

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.10 - phase: 0
(330 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
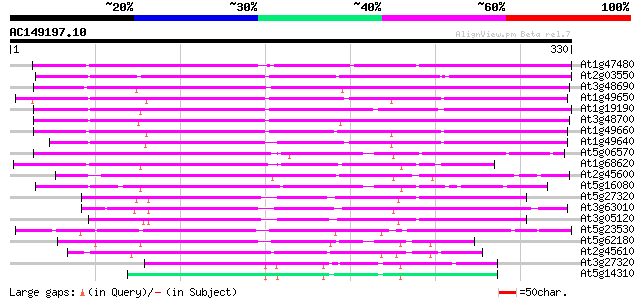
Score E
Sequences producing significant alignments: (bits) Value
At1g47480 unknown protein 247 7e-66
At2g03550 putative esterase 239 1e-63
At3g48690 unknown protein 236 2e-62
At1g49650 unknown protein 236 2e-62
At1g19190 unknown protein 236 2e-62
At3g48700 unknown protein 230 9e-61
At1g49660 unknown protein 227 6e-60
At1g49640 hypothetical protein 226 1e-59
At5g06570 unknown protein 133 2e-31
At1g68620 putative carboxylesterase 129 2e-30
At2g45600 unknown protein 127 1e-29
At5g16080 unknown protein 125 2e-29
At5g27320 Unknown protein 120 1e-27
At3g63010 unknown protein 118 4e-27
At3g05120 unknown protein 115 3e-26
At5g23530 unknown protein 112 4e-25
At5g62180 putative protein 103 2e-22
At2g45610 unknown protein 99 2e-21
At3g27320 esterase, putative 81 9e-16
At5g14310 unknown protein 80 1e-15
>At1g47480 unknown protein
Length = 314
Score = 247 bits (630), Expect = 7e-66
Identities = 130/317 (41%), Positives = 189/317 (59%), Gaps = 11/317 (3%)
Query: 14 KEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPK 73
K++ E+ L + +DG++ER + PP L DP TG+ SKDI I +S+RIY P
Sbjct: 6 KQVSLELLPWLVVHTDGTVERLAGTEVCPPGL-DPITGVFSKDIIIEPKTGLSARIYRPF 64
Query: 74 ITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTC 133
P K P+++YFHGG F+ ST YH L +QANVI VS+ Y LAPE+PLPT
Sbjct: 65 SIQPGQKIPLMLYFHGGAFLISSTSFPSYHTSLNKIVNQANVIAVSVNYRLAPEHPLPTA 124
Query: 134 YHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLP 193
Y D W ALK I + IN EPW+ ++ + + LF+ GDSAGANI+H++A +A +
Sbjct: 125 YEDSWTALKNIQA-----IN--EPWINDYADLDSLFLVGDSAGANISHHLAFRAKQSDQT 177
Query: 194 CDVKILGAIIIHPYFYSANPIGSEPIIEPENNIIHTFWHFAYPNAPFGIDNPRFNPLGEG 253
+K +G +IHPYF+ PIG+E E ++ +W F P+ G D+P NP +G
Sbjct: 178 LKIKGIG--MIHPYFWGTQPIGAEIKDEARKQMVDGWWEFVCPSEK-GSDDPWINPFADG 234
Query: 254 APSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPE 313
+P L LGC R+++ VA KD L ERG Y+E + S WKGK+E E K++ HV+ + +P+
Sbjct: 235 SPDLGGLGCERVMITVAEKDILNERGKMYYERLVKSEWKGKVEIMETKEKDHVFHIFEPD 294
Query: 314 SESAKIFIQRLVGFVQE 330
+ A ++ L F+ +
Sbjct: 295 CDEAMEMVRCLALFINQ 311
>At2g03550 putative esterase
Length = 312
Score = 239 bits (611), Expect = 1e-63
Identities = 129/317 (40%), Positives = 187/317 (58%), Gaps = 11/317 (3%)
Query: 16 IVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLP-KI 74
I + + R++ G IER PPSL P G+ SKDI +S RIYLP K+
Sbjct: 5 IAFDRSPMFRVYKSGRIERLLGETTVPPSLT-PQNGVVSKDIIHSPEKNLSLRIYLPEKV 63
Query: 75 TNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCY 134
T + K PIL+YFHGG F+ E+ FS YH L + + AN + +S+ Y APE+P+P Y
Sbjct: 64 T--VKKLPILIYFHGGGFIIETAFSPPYHTFLTSAVAAANCLAISVNYRRAPEFPVPIPY 121
Query: 135 HDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPC 194
D W +LKW+ +H PE W+ +HG+F K+F+ GDSAG NI+H++ ++A E L C
Sbjct: 122 EDSWDSLKWVLTHITG--TGPETWINKHGDFGKVFLAGDSAGGNISHHLTMRAKKEKL-C 178
Query: 195 DVKILGAIIIHPYFYSANPIGSEPIIE-PENNIIHTFWHFAYPNAPFGIDNPRFNPLGEG 253
D I G I+IHPYF+S PI + + + + W A PN+ G+D+P N +G
Sbjct: 179 DSLISGIILIHPYFWSKTPIDEFEVRDVGKTKGVEGSWRVASPNSKQGVDDPWLNVVGSD 238
Query: 254 APSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPE 313
PS LGC R++V VAG D +G Y E +K SGW+G++E E K+EGHV+ L P
Sbjct: 239 -PS--GLGCGRVLVMVAGDDLFVRQGWCYAEKLKKSGWEGEVEVMETKNEGHVFHLKNPN 295
Query: 314 SESAKIFIQRLVGFVQE 330
S++A+ +++L F+ +
Sbjct: 296 SDNARQVVKKLEEFINK 312
>At3g48690 unknown protein
Length = 324
Score = 236 bits (601), Expect = 2e-62
Identities = 125/321 (38%), Positives = 186/321 (57%), Gaps = 9/321 (2%)
Query: 15 EIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPK- 73
EI + +L+I+ G IER PPS ++P G+ SKD+ + +S RIYLP+
Sbjct: 4 EIAVDCSPLLKIYKSGRIERLMGEATVPPS-SEPQNGVVSKDVVYSADNNLSVRIYLPEK 62
Query: 74 -ITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPT 132
SK P+LVYFHGG F+ E+ FS YH L T S +N + VS++Y APE+P+
Sbjct: 63 AAAETDSKLPLLVYFHGGGFIIETAFSPTYHTFLTTSVSASNCVAVSVDYRRAPEHPISV 122
Query: 133 CYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENL 192
+ D W ALKW+ +H + E WL +H +F+++F+ GDSAGANI H++A++A E L
Sbjct: 123 PFDDSWTALKWVFTHITGS--GQEDWLNKHADFSRVFLSGDSAGANIVHHMAMRAAKEKL 180
Query: 193 PC---DVKILGAIIIHPYFYSANPIGSEPIIEPENNI-IHTFWHFAYPNAPFGIDNPRFN 248
D I G I++HPYF+S PI + + + I FW A PN+ G D+P N
Sbjct: 181 SPGLNDTGISGIILLHPYFWSKTPIDEKDTKDETLRMKIEAFWMMASPNSKDGTDDPLLN 240
Query: 249 PLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQ 308
+ + L LGC +++V VA KD L +G Y ++ SGWKG++E E + E HV+
Sbjct: 241 VVQSESVDLSGLGCGKVLVMVAEKDALVRQGWGYAAKLEKSGWKGEVEVVESEGEDHVFH 300
Query: 309 LVKPESESAKIFIQRLVGFVQ 329
L+KPE ++A + + GF++
Sbjct: 301 LLKPECDNAIEVMHKFSGFIK 321
>At1g49650 unknown protein
Length = 374
Score = 236 bits (601), Expect = 2e-62
Identities = 125/334 (37%), Positives = 197/334 (58%), Gaps = 15/334 (4%)
Query: 4 TTPTTPKTI----TKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQI 59
+TPTT + I + EI++E +R++ DG IER + P SLN P + SKD+
Sbjct: 45 STPTTLRCICSHSSSEIISEHPPFVRVYKDGRIERLSGTETVPASLN-PRNDVVSKDVVY 103
Query: 60 PHNPTISSRIYLPKITNPLS---KFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVI 116
+S R++LP + L+ K P+L+YFHGG ++ ES FS YH L AN +
Sbjct: 104 SPGHNLSVRLFLPHKSTQLAAGNKLPLLIYFHGGAWINESPFSPIYHNFLTEVVKSANCL 163
Query: 117 IVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAG 176
VS++Y APE P+P Y D W+A++WI SHS + E W+ ++ +F ++F+ GDSAG
Sbjct: 164 AVSVQYRRAPEDPVPAAYEDTWSAIQWIFSHSCG--SGEEDWINKYADFERVFLAGDSAG 221
Query: 177 ANIAHNIAIQAGLENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPE--NNIIHTFWHFA 234
NI+H++A++AG E L +K G +I+HP + +P+ + + E + + +
Sbjct: 222 GNISHHMAMRAGKEKLKPRIK--GTVIVHPAIWGKDPVDEHDVQDREIRDGVAEVWEKIV 279
Query: 235 YPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGK 294
PN+ G D+P FN +G G+ + +GC +++V VAGKD +G+ Y +K SGWKG+
Sbjct: 280 SPNSVDGADDPWFNVVGSGS-NFSGMGCDKVLVEVAGKDVFWRQGLAYAAKLKKSGWKGE 338
Query: 295 LEFFEEKDEGHVYQLVKPESESAKIFIQRLVGFV 328
+E EE+DE H + L+ P SE+A F++R V F+
Sbjct: 339 VEVIEEEDEEHCFHLLNPSSENAPSFMKRFVEFI 372
>At1g19190 unknown protein
Length = 318
Score = 236 bits (601), Expect = 2e-62
Identities = 133/320 (41%), Positives = 183/320 (56%), Gaps = 12/320 (3%)
Query: 15 EIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKI 74
EI + RIF +G IER F PPSLN P G+ SKD +S RIYLP+
Sbjct: 4 EIAFDYSPRFRIFKNGGIERLVPETFVPPSLN-PENGVVSKDAVYSPEKNLSLRIYLPQN 62
Query: 75 T---NPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLP 131
+ K P+LVYFHGG F+ E+ FS YH L + S + I VS+EY APE+P+P
Sbjct: 63 SVYETGEKKIPLLVYFHGGGFIMETAFSPIYHTFLTSAVSATDCIAVSVEYRRAPEHPIP 122
Query: 132 TCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLEN 191
T Y D W A++WI +H + PE WL +H +F+K+F+ GDSAGANIAH++AI+ E
Sbjct: 123 TLYEDSWDAIQWIFTHITR--SGPEDWLNKHADFSKVFLAGDSAGANIAHHMAIRVDKEK 180
Query: 192 LPCD-VKILGAIIIHPYFYSANPIGSEPIIEPENNIIHTFWHFAYPNAPFGIDNPRFNPL 250
LP + KI G I+ HPYF S I E + W A P++ G+++P N +
Sbjct: 181 LPPENFKISGMILFHPYFLSKALI--EEMEVEAMRYYERLWRIASPDSGNGVEDPWINVV 238
Query: 251 GEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLV 310
G L LGC R++V VAG D L G Y ++ SGW GK++ E K+EGHV+ L
Sbjct: 239 GS---DLTGLGCRRVLVMVAGNDVLARGGWSYVAELEKSGWIGKVKVMETKEEGHVFHLR 295
Query: 311 KPESESAKIFIQRLVGFVQE 330
P+SE+A+ ++ F++E
Sbjct: 296 DPDSENARRVLRNFAEFLKE 315
>At3g48700 unknown protein
Length = 329
Score = 230 bits (586), Expect = 9e-61
Identities = 127/326 (38%), Positives = 180/326 (54%), Gaps = 14/326 (4%)
Query: 15 EIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKI 74
EI + +L I+ G IER PPS N P G+ SKD+ + +S RIYLP+
Sbjct: 4 EIAADYSPMLIIYKSGRIERLVGETTVPPSSN-PQNGVVSKDVVYSPDNNLSLRIYLPEK 62
Query: 75 -----TNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYP 129
T K P+LVYFHGG F+ E+ FS YH L S ++ + VS++Y APE+P
Sbjct: 63 AATAETEASVKLPLLVYFHGGGFLVETAFSPTYHTFLTAAVSASDCVAVSVDYRRAPEHP 122
Query: 130 LPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGL 189
+PT Y D W ALKW+ SH + E WL +H +F+K+F+ GDSAGANI H++ ++A
Sbjct: 123 IPTSYDDSWTALKWVFSHIAG--SGSEDWLNKHADFSKVFLAGDSAGANITHHMTMKAAK 180
Query: 190 ENLP----CDVKILGAIIIHPYFYSANPIGSEPIIEPE-NNIIHTFWHFAYPNAPFGIDN 244
+ L + I G I++HPYF+S P+ + + I + W A PN+ G D+
Sbjct: 181 DKLSPESLNESGISGIILVHPYFWSKTPVDDKETTDVAIRTWIESVWTLASPNSKDGSDD 240
Query: 245 PRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGK-LEFFEEKDE 303
P N + + L LGC +++V VA KD L +G YWE + S W G+ L+ E K E
Sbjct: 241 PFINVVQSESVDLSGLGCGKVLVMVAEKDALVRQGWGYWEKLGKSRWNGEVLDVVETKGE 300
Query: 304 GHVYQLVKPESESAKIFIQRLVGFVQ 329
GHV+ L P SE A + R GF++
Sbjct: 301 GHVFHLRDPNSEKAHELVHRFAGFIK 326
>At1g49660 unknown protein
Length = 319
Score = 227 bits (579), Expect = 6e-60
Identities = 120/319 (37%), Positives = 185/319 (57%), Gaps = 10/319 (3%)
Query: 15 EIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKI 74
EI +E RI+ DG +ER + P SL DP + SKD+ +S R++LP
Sbjct: 4 EIASEFLPFCRIYKDGRVERLIGTDTIPASL-DPTYDVVSKDVIYSPENNLSVRLFLPHK 62
Query: 75 TNPLS---KFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLP 131
+ L+ K P+L+Y HGG ++ ES FS YH +L AN + VS++Y APE P+P
Sbjct: 63 STKLTAGNKLPLLIYIHGGAWIIESPFSPLYHNYLTEVVKSANCLAVSVQYRRAPEDPVP 122
Query: 132 TCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLEN 191
Y D W+A++WI +HSN + P W+ +H +F K+F+GGDSAG NI+H++A++AG E
Sbjct: 123 AAYEDVWSAIQWIFAHSNG--SGPVDWINKHADFGKVFLGGDSAGGNISHHMAMKAGKEK 180
Query: 192 LPCDVKILGAIIIHPYFYSANPIGSEPIIEPE--NNIIHTFWHFAYPNAPFGIDNPRFNP 249
D+KI G ++HP F+ +P+ + + E + I + A PN+ G D+P FN
Sbjct: 181 -KLDLKIKGIAVVHPAFWGTDPVDEYDVQDKETRSGIAEIWEKIASPNSVNGTDDPLFNV 239
Query: 250 LGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQL 309
G G+ LGC +++V VAGKD +G+ Y ++ W+G +E EE+ E HV+ L
Sbjct: 240 NGSGS-DFSGLGCDKVLVAVAGKDVFVRQGLAYAAKLEKCEWEGTVEVVEEEGEDHVFHL 298
Query: 310 VKPESESAKIFIQRLVGFV 328
P+S+ A F+++ V F+
Sbjct: 299 QNPKSDKALKFLKKFVEFI 317
>At1g49640 hypothetical protein
Length = 315
Score = 226 bits (577), Expect = 1e-59
Identities = 114/311 (36%), Positives = 188/311 (59%), Gaps = 17/311 (5%)
Query: 24 LRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKITNPL----S 79
+RI +G +ER + P SLN P + SKD+ + +S R++LP + L +
Sbjct: 14 IRIHKNGRVERLSGNDIKPTSLN-PQNDVVSKDVMYSSDHNLSVRMFLPNKSRKLDTAGN 72
Query: 80 KFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWA 139
K P+L+YFHGG ++ +S FS YH +L AN + VS++Y LAPE+P+P Y D W+
Sbjct: 73 KIPLLIYFHGGAYIIQSPFSPVYHNYLTEVVITANCLAVSVQYRLAPEHPVPAAYDDSWS 132
Query: 140 ALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKIL 199
A++WI SHS++ W+ E+ +F+++FI GDSAGANI+H++ I+AG E L +K
Sbjct: 133 AIQWIFSHSDD-------WINEYADFDRVFIAGDSAGANISHHMGIRAGKEKLSPTIK-- 183
Query: 200 GAIIIHPYFYSANPIGSEPIIEPE--NNIIHTFWHFAYPNAPFGIDNPRFNPLGEGAPSL 257
G +++HP F+ PI + + E N I + + + PN+ G+++P FN +G G+ +
Sbjct: 184 GIVMVHPGFWGKEPIDEHDVQDGEVRNKIAYIWENIVSPNSVDGVNDPWFNVVGSGS-DV 242
Query: 258 EKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPESESA 317
++GC +++V VAGKD +G+ Y ++ S WKG +E EE++EGH + L S++A
Sbjct: 243 SEMGCEKVLVAVAGKDVFWRQGLAYAAKLEKSQWKGSVEVIEEEEEGHCFHLHNHNSQNA 302
Query: 318 KIFIQRLVGFV 328
+Q+ + F+
Sbjct: 303 SKLMQKFLEFI 313
>At5g06570 unknown protein
Length = 329
Score = 133 bits (334), Expect = 2e-31
Identities = 91/327 (27%), Positives = 164/327 (49%), Gaps = 28/327 (8%)
Query: 15 EIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPT-ISSRIYLPK 73
++ + +L++ S+G++ R + + N I H P + R+Y P
Sbjct: 9 QVAEDCMGLLQLLSNGTVLRSESIDLITQQIPFKNNQTVLFKDSIYHKPNNLHLRLYKPI 68
Query: 74 ITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTC 133
+ + P++V+FHGG F F S +H T AS N ++VS +Y LAPE+ LP
Sbjct: 69 SASNRTALPVVVFFHGGGFCFGSRSWPHFHNFCLTLASSLNALVVSPDYRLAPEHRLPAA 128
Query: 134 YHDCWAALKWISSHS-NNNINNPEPWLIEHG---NFNKLFIGGDSAGANIAHNIAIQAGL 189
+ D A L W+ + ++ +N+ W E G +F+++F+ GDS+G NIAH +A++ G
Sbjct: 129 FEDAEAVLTWLWDQAVSDGVNH---W-FEDGTDVDFDRVFVVGDSSGGNIAHQLAVRFGS 184
Query: 190 ENLP-CDVKILGAIIIHPYFYSANPIGSEPIIEPEN---------NIIHTFWHFAYPNAP 239
++ V++ G +++ P+F G E EN +++ FW + PN
Sbjct: 185 GSIELTPVRVRGYVLMGPFF------GGEERTNSENGPSEALLSLDLLDKFWRLSLPNGA 238
Query: 240 FGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFE 299
D+ NP G +P+LE + ++V V G + LR+R Y +K G K ++++ E
Sbjct: 239 TR-DHHMANPFGPTSPTLESISLEPMLVIVGGSELLRDRAKEYAYKLKKMGGK-RVDYIE 296
Query: 300 EKDEGHVYQLVKPESESAKIFIQRLVG 326
+++ H + P SE+A+ + R++G
Sbjct: 297 FENKEHGFYSNYPSSEAAE-QVLRIIG 322
>At1g68620 putative carboxylesterase
Length = 336
Score = 129 bits (324), Expect = 2e-30
Identities = 86/292 (29%), Positives = 147/292 (49%), Gaps = 22/292 (7%)
Query: 3 STTPTTPKTITKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHN 62
+TT I +V E+ +++++ DG +ER + P PSL G++ D+ I
Sbjct: 10 TTTNPNNSNIHGPVVDEVEGLIKVYKDGHVERSQLLPCVDPSL-PLELGVTCSDVVIDKL 68
Query: 63 PTISSRIYLPKIT--NPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSI 120
+ +R+Y+P T + +SK P++VYFHGG F S YHE L ++++ +++S+
Sbjct: 69 TNVWARLYVPMTTTKSSVSKLPLIVYFHGGGFCVGSASWLCYHEFLARLSARSRCLVMSV 128
Query: 121 EYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIA 180
Y LAPE PLP Y D A+ W++ N+N+ W + +F ++F+ GDSAG NIA
Sbjct: 129 NYRLAPENPLPAAYEDGVNAILWLNKARNDNL-----W-AKQCDFGRIFLAGDSAGGNIA 182
Query: 181 HNIAIQ-AGLENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPENNIIHT------FWHF 233
+A + A E+L +KI G I+I P++ SE + + + T +W
Sbjct: 183 QQVAARLASPEDLA--LKIEGTILIQPFYSGEERTESERRVGNDKTAVLTLASSDAWWRM 240
Query: 234 AYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEG 285
+ P ++P P+ ++ +R +VCVA D L + + +G
Sbjct: 241 SLPRGA-NREHPYCKPV---KMIIKSSTVTRTLVCVAEMDLLMDSNMEMCDG 288
>At2g45600 unknown protein
Length = 329
Score = 127 bits (318), Expect = 1e-29
Identities = 96/312 (30%), Positives = 146/312 (46%), Gaps = 25/312 (8%)
Query: 28 SDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKITNPLSKFPILVYF 87
SDGS+ R + P PP+ SKDI + RI+ P+ P SK PILVYF
Sbjct: 20 SDGSLTRHRDFPKLPPTEQ-------SKDIPLNQTNNTFIRIFKPRNIPPESKLPILVYF 72
Query: 88 HGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSH 147
HGG F+ S S +HE A + II+S+EY LAPE+ LP Y D A+ W+
Sbjct: 73 HGGGFILYSAASAPFHESCTKMADRLQTIILSVEYRLAPEHRLPAAYEDAVEAILWLRDQ 132
Query: 148 SNNNIN--NPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIH 205
+ IN + + WL + +F+K ++ G S+G NI +N+A++ +L VKI G I+
Sbjct: 133 ARGPINGGDCDTWLKDGVDFSKCYVMGSSSGGNIVYNVALRVVDTDL-SPVKIQGLIMNQ 191
Query: 206 PYFYSANPIGSEPIIEPEN----NIIHTFWHFAYPNAPFGIDNPRF--NPLGEGAP-SLE 258
+F P SE ++ + H W P+ G+D NP+ P +
Sbjct: 192 AFFGGVEPSDSESRLKDDKICPLPATHLLWSLCLPD---GVDRDHVYSNPIKSSGPQEKD 248
Query: 259 KLG-CSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPESESA 317
K+G ++ G D L +R E +K G + F +KD H +L + A
Sbjct: 249 KMGRFPSTLINGYGGDPLVDRQRHVAEMLKGRGVHVETRF--DKDGFHACELF--DGNKA 304
Query: 318 KIFIQRLVGFVQ 329
K + + F++
Sbjct: 305 KALYETVEAFMK 316
>At5g16080 unknown protein
Length = 344
Score = 125 bits (315), Expect = 2e-29
Identities = 93/316 (29%), Positives = 155/316 (48%), Gaps = 32/316 (10%)
Query: 16 IVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKIT 75
+V E+ ++++F+DG +ERP P P+++ P++ ++ DI++ ++ +R+Y+P
Sbjct: 28 VVEEIEGLIKVFNDGCVERPPIVPIVSPTIH-PSSKATAFDIKLSNDTW--TRVYIPDAA 84
Query: 76 --NPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTC 133
+P P+LVYFHGG F S YH+ L + A +A +IVS+ Y LAPE+ LP
Sbjct: 85 AASPSVTLPLLVYFHGGGFCVGSAAWSCYHDFLTSLAVKARCVIVSVNYRLAPEHRLPAA 144
Query: 134 YHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLP 193
Y D + W+ + WL N + +F+ GDSAGANIA+ +A++
Sbjct: 145 YDDGVNVVSWLVKQQISTGGGYPSWL-SKCNLSNVFLAGDSAGANIAYQVAVRIMASGKY 203
Query: 194 CD-VKILGAIIIHPYFYSANPIGSEPIIEPENNIIHT------------FWHFAYPNAPF 240
+ + + G I+IHP+F G E E HT +W A P
Sbjct: 204 ANTLHLKGIILIHPFF------GGESRTSSEKQQHHTKSSALTLSASDAYWRLALPRGA- 256
Query: 241 GIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEE 300
D+P NPL A + KL + +V +A D L+ER + + +++ G ++E
Sbjct: 257 SRDHPWCNPLMSSAGA--KLPTT--MVFMAEFDILKERNLEMCKVMRSHG--KRVEGIVH 310
Query: 301 KDEGHVYQLVKPESES 316
GH + ++ S S
Sbjct: 311 GGVGHAFHILDNSSVS 326
>At5g27320 Unknown protein
Length = 344
Score = 120 bits (300), Expect = 1e-27
Identities = 81/279 (29%), Positives = 132/279 (47%), Gaps = 31/279 (11%)
Query: 43 PSLNDPNTGISSKDIQIPHNPTISSRIYLPK----------ITNPLSK--FPILVYFHGG 90
P+ +P G+ S D+ I + SR+Y P + NP+ P++V+FHGG
Sbjct: 54 PANANPVNGVFSFDVIIDRQTNLLSRVYRPADAGTSPSITDLQNPVDGEIVPVIVFFHGG 113
Query: 91 VFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNN 150
F S S Y + ++VS+ Y APE P Y D WA LKW++S S
Sbjct: 114 SFAHSSANSAIYDTLCRRLVGLCGAVVVSVNYRRAPENRYPCAYDDGWAVLKWVNSSS-- 171
Query: 151 NINNPEPWLIEHGNFN-KLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHPYFY 209
WL + ++F+ GDS+G NI HN+A++A + + +LG I+++P F
Sbjct: 172 -------WLRSKKDSKVRIFLAGDSSGGNIVHNVAVRA----VESRIDVLGNILLNPMFG 220
Query: 210 SANPIGSEPIIEPENNII----HTFWHFAYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRI 265
SE ++ + + +W P ++P +P G + SLE L +
Sbjct: 221 GTERTESEKRLDGKYFVTVRDRDWYWRAFLPEGE-DREHPACSPFGPRSKSLEGLSFPKS 279
Query: 266 IVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEG 304
+V VAG D +++ + Y EG+K +G + KL + E+ G
Sbjct: 280 LVVVAGLDLIQDWQLKYAEGLKKAGQEVKLLYLEQATIG 318
>At3g63010 unknown protein
Length = 358
Score = 118 bits (296), Expect = 4e-27
Identities = 88/305 (28%), Positives = 145/305 (46%), Gaps = 36/305 (11%)
Query: 43 PSLNDPNTGISSKDIQIPHNPTISSRIYLP------------KITNPLSK---FPILVYF 87
P+ + P G+ S D + + +RIY P ++T PLS P+L++F
Sbjct: 54 PANSFPLDGVFSFD-HVDSTTNLLTRIYQPASLLHQTRHGTLELTKPLSTTEIVPVLIFF 112
Query: 88 HGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSH 147
HGG F S S Y + + V++VS++Y +PE+ P Y D W AL W+ S
Sbjct: 113 HGGSFTHSSANSAIYDTFCRRLVTICGVVVVSVDYRRSPEHRYPCAYDDGWNALNWVKS- 171
Query: 148 SNNNINNPEPWLIEHGNFN-KLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHP 206
WL + N +++ GDS+G NIAHN+A++A E VK+LG I++HP
Sbjct: 172 --------RVWLQSGKDSNVYVYLAGDSSGGNIAHNVAVRATNEG----VKVLGNILLHP 219
Query: 207 YFYSANPIGSEPIIEPEN--NIIHTFWHF-AYPNAPFGIDNPRFNPLGEGAPSLEKLGCS 263
F SE ++ + I W++ AY D+P NP G SL+ +
Sbjct: 220 MFGGQERTQSEKTLDGKYFVTIQDRDWYWRAYLPEGEDRDHPACNPFGPRGQSLKGVNFP 279
Query: 264 RIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPESESAKIFIQR 323
+ +V VAG D +++ + Y +G+K +G + L + ++ G + P ++ ++
Sbjct: 280 KSLVVVAGLDLVQDWQLAYVDGLKKTGLEVNLLYLKQATIGFYF---LPNNDHFHCLMEE 336
Query: 324 LVGFV 328
L FV
Sbjct: 337 LNKFV 341
>At3g05120 unknown protein
Length = 345
Score = 115 bits (288), Expect = 3e-26
Identities = 81/277 (29%), Positives = 126/277 (45%), Gaps = 33/277 (11%)
Query: 47 DPNTGISSKDIQIPHNPTISSRIYLPKITNP--------LSK------FPILVYFHGGVF 92
+P G+ S D+ I + SR+Y P + L K P++++FHGG F
Sbjct: 58 NPVDGVFSFDVLIDRRINLLSRVYRPAYADQEQPPSILDLEKPVDGDIVPVILFFHGGSF 117
Query: 93 MFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNNNI 152
S S Y + ++VS+ Y APE P P Y D W AL W++S S
Sbjct: 118 AHSSANSAIYDTLCRRLVGLCKCVVVSVNYRRAPENPYPCAYDDGWIALNWVNSRS---- 173
Query: 153 NNPEPWLIEHGNFN-KLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHPYFYSA 211
WL + +F+ GDS+G NIAHN+A++AG + +LG I+++P F
Sbjct: 174 -----WLKSKKDSKVHIFLAGDSSGGNIAHNVALRAG----ESGIDVLGNILLNPMFGGN 224
Query: 212 NPIGSEPIIEPENNII----HTFWHFAYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIV 267
SE ++ + + +W P ++P NP SLE + + +V
Sbjct: 225 ERTESEKSLDGKYFVTVRDRDWYWKAFLPEGE-DREHPACNPFSPRGKSLEGVSFPKSLV 283
Query: 268 CVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEG 304
VAG D +R+ + Y EG+K +G + KL E+ G
Sbjct: 284 VVAGLDLIRDWQLAYAEGLKKAGQEVKLMHLEKATVG 320
>At5g23530 unknown protein
Length = 335
Score = 112 bits (279), Expect = 4e-25
Identities = 101/338 (29%), Positives = 150/338 (43%), Gaps = 32/338 (9%)
Query: 4 TTPTTPKTITKEIVTEMGNILRIFSDGSIERPKQSPF---APPSLNDPNTGISSKDIQIP 60
T P + I T N R DG+I R F APP+ N +S+ D +
Sbjct: 12 TLPLKTRIALTVISTMTDNAQR--PDGTINRRFLRLFDFRAPPNPKPVNI-VSTSDFVVD 68
Query: 61 HNPTISSRIYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSI 120
+ + R+Y P ++ K P++V+FHGG F F S + Y + FA + ++S+
Sbjct: 69 QSRDLWFRLYTPHVSG--DKIPVVVFFHGGGFAFLSPNAYPYDNVCRRFARKLPAYVISV 126
Query: 121 EYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIA 180
Y LAPE+ P Y D + ALK+I N L + + ++ F GDSAG NIA
Sbjct: 127 NYRLAPEHRYPAQYDDGFDALKYIE-------ENHGSILPANADLSRCFFAGDSAGGNIA 179
Query: 181 HNIAIQAGLE--NLPCDVKILGAIIIHPYFYSANPIGSE------PIIEPENNIIHTFWH 232
HN+AI+ E + VK++G I I P+F +E P++ P+ T W
Sbjct: 180 HNVAIRICREPRSSFTAVKLIGLISIQPFFGGEERTEAEKQLVGAPLVSPD----RTDW- 234
Query: 233 FAYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWK 292
+ D+ N G A + L +V VAG D L++ Y+E +K G K
Sbjct: 235 -CWKAMGLNRDHEAVNVGGPNAVDISGLDYPETMVVVAGFDPLKDWQRSYYEWLKLCGKK 293
Query: 293 GKLEFFEEKDEGHVYQLVKPESESAKIFIQRLVGFVQE 330
L E + H + + PE A I R+ FV E
Sbjct: 294 ATL--IEYPNMFHAFYIF-PELPEAGQLIMRIKDFVDE 328
>At5g62180 putative protein
Length = 327
Score = 103 bits (256), Expect = 2e-22
Identities = 82/268 (30%), Positives = 130/268 (47%), Gaps = 40/268 (14%)
Query: 29 DGSIERPKQSPFAPPSLNDPN--TGISSKDIQIPHNPTISSRIYLPKIT----NPLS-KF 81
DGSI R + + DP+ SKD+ + + R+YLP N S K
Sbjct: 21 DGSITRDLSNFPCTAATPDPSPLNPAVSKDLPVNQLKSTWLRLYLPSSAVNEGNVSSQKL 80
Query: 82 PILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAAL 141
PI+VY+HGG F+ S + +H+ A N I+VS Y LAPE+ LP Y D AL
Sbjct: 81 PIVVYYHGGGFILCSVDMQLFHDFCSEVARDLNAIVVSPSYRLAPEHRLPAAYDDGVEAL 140
Query: 142 KWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQA--GLENLPCDVKIL 199
WI + + + W+ H +F+ +F+ G SAG N+A+N+ +++ + +L ++I
Sbjct: 141 DWIKT-------SDDEWIKSHADFSNVFLMGTSAGGNLAYNVGLRSVDSVSDL-SPLQIR 192
Query: 200 GAIIIHPYFYSANPIGSEPIIEPENNIIH----------TFWHFAYPNAPFGIDNPR--F 247
G I+ HP+F G E E E +++ W + P G+D
Sbjct: 193 GLILHHPFF------GGEERSESEIRLMNDQVCPPIVTDVMWDL---SLPVGVDRDHEYS 243
Query: 248 NP-LGEGAPSLEKLGCSR-IIVCVAGKD 273
NP +G+G+ LEK+G R ++ + G+D
Sbjct: 244 NPTVGDGSEKLEKIGRLRWKVMMIGGED 271
>At2g45610 unknown protein
Length = 324
Score = 99.4 bits (246), Expect = 2e-21
Identities = 77/255 (30%), Positives = 120/255 (46%), Gaps = 20/255 (7%)
Query: 35 PKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIY----LPKITNPLSKFPILVYFHGG 90
P+ P P P +SKD+ I H +S RI+ LP N +++ PI+++ HG
Sbjct: 32 PRVEPDPDPC---PGKLAASKDVTINHETGVSVRIFRPTNLPSNDNAVARLPIIIHLHGS 88
Query: 91 VFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNN 150
++ S AS+ VI+VS+ Y L PE+ LP Y D AL W+ +
Sbjct: 89 GWILYPANSAANDRCCSQMASELTVIVVSVHYRLPPEHRLPAQYDDALDALLWVKQQVVD 148
Query: 151 NINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHPYFYS 210
+ N EPWL ++ +F++ +I G S GANIA +A+++ L++ ++I G + P F
Sbjct: 149 STNG-EPWLKDYADFSRCYICGSSNGANIAFQLALRS-LDHDLTPLQIDGCVFYQPLFGG 206
Query: 211 ANPIGSE--PIIEPENNI--IHTFWHFAYPNAPFGIDNPR--FNPLGEGAPSLEKLG-CS 263
SE +P + + W + P G+D NPLG P EK+G
Sbjct: 207 KTRTKSELKNFADPVMPVPAVDAMWEL---SLPVGVDRDHRYCNPLGY-LPQKEKVGRLG 262
Query: 264 RIIVCVAGKDKLRER 278
R +V G D +R
Sbjct: 263 RCLVIGYGGDTSLDR 277
>At3g27320 esterase, putative
Length = 460
Score = 80.9 bits (198), Expect = 9e-16
Identities = 66/248 (26%), Positives = 110/248 (43%), Gaps = 46/248 (18%)
Query: 80 KFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWA 139
K P+++ FHGG ++ S S + A ++I++++ Y LAPE P D +
Sbjct: 165 KLPVMLQFHGGGWVSGSNDSVANDFFCRRMAKHCDIIVLAVGYRLAPENRYPAACEDGFK 224
Query: 140 ALKWISSHSN--------NNINNP----------------------EPWLIEHGNFNKLF 169
LKW+ +N N P EPWL H + ++
Sbjct: 225 VLKWLGKQANLAECNKSMGNSRRPGGEVKKSEVNKHIVDAFGASLVEPWLANHADPSRCV 284
Query: 170 IGGDSAGANIAHNI---AIQAGLENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPENNI 226
+ G S GANIA + AI+ G +NL VK++ ++++P+F + P SE I+ N+
Sbjct: 285 LLGVSCGANIADYVARKAIEVG-QNLD-PVKVVAQVLMYPFFIGSVPTQSE--IKQANSY 340
Query: 227 IH------TFWHFAYPNAPFGIDNPRFNPLGEG-APSLEKLGCSRIIVCVAGKDKLRERG 279
+ W P F +D+ NPL G +P L+ + + VA D +R+R
Sbjct: 341 FYDKPMCILAWKLFLPEEEFSLDHQAANPLVPGRSPPLKFM--PPTLTIVAEHDWMRDRA 398
Query: 280 VWYWEGVK 287
+ Y E ++
Sbjct: 399 IAYSEELR 406
Score = 29.6 bits (65), Expect = 2.4
Identities = 18/45 (40%), Positives = 27/45 (60%), Gaps = 4/45 (8%)
Query: 30 GSIERPKQSPFAP-PSLNDPNTGISSKDIQIPHNPTISSRIYLPK 73
G RP++S AP P D G+++KDI I ++S RI+LP+
Sbjct: 38 GVTTRPEESVAAPNPLFTD---GVATKDIHIDPLTSLSVRIFLPE 79
>At5g14310 unknown protein
Length = 446
Score = 80.5 bits (197), Expect = 1e-15
Identities = 64/257 (24%), Positives = 104/257 (39%), Gaps = 43/257 (16%)
Query: 70 YLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYP 129
Y P K P+++ FHGG ++ S+ S + A +VI++++ Y LAPE
Sbjct: 140 YAPSAKRNSRKLPVMLQFHGGGWVSGSSDSAANDFFCRRIAKVCDVIVLAVGYRLAPENR 199
Query: 130 LPTCYHDCWAALKWISSHSN----------NNINNPE--------------------PWL 159
P + D L W+ +N +N E PWL
Sbjct: 200 YPAAFEDGVKVLHWLGKQANLADCCKSLGNRRVNGVEVKKLNVQGQIVDAFGASMVEPWL 259
Query: 160 IEHGNFNKLFIGGDSAGANIAHNI---AIQAGLENLPCDVKILGAIIIHPYFYSANPIGS 216
H + ++ + G S G NIA + A++AG L VK++ ++++P+F NP S
Sbjct: 260 AAHADPSRCVLLGVSCGGNIADYVARKAVEAG--KLLEPVKVVAQVLMYPFFIGNNPTQS 317
Query: 217 EPIIEPENNIIH------TFWHFAYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVA 270
E I+ N+ + W P F D+P NPL + VA
Sbjct: 318 E--IKLANSYFYDKPVSVLAWKLFLPEKEFDFDHPAANPLAHNRSGPPLKLMPPTLTVVA 375
Query: 271 GKDKLRERGVWYWEGVK 287
D +R+R + Y E ++
Sbjct: 376 EHDWMRDRAIAYSEELR 392
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,403,970
Number of Sequences: 26719
Number of extensions: 393828
Number of successful extensions: 950
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 853
Number of HSP's gapped (non-prelim): 29
length of query: 330
length of database: 11,318,596
effective HSP length: 100
effective length of query: 230
effective length of database: 8,646,696
effective search space: 1988740080
effective search space used: 1988740080
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149197.10


