPLLQPPSSKKKKNRTN-VFRVLRTVF 63
Query: 67 RSFPIIVP---SCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPF 123
RSFPI +CK+P ++ + H RITGTLFGYRK RV+L+ QE+ KC P
Sbjct: 64 RSFPIFTTPSVACKIPVIHPGLGLPDPH-HNTSRITGTLFGYRKGRVSLSIQENPKCLPS 122
Query: 124 LLLELAIPTGKLLQDMGMGLNRIALECEKHSSND--------KTKIVDEPIWTLFCNGKK 175
L++ELA+ T L +++ G+ RIALE EK D KT I++EP+WT++C G+K
Sbjct: 123 LVVELAMQTTTLQKELSTGMVRIALETEKQPRADNNNSKTEKKTDILEEPLWTMYCKGEK 182
Query: 176 MGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDM-SDPQDGELSYMRAHFERVIGSKDSE 234
GYGVKR+ T++DL V+++L VS+ G LP + S+ DGE++YMRA+FERVIGSKDSE
Sbjct: 183 TGYGVKREATEEDLNVMELLRPVSMGAGVLPGNSESEGPDGEMAYMRAYFERVIGSKDSE 242
Query: 235 TYYMMMPDGNSNGPELSVFFVRV 257
T+YM+ P+GN NGPELS FFVRV
Sbjct: 243 TFYMLSPEGN-NGPELSFFFVRV 264
>At5g42680 putative protein
Length = 238
Score = 161 bits (407), Expect = 4e-40
Identities = 86/214 (40%), Positives = 133/214 (61%), Gaps = 9/214 (4%)
Query: 47 NSKSKQNSTNKLFGKFRSMFRSFPIIVPSCKMPTM-NGNHRTSETIIHGGIRITGTLFGY 105
+SKS + S+ + G MF+ P++ CKM + + HR + TGT+FG+
Sbjct: 28 DSKSSKKSSGSIGGGVLKMFKLIPMLSSGCKMVNLLSRGHRRP---LLKDYATTGTIFGF 84
Query: 106 RKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEP 165
RK RV LA QED C P ++EL + T L ++M RIALE E +S + K+++E
Sbjct: 85 RKGRVFLAIQEDPHCLPIFIIELPMLTSALQKEMASETVRIALESETKTS--RKKVLEEY 142
Query: 166 IWTLFCNGKKMGYGVKR-DPTDDDLYVIQMLHSVSVAVGELP--SDMSDPQDGELSYMRA 222
+W ++CNG+K+GY ++R + +++++YVI L VS+ G LP + +GE++YMRA
Sbjct: 143 VWGIYCNGRKIGYSIRRKNMSEEEMYVIDALRGVSMGAGVLPCKNQYDQETEGEMTYMRA 202
Query: 223 HFERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 256
F+RVIGSKDSE YM+ P+G+ G ELS++F+R
Sbjct: 203 RFDRVIGSKDSEALYMINPEGSGQGTELSIYFLR 236
>At2g41660 unknown protein
Length = 297
Score = 159 bits (401), Expect = 2e-39
Identities = 104/279 (37%), Positives = 148/279 (52%), Gaps = 38/279 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQP---------ANSKSKQNSTNKLFGKFRS 64
P SP SP+S P + L P ++S S ++ K KF
Sbjct: 22 PVTSPARSSHVRSPSSSALIPSIPEHELFLVPCRRCSYVPLSSSSSASHNIGKFHLKFSL 81
Query: 65 MFRSFPIIV--PSCKMPTMNGNHRTSETIIHG------------GIRITGTLFGYRKARV 110
+ RSF I+ P+CKM ++ +S ++ + G R+TGTL+G+++ V
Sbjct: 82 LLRSFINIINIPACKMLSLPSPPSSSSSVSNQLISLVTGGSSSLGRRVTGTLYGHKRGHV 141
Query: 111 NLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEK-HSSNDKTKIVDEPIWTL 169
+ Q + + P LLL+LA+ T L+++M GL RIALECEK H S TK+ EP WT+
Sbjct: 142 TFSVQYNQRSDPVLLLDLAMSTATLVKEMSSGLVRIALECEKRHRSG--TKLFQEPKWTM 199
Query: 170 FCNGKKMGYGVKRDP--TDDDLYVIQMLHSVSVAVGELPSDM---------SDPQDGELS 218
+CNG+K GY V R TD D V+ + V+V G +P+ S + GEL
Sbjct: 200 YCNGRKCGYAVSRGGACTDTDWRVLNTVSRVTVGAGVIPTPKTIDDVSGVGSGTELGELL 259
Query: 219 YMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
YMR FERV+GS+DSE +YMM PD N GPELS+F +R+
Sbjct: 260 YMRGKFERVVGSRDSEAFYMMNPDKN-GGPELSIFLLRI 297
>At3g25640 unknown protein (At3g25640)
Length = 267
Score = 144 bits (364), Expect = 4e-35
Identities = 72/173 (41%), Positives = 108/173 (61%), Gaps = 10/173 (5%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHS 154
G R+ GTLFG R+ V A Q+D P +L++L PT L+++M GL RIALE +
Sbjct: 95 GFRVVGTLFGNRRGHVYFAVQDDPTRLPAVLIQLPTPTSVLVREMASGLVRIALETAAYK 154
Query: 155 SNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMS---- 210
++ K K+++E W +CNGKK GY +++ + + V++ + +++ G LP+ +
Sbjct: 155 TDSKKKLLEESTWRTYCNGKKCGYAARKECGEAEWKVLKAVGPITMGAGVLPATTTTVDE 214
Query: 211 ------DPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
+ GEL YMRA FERV+GS+DSE +YMM PD +S GPELSV+F+RV
Sbjct: 215 EGNGAVGSEKGELMYMRARFERVVGSRDSEAFYMMNPDVSSGGPELSVYFLRV 267
>At1g21050 unknown protein
Length = 245
Score = 142 bits (357), Expect = 2e-34
Identities = 78/195 (40%), Positives = 123/195 (63%), Gaps = 12/195 (6%)
Query: 66 FRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSK-CHPFL 124
+++ + V S + + +H +S ++GT FG+R+ RV+ Q+ + P L
Sbjct: 58 YQNDTLSVSSTSSSSSSSDHSSSS---QSSSTVSGTFFGHRRGRVSFCLQDAAVGSSPLL 114
Query: 125 LLELAIPTGKLLQDMGM-GLNRIALECEKHSSNDK--TKIVDEPIWTLFCNGKKMGYGVK 181
LLELA+PT L ++M G+ RIALEC++ S++ + I D P+W++FCNG+KMG+ V+
Sbjct: 115 LLELAVPTAALAKEMDEEGVLRIALECDRRRSSNSRSSSIFDVPVWSMFCNGRKMGFAVR 174
Query: 182 RDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMP 241
R T++D ++M+ SVSV G +PS+ D ++ Y+RA FERV GS DSE+++MM P
Sbjct: 175 RKVTENDAVFLRMMQSVSVGAGVVPSEEED----QMLYLRARFERVTGSSDSESFHMMNP 230
Query: 242 DGNSNGPELSVFFVR 256
G S G ELS+F +R
Sbjct: 231 -GGSYGQELSIFLLR 244
>At5g23100 putative protein
Length = 277
Score = 137 bits (346), Expect = 4e-33
Identities = 75/192 (39%), Positives = 107/192 (55%), Gaps = 30/192 (15%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEK-- 152
G R+ GTLFG R+ V+ + Q+D P L+ELA P L+++M GL RIALEC+K
Sbjct: 87 GSRVVGTLFGSRRGHVHFSIQKDPNSPPAFLIELATPISGLVKEMASGLVRIALECDKGK 146
Query: 153 -------------HSSNDKTK----------IVDEPIWTLFCNGKKMGYGVKRDPTDDDL 189
H DKTK +V+EP+W +CNGKK G+ +R+ + +
Sbjct: 147 EEEEGEEKNGTLRHGGGDKTKTTTTAAVSRRLVEEPMWRTYCNGKKCGFATRRECGEKEK 206
Query: 190 YVIQMLHSVSVAVGELPSDM----SDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNS 245
V++ L VS+ G LP G++ YMRA FER++GS+DSE +YMM PD N
Sbjct: 207 KVLKALEMVSMGAGVLPETEEIGGGGGGGGDIMYMRAKFERIVGSRDSEAFYMMNPDSN- 265
Query: 246 NGPELSVFFVRV 257
PELS++ +R+
Sbjct: 266 GAPELSIYLLRI 277
>At2g37880 unknown protein
Length = 247
Score = 132 bits (331), Expect = 2e-31
Identities = 85/248 (34%), Positives = 131/248 (52%), Gaps = 27/248 (10%)
Query: 15 PPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSMFRSFPIIVP 74
P +PTT + AS ++R I++Q + S S +S + +
Sbjct: 18 PTNPTTVSTTSTGASKKRLSTSLRDDIDVQDSASSSASSS---------EATSAVDYSIS 68
Query: 75 SCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDS-KCHPFLLLELAIPTG 133
+ +P R S+T++ GT+FG RK V Q D P LLLEL+I T
Sbjct: 69 AVTVP-----QRPSKTMV------IGTIFGRRKGHVWFCVQHDRLSVKPILLLELSIATS 117
Query: 134 KLLQDMGMGLNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQ 193
+L+ +MG GL R+ALEC + P+WT+FCNG+K+G+ V+R ++ +++
Sbjct: 118 QLVHEMGSGLVRVALECPTRPELKSCLLRSVPVWTMFCNGRKLGFAVRRSANEETRMMLK 177
Query: 194 MLHSVSVAVGELPSDM----SDPQD-GELSYMRAHFERVIGSKDSETYYMMMPDGNSNGP 248
L S++V G LPS SD D E+ YMRA++E V+GS DSE+++++ PD NS
Sbjct: 178 RLESMTVGAGVLPSGSGLGGSDESDTDEVMYMRANYEHVVGSSDSESFHLINPDANS-AQ 236
Query: 249 ELSVFFVR 256
ELS+F +R
Sbjct: 237 ELSIFLLR 244
>At1g76610 hypothetical protein
Length = 226
Score = 130 bits (328), Expect = 5e-31
Identities = 67/164 (40%), Positives = 106/164 (63%), Gaps = 13/164 (7%)
Query: 95 GIRITGTLFGYRKARVNLAFQEDSK--CHPFLLLELAIPTGKLLQDMGMGLNRIALECEK 152
G+ +TGT +G+R+ V+ Q+D++ P LLLELA+PT L ++M G RIAL +
Sbjct: 73 GVVVTGTFYGHRRGHVSFCLQDDTRPSSPPLLLLELAVPTAALAREMEEGFLRIAL---R 129
Query: 153 HSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMSDP 212
SN ++ I + P+W+++CNG+K G+ V+R+ T++D+ ++++ SVSV G +P
Sbjct: 130 SKSNRRSSIFNVPVWSMYCNGRKFGFAVRRETTENDVGFLRLMQSVSVGAGVIP------ 183
Query: 213 QDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVR 256
+GE Y+RA FERV GS DSE+++ M+ G G ELS+F R
Sbjct: 184 -NGETLYLRAKFERVTGSSDSESFH-MVNQGGGYGQELSIFLSR 225
>At2g22460 unknown protein
Length = 245
Score = 124 bits (311), Expect = 5e-29
Identities = 67/159 (42%), Positives = 93/159 (58%), Gaps = 3/159 (1%)
Query: 98 ITGTLFGYRKARVNLAFQEDSKCHPF-LLLELAIPTGKLLQDMGMGLNRIALECEKHSSN 156
+TGT+FGYRK ++N Q K LLLELA+PT L ++M G RI LE +
Sbjct: 85 VTGTIFGYRKGKINFCIQTPRKSTNLDLLLELAVPTTVLAREMREGALRIVLE-RNNEKQ 143
Query: 157 DKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQDGE 216
D + +P W ++CNGK++GY KR P+ DD+ + L V V G + D E
Sbjct: 144 DDDSFLSKPFWNMYCNGKRVGYARKRSPSQDDMTALTALSKVVVGAGVVTGKELGRFDDE 203
Query: 217 LSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFV 255
L Y+RA F RV GSK+SE+++++ P GN G ELS+F V
Sbjct: 204 LMYLRASFRRVNGSKESESFHLIDPAGNI-GQELSIFIV 241
>At5g65340 putative protein
Length = 253
Score = 113 bits (282), Expect = 1e-25
Identities = 65/209 (31%), Positives = 116/209 (55%), Gaps = 24/209 (11%)
Query: 71 IIVPSCKMPTMNGNHRTSETIIHGGIR--------------ITGTLFGYRKARVNLAFQE 116
+++PSC ++ N + + H I+ TGT+FG+R+ +VN Q
Sbjct: 42 LLIPSCYCTIVDPNDSQEDKLSHRQIKPRTSSASSTATNSTFTGTIFGFRRGKVNFCIQA 101
Query: 117 DSK--CHPFL-LLELAIPTGKLLQDMGMGLNRIALECEKHSSNDK-----TKIVDEPIWT 168
+ +P + LLEL +PT L ++M G+ RIALE + D + ++ P+W
Sbjct: 102 TNSKTLNPIIVLLELTVPTEVLAREMQGGVLRIALESNNNDGYDSHEDSSSSLLTTPLWN 161
Query: 169 LFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGEL-PSDMSDPQDGELSYMRAHFERV 227
++CNG+K+G+ +KR+P+ +L +++L V+ G + +++ + + Y+RA F+RV
Sbjct: 162 MYCNGRKVGFAIKREPSKSELAALKVLTPVAEGAGVVNGEEINREKSDHMMYLRASFKRV 221
Query: 228 IGSKDSETYYMMMPDGNSNGPELSVFFVR 256
GS DSE+++++ P G G ELS+FF R
Sbjct: 222 FGSFDSESFHLVDPRG-IIGQELSIFFSR 249
>At3g52460 putative protein
Length = 300
Score = 35.8 bits (81), Expect = 0.023
Identities = 17/44 (38%), Positives = 19/44 (42%)
Query: 2 GSLHESQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQP 45
G S+ QPPP P QPPP T PP + P QP
Sbjct: 16 GPGQNSERDINQPPPPPPQSQPPPPQTQQQTYPPVMGYPGYHQP 59
>At3g24550 protein kinase, putative
Length = 652
Score = 35.0 bits (79), Expect = 0.040
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 6/45 (13%)
Query: 7 SQTTT-----GQPPPSPTTGQPPPSPASVNTC-PPTVRMPINLQP 45
S TTT PPP+ T PPPSP++ +T PP+ +P +L P
Sbjct: 22 STTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLPP 66
Score = 32.7 bits (73), Expect = 0.20
Identities = 18/38 (47%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 15 PPSPTTGQPPPSPASVNTCPP------TVRMPINLQPA 46
PPSPTT P SP S N PP T R P N +P+
Sbjct: 90 PPSPTTPSNPRSPPSPNQGPPNTPSGSTPRTPSNTKPS 127
Score = 29.6 bits (65), Expect = 1.7
Identities = 14/31 (45%), Positives = 17/31 (54%), Gaps = 3/31 (9%)
Query: 9 TTTGQPPPSPTTGQ---PPPSPASVNTCPPT 36
TT PPPSP+T PP SP + PP+
Sbjct: 38 TTPSSPPPSPSTNSTSPPPSSPLPPSLPPPS 68
Score = 29.3 bits (64), Expect = 2.2
Identities = 14/37 (37%), Positives = 17/37 (45%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKS 50
PPPSP PP P + P T P P+N +S
Sbjct: 65 PPPSPPGSLTPPLPQPSPSAPITPSPPSPTTPSNPRS 101
Score = 28.1 bits (61), Expect = 4.8
Identities = 16/48 (33%), Positives = 21/48 (43%), Gaps = 3/48 (6%)
Query: 9 TTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTN 56
TT PPSP T +P + PP P + P+ S NST+
Sbjct: 9 TTPSPSPPSPPTNSTTTTPPPAASSPPPTTTPSSPPPSPS---TNSTS 53
Score = 27.7 bits (60), Expect = 6.3
Identities = 18/51 (35%), Positives = 22/51 (42%), Gaps = 4/51 (7%)
Query: 7 SQTTTGQPPPSP-TTGQPPPSPASVNTCP---PTVRMPINLQPANSKSKQN 53
S +T PP SP PPPSP T P P+ PI P + + N
Sbjct: 48 STNSTSPPPSSPLPPSLPPPSPPGSLTPPLPQPSPSAPITPSPPSPTTPSN 98
>At3g22070 unknown protein
Length = 146
Score = 35.0 bits (79), Expect = 0.040
Identities = 15/27 (55%), Positives = 16/27 (58%)
Query: 9 TTTGQPPPSPTTGQPPPSPASVNTCPP 35
TTT PPPS PPP PA V+ PP
Sbjct: 99 TTTAPPPPSTDIPIPPPPPAPVSASPP 125
>At1g69780 homeobox gene 13 protein
Length = 294
Score = 35.0 bits (79), Expect = 0.040
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query: 5 HESQTTTGQPPPSPTTGQP--PPSPASVNTCPPTVRMPINLQPANSKSK-QNSTNKLF 59
++S T G PPP T G+ PPSPA+ T T++ N S K +NS + +F
Sbjct: 218 NDSTLTGGHPPPPQTVGRHFFPPSPATATTTTTTMQFFQNSSSGQSMVKEENSISNMF 275
>At1g68690 protein kinase, putative
Length = 708
Score = 34.7 bits (78), Expect = 0.052
Identities = 27/80 (33%), Positives = 35/80 (43%), Gaps = 3/80 (3%)
Query: 6 ESQTTTGQPPPS--PTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFR 63
E T + PPS PT PPPSP S + P+ P + + + S N F
Sbjct: 178 ERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSPNSPPPTFS 237
Query: 64 SMFRSFP-IIVPSCKMPTMN 82
S RS P I+VP P+ N
Sbjct: 238 SPPRSPPEILVPGSNNPSQN 257
Score = 31.2 bits (69), Expect = 0.57
Identities = 18/55 (32%), Positives = 24/55 (42%), Gaps = 6/55 (10%)
Query: 7 SQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQP------ANSKSKQNST 55
S + PPP P +P P S N+ PPT P P +N+ S+ N T
Sbjct: 206 SDRPSQSPPPPPEDTKPQPPRRSPNSPPPTFSSPPRSPPEILVPGSNNPSQNNPT 260
Score = 29.3 bits (64), Expect = 2.2
Identities = 12/28 (42%), Positives = 15/28 (52%)
Query: 10 TTGQPPPSPTTGQPPPSPASVNTCPPTV 37
T+ PP +P + PP P V T PP V
Sbjct: 36 TSPLPPSAPPPNRAPPPPPPVTTSPPPV 63
Score = 28.5 bits (62), Expect = 3.7
Identities = 15/29 (51%), Positives = 16/29 (54%), Gaps = 2/29 (6%)
Query: 14 PPPS--PTTGQPPPSPASVNTCPPTVRMP 40
PPPS PT PPPSP S + PT P
Sbjct: 157 PPPSDRPTQSPPPPSPPSPPSERPTQSPP 185
>At5g19810 proline-rich protein
Length = 249
Score = 34.3 bits (77), Expect = 0.067
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 5/37 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRM-----PINLQP 45
PPP P PPP P +++ PP V + P+NL P
Sbjct: 71 PPPPPVNLSPPPPPVNLSPPPPPVLLSPPPPPVNLSP 107
Score = 33.9 bits (76), Expect = 0.088
Identities = 15/37 (40%), Positives = 19/37 (50%), Gaps = 5/37 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRM-----PINLQP 45
PPP P PPP P ++ PP V + P+NL P
Sbjct: 80 PPPPPVNLSPPPPPVLLSPPPPPVNLSPPPPPVNLSP 116
Score = 33.5 bits (75), Expect = 0.12
Identities = 14/32 (43%), Positives = 18/32 (55%), Gaps = 4/32 (12%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQP 45
PPP P PPP P +++ PP P+NL P
Sbjct: 62 PPPPPVNLSPPPPPVNLSPPPP----PVNLSP 89
Score = 33.1 bits (74), Expect = 0.15
Identities = 15/37 (40%), Positives = 19/37 (50%), Gaps = 5/37 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRM-----PINLQP 45
PPP P PPP P ++ PP V + P+NL P
Sbjct: 107 PPPPPVNLSPPPPPVLLSPPPPPVLLSPPPPPVNLSP 143
Score = 32.0 bits (71), Expect = 0.33
Identities = 14/37 (37%), Positives = 19/37 (50%), Gaps = 5/37 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRM-----PINLQP 45
PPP P PP P +++ PP V + P+NL P
Sbjct: 44 PPPPPVNISSPPPPVNLSPPPPPVNLSPPPPPVNLSP 80
Score = 32.0 bits (71), Expect = 0.33
Identities = 13/27 (48%), Positives = 14/27 (51%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMP 40
PPP P PPP P + PPTV P
Sbjct: 143 PPPPPVLLSPPPPPVLFSPPPPTVTRP 169
Score = 32.0 bits (71), Expect = 0.33
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 6/41 (14%)
Query: 11 TGQPPPSPTTG-QPPPSPASVNTCPPTVRM-----PINLQP 45
T P P+P PPP P ++++ PP V + P+NL P
Sbjct: 31 TSAPEPAPLVDLSPPPPPVNISSPPPPVNLSPPPPPVNLSP 71
Score = 30.8 bits (68), Expect = 0.75
Identities = 14/37 (37%), Positives = 19/37 (50%), Gaps = 5/37 (13%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRM-----PINLQP 45
PPP P PPP P +++ PP V + P+ L P
Sbjct: 98 PPPPPVNLSPPPPPVNLSPPPPPVLLSPPPPPVLLSP 134
Score = 29.6 bits (65), Expect = 1.7
Identities = 11/24 (45%), Positives = 14/24 (57%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTV 37
PPP P PPP P +++ PP V
Sbjct: 125 PPPPPVLLSPPPPPVNLSPPPPPV 148
Score = 28.9 bits (63), Expect = 2.8
Identities = 11/24 (45%), Positives = 13/24 (53%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTV 37
PPP P PPP P ++ PP V
Sbjct: 134 PPPPPVNLSPPPPPVLLSPPPPPV 157
Score = 28.1 bits (61), Expect = 4.8
Identities = 10/21 (47%), Positives = 12/21 (56%)
Query: 15 PPSPTTGQPPPSPASVNTCPP 35
PP PT +PPP P + PP
Sbjct: 161 PPPPTVTRPPPPPTITRSPPP 181
>At3g11030 unknown protein
Length = 451
Score = 34.3 bits (77), Expect = 0.067
Identities = 16/39 (41%), Positives = 20/39 (51%)
Query: 13 QPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSK 51
+PPP P T PPPSP + PP+ P PA + K
Sbjct: 63 EPPPPPPTSPPPPSPPPPSPPPPSPPPPSPPPPAFAVGK 101
Score = 27.7 bits (60), Expect = 6.3
Identities = 13/26 (50%), Positives = 14/26 (53%), Gaps = 1/26 (3%)
Query: 11 TGQPPPSPTT-GQPPPSPASVNTCPP 35
T PPPSP PPPSP + PP
Sbjct: 70 TSPPPPSPPPPSPPPPSPPPPSPPPP 95
>At3g24400 protein kinase, putative
Length = 694
Score = 33.9 bits (76), Expect = 0.088
Identities = 15/35 (42%), Positives = 16/35 (44%)
Query: 11 TGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQP 45
T PPP P PPP P V PP +P L P
Sbjct: 10 TPSPPPQPLPIPPPPQPLPVTPPPPPTALPPALPP 44
Score = 30.8 bits (68), Expect = 0.75
Identities = 17/51 (33%), Positives = 22/51 (42%)
Query: 4 LHESQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNS 54
L S TT PPPSP+ PP +P+ + P P PA + S
Sbjct: 124 LPPSPTTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRS 174
Score = 30.4 bits (67), Expect = 0.97
Identities = 12/26 (46%), Positives = 14/26 (53%)
Query: 15 PPSPTTGQPPPSPASVNTCPPTVRMP 40
PPSPTT PP +P+ PP P
Sbjct: 84 PPSPTTPSPPLTPSPTTPSPPLTPSP 109
Score = 29.3 bits (64), Expect = 2.2
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 5/41 (12%)
Query: 10 TTGQPP--PSPTTGQPP--PS-PASVNTCPPTVRMPINLQP 45
TT PP PSPTT PP PS P ++ PP P+ P
Sbjct: 88 TTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSPLPPSP 128
Score = 28.9 bits (63), Expect = 2.8
Identities = 13/33 (39%), Positives = 15/33 (45%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQPA 46
PPP P PPP P ++ P P L PA
Sbjct: 22 PPPQPLPVTPPPPPTALPPALPPPPPPTALPPA 54
Score = 28.5 bits (62), Expect = 3.7
Identities = 12/32 (37%), Positives = 16/32 (49%)
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQP 45
PPP P T PP P++ + PP P+ P
Sbjct: 56 PPPPPPTTVPPIPPSTPSPPPPLTPSPLPPSP 87
Score = 28.1 bits (61), Expect = 4.8
Identities = 13/32 (40%), Positives = 15/32 (46%), Gaps = 5/32 (15%)
Query: 14 PPPSPTTG-----QPPPSPASVNTCPPTVRMP 40
PPP P T PPP P +V PP+ P
Sbjct: 43 PPPPPPTALPPALPPPPPPTTVPPIPPSTPSP 74
Score = 27.7 bits (60), Expect = 6.3
Identities = 16/45 (35%), Positives = 21/45 (46%), Gaps = 2/45 (4%)
Query: 3 SLHESQTTTGQPPPSPT--TGQPPPSPASVNTCPPTVRMPINLQP 45
S+ T PP SP + PPPSPA+ +T P + P P
Sbjct: 139 SIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTP 183
Score = 27.7 bits (60), Expect = 6.3
Identities = 11/27 (40%), Positives = 16/27 (58%)
Query: 9 TTTGQPPPSPTTGQPPPSPASVNTCPP 35
+T + PP P+T PPP S++ PP
Sbjct: 169 STPPRSPPPPSTPTPPPRVGSLSPPPP 195
Score = 27.7 bits (60), Expect = 6.3
Identities = 14/27 (51%), Positives = 14/27 (51%), Gaps = 3/27 (11%)
Query: 14 PPPSPTTGQPPPS---PASVNTCPPTV 37
PPPSP T PP P S T PP V
Sbjct: 161 PPPSPATPSTPPRSPPPPSTPTPPPRV 187
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,552,870
Number of Sequences: 26719
Number of extensions: 303715
Number of successful extensions: 3388
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 1809
Number of HSP's gapped (non-prelim): 1106
length of query: 257
length of database: 11,318,596
effective HSP length: 97
effective length of query: 160
effective length of database: 8,726,853
effective search space: 1396296480
effective search space used: 1396296480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149197.3


