

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.1 + phase: 0 /pseudo
(610 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
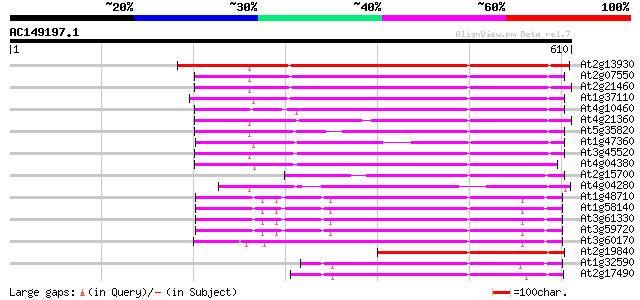
Score E
Sequences producing significant alignments: (bits) Value
At2g13930 putative retroelement pol polyprotein 350 2e-96
At2g07550 putative retroelement pol polyprotein 314 1e-85
At2g21460 putative retroelement pol polyprotein 308 4e-84
At1g37110 307 1e-83
At4g10460 putative retrotransposon 304 8e-83
At4g21360 putative transposable element 304 1e-82
At5g35820 copia-like retrotransposable element 302 4e-82
At1g47360 polyprotein, putative 300 1e-81
At3g45520 copia-like polyprotein 296 3e-80
At4g04380 putative polyprotein 279 4e-75
At2g15700 copia-like retroelement pol polyprotein 253 2e-67
At4g04280 putative transposon protein 228 8e-60
At1g48710 hypothetical protein 224 8e-59
At1g58140 hypothetical protein 224 1e-58
At3g61330 copia-type polyprotein 223 3e-58
At3g59720 copia-type reverse transcriptase-like protein 223 3e-58
At3g60170 putative protein 213 2e-55
At2g19840 copia-like retroelement pol polyprotein 196 4e-50
At1g32590 hypothetical protein, 5' partial 188 9e-48
At2g17490 putative retroelement pol polyprotein 187 2e-47
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 350 bits (897), Expect = 2e-96
Identities = 187/444 (42%), Positives = 267/444 (60%), Gaps = 24/444 (5%)
Query: 183 LILAMTLLYLLSPLIRGVKLLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*I 242
++LA +++ I +LDT +FHM P +D F F+ + G V MGND V I
Sbjct: 267 VLLATDETLVVTDSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGI 326
Query: 243 GIVHFKTNDGVVRTLTN-----------------ESLVCKYSAEGGVLMVSKGYLVLLKA 285
G + + +DG LT+ E C + ++ G+L + KG +LK
Sbjct: 327 GSIKIRNSDGSQVILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCSTILKG 386
Query: 286 SRIDSLYVLQGIVVTGSAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGID 345
+ D+LY+L G+ G + SS KD T LWH LGHM++KGM +L K+G L ++ I
Sbjct: 387 QKRDTLYILDGVTEEGES--HSSAEVKDETALWHSRLGHMSQKGMEILVKKGCLRREVIK 444
Query: 346 KLEFCKHYVFWKQKKVSFSTATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFP 404
+LEFC+ V+ KQ +VSF+ A H TK L Y+HSDLWG P S G +Y ++ +DD+
Sbjct: 445 ELEFCEDCVYGKQHRVSFAPAQHVTKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYS 504
Query: 405 RKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIAR 464
RKVW+YFLR K+E F F +W+ +VE Q+ + VKKL TDN LE+C+ F +FC GI R
Sbjct: 505 RKVWIYFLRKKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVR 564
Query: 465 HKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSAL 524
HKT PQQNG+AER+ RT++++ R MLS +G+ + W EAASTA +L+NRSP +A+
Sbjct: 565 HKTCAYTPQQNGIAERLNRTIMDKVRSMLSRSGM--EKKFWAEAASTAVYLINRSPSTAI 622
Query: 525 DFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSD 584
+F +PE+ W+G L D S+LR FGC AY + GKL PR+ + IF SY KGY++W +
Sbjct: 623 NFDLPEEKWTGALPDLSSLRKFGCLAYIHADQGKLNPRSKKGIFTSYPEGVKGYKVWVLE 682
Query: 585 PKSQKLILSRDVTFNEDALLSSGK 608
K K ++SR+V F E + K
Sbjct: 683 DK--KCVISRNVIFREQVMFKDLK 704
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 314 bits (804), Expect = 1e-85
Identities = 176/420 (41%), Positives = 247/420 (57%), Gaps = 24/420 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT ++HM R+ F F G V MGN +V +G + K +DG+ LTN
Sbjct: 310 ILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIRVKNSDGLTIVLTNVR 369
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
E K+ +E G+L + G VLL R D+LY+L V +
Sbjct: 370 YIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRRYDTLYLLNWKPVASESL 429
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
+ + + D T LWH L HM++K M +L ++G L K+ + L+ C+ ++ K K+ SFS
Sbjct: 430 --AVVKRADDTVLWHQRLCHMSQKNMEILVRKGFLDKKKVSSLDVCEDCIYGKAKRKSFS 487
Query: 365 TATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
A H TK L+YIHSDLWG P S G +Y M+IIDDF RKVWVYF++ K+E F F
Sbjct: 488 LAHHDTKEKLEYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYFMKTKDEAFEKFV 547
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W LVE QT + VK L TDN LEFC+ F+ FC + GI RH+T PQQNGVAERM R
Sbjct: 548 EWVNLVENQTDRRVKTLRTDNGLEFCNKLFDGFCESIGIHRHRTCAYTPQQNGVAERMNR 607
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
T++E+ R MLS++GL + W EA T L+N++P SAL+F++P+ WSGN YS L
Sbjct: 608 TIMEKVRSMLSDSGL--PKRFWAEATHTTVLLINKTPSSALNFEIPDKKWSGNPPVYSYL 665
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
R +GC A+ +DGKL PRA + + + Y KGY++W D +K ++SR++ F E+A+
Sbjct: 666 RRYGCVAFVHTDDGKLEPRAKKGVLIGYPVGVKGYKVWILD--ERKCVVSRNIIFQENAV 723
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 308 bits (790), Expect = 4e-84
Identities = 180/427 (42%), Positives = 245/427 (57%), Gaps = 24/427 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
++DT ++HM R+ F G V MGN KV IG + K G+V LTN
Sbjct: 288 VMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNVR 347
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
E + E G L + G VLL R +LY+LQ VT +
Sbjct: 348 YIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYTLYLLQWRPVTEESL 407
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
S + ++D T LWH LGHM++K M LL K+G L K+ + KLE C+ ++ K K++ F+
Sbjct: 408 --SVVKRQDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKLETCEDCIYGKAKRIGFN 465
Query: 365 TATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
A H T+ L+Y+HSDLWG PS S G +Y ++ IDD+ RKV +YFL+ K+E F F
Sbjct: 466 LAQHDTREKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDEAFDKFV 525
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W LVE QT K +K L TDN LEFC+ F+EFC+ GI H+T PQQNGVAERM R
Sbjct: 526 EWANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNGVAERMNR 585
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
TL+E+ R MLS++GL + W EA T L+N++P SAL+++VP+ WSG YS L
Sbjct: 586 TLMEKVRSMLSDSGL--PKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSPIYSYL 643
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
R FGC A+ +DGKL PRA + I + Y KGY++W + K K ++SR+V F E+A
Sbjct: 644 RRFGCIAFVHTDDGKLNPRAKKGILVGYPIGVKGYKIWLLEEK--KCVVSRNVIFQENAS 701
Query: 604 LSSGKQS 610
QS
Sbjct: 702 YKDMMQS 708
>At1g37110
Length = 1356
Score = 307 bits (787), Expect = 1e-83
Identities = 169/426 (39%), Positives = 247/426 (57%), Gaps = 24/426 (5%)
Query: 196 LIRGVKLLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVR 255
+++ + +LD+ HM RD F +F+ +L+G+D + G + T+ G ++
Sbjct: 303 MVKDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHSVESQGQGTIRIDTHGGTIK 362
Query: 256 TLTNESLV---------------CKYSAEGGVLMVS--KGYLVLLKASRIDSLYVLQGIV 298
L N V Y EGG V K L+ S + LYVL G
Sbjct: 363 ILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNNKTALRGSLSNGLYVLDGST 422
Query: 299 VTGSAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQ 358
V + ++ K T LWH LGHM+ + +L+ +G + ++ I++LEFC+H V K
Sbjct: 423 VMSE--LCNAETDKVKTALWHSRLGHMSMNNLKVLAGKGLIDRKEINELEFCEHCVMGKS 480
Query: 359 KKVSFSTATHRTKGILDYIHSDLWGPSKVT-SYGGRRYMMTIIDDFPRKVWVYFLRYKNE 417
KKVSF+ H ++ L Y+H+DLWG VT S G++Y ++IIDD RKVW+YFL+ K+E
Sbjct: 481 KKVSFNVGKHTSEDALSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVWLYFLKSKDE 540
Query: 418 TFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGV 477
TF F +W+ LVE Q K VK L TDN LEFC+S F+ +C HGI RH+T PQQNGV
Sbjct: 541 TFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHRTCTYTPQQNGV 600
Query: 478 AERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNL 537
AERM RT++E+ RC+L+ +G+ W EAA+TA +L+NRSP SA++ VPE++W
Sbjct: 601 AERMNRTIMEKVRCLLNKSGV--EEVFWAEAAATAAYLINRSPASAINHNVPEEMWLNRK 658
Query: 538 VDYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVT 597
Y +LR FG AY + GKL PRA++ FL Y + +KGY++W + +K ++SR+V
Sbjct: 659 PGYKHLRKFGSIAYVHQDQGKLKPRALKGFFLGYPAGTKGYKVWLLE--EEKCVISRNVV 716
Query: 598 FNEDAL 603
F E +
Sbjct: 717 FQESVV 722
>At4g10460 putative retrotransposon
Length = 1230
Score = 304 bits (779), Expect = 8e-83
Identities = 168/406 (41%), Positives = 242/406 (59%), Gaps = 15/406 (3%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNES 261
++DT N+HM ++ F G V MGN K I N + TL
Sbjct: 313 VMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKSTSKFRVKYIPDMDRNLLSMGTLEEHG 372
Query: 262 LVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAAVSSSMP---KKDVTKLW 318
+ ++ GVL+V +G LL SR + LY+LQG VS SM + D T LW
Sbjct: 373 Y--SFESKNGVLVVKEGTRTLLIGSRHEKLYLLQG-----KPEVSHSMTVERRNDDTVLW 425
Query: 319 HMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGILDYIH 378
H LGH+++K M +L K+G L + + KLE C+ ++ K +++SF ATH T+ L+Y+H
Sbjct: 426 HRRLGHISQKNMDILVKKGYLDGKKVSKLELCEDCIYGKARRLSFVVATHNTEDKLNYVH 485
Query: 379 SDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNV 437
SDLWG PS S G +Y ++ ID + RK WVYFL++K+E F TF +W ++VE QTG+ +
Sbjct: 486 SDLWGAPSVPLSLGKCQYFISFIDVYSRKTWVYFLKHKDEAFGTFAEWSVMVENQTGRKI 545
Query: 438 KKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAG 497
K L DN LEFC+ FN+FC GI RH+T PQQNGVAERM T++E+ R MLS +G
Sbjct: 546 KILRIDNGLEFCNQQFNDFCKEKGIVRHQTCAYTPQQNGVAERMNHTIMEKVRRMLSYSG 605
Query: 498 L*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDG 557
L + W EA +T L+N++P SA++F++ + WSG Y+ L+ FGC A+ ++G
Sbjct: 606 L--PKTFWAEATNTVVTLINKTPSSAVNFEISDKRWSGKSPVYNYLKRFGCVAFTYADEG 663
Query: 558 KLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
KL PRA + +FL Y S KGY++W + +K +SR+VTF E+A+
Sbjct: 664 KLVPRAKKGVFLGYLSGEKGYKVWLLE--ERKCSVSRNVTFQENAV 707
>At4g21360 putative transposable element
Length = 1308
Score = 304 bits (778), Expect = 1e-82
Identities = 171/427 (40%), Positives = 247/427 (57%), Gaps = 34/427 (7%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
++D+ +HM D F+ F + ++L+G+D + G V T+ G +R L N
Sbjct: 310 VIDSGCTYHMTSRMDWFSEFNENETTMILLGDDHTVESKGSGTVKVNTHGGSIRVLKNVR 369
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQG-IVVTGSA 303
+ L K+ G + K L + ++ LYVL G VV +
Sbjct: 370 FVPNLRRNLISTGTLDKLGYKHEGGDGKVRFYKENKTALCGNLVNGLYVLDGHTVVNENC 429
Query: 304 AVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSF 363
V S K T+LWH LGHM+ M +L+++G L K+ I +L FC++ V K KK+SF
Sbjct: 430 NVEGSNEK---TELWHCRLGHMSLNNMKILAEKGLLEKKDIKELSFCENCVMGKSKKLSF 486
Query: 364 STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
+ H T +L YIH+DLWG ++Y ++IIDD RKVW+ FL+ K+ETF F
Sbjct: 487 NVGKHITDEVLGYIHADLWG---------KQYFLSIIDDKSRKVWLMFLKTKDETFERFC 537
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W+ LVE Q K VK L TDN LEFC+ F+EFC +GI RH+T PQQNGVA+RM R
Sbjct: 538 EWKELVENQVNKKVKILRTDNGLEFCNLKFDEFCKQNGIERHRTCTYTPQQNGVAKRMNR 597
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
TL+E+ RC+L+ +GL W EAA+TA +LVNRSP SA+D VPE++W Y +L
Sbjct: 598 TLMEKVRCLLNESGL--EEVFWAEAAATAAYLVNRSPASAVDHNVPEELWLDKKPGYKHL 655
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
R FGC AY ++ GKL PRA++ +FL Y +KGY++W D +K ++SR++ FNE+ +
Sbjct: 656 RRFGCIAYVHLDQGKLKPRALKGVFLGYPQGTKGYKVWLLD--EEKCVISRNIVFNENQV 713
Query: 604 LSSGKQS 610
++S
Sbjct: 714 YKDIRES 720
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 302 bits (773), Expect = 4e-82
Identities = 168/420 (40%), Positives = 234/420 (55%), Gaps = 40/420 (9%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
+LDT +FHM +D F+ G V MGND +V IG V K DG LT+
Sbjct: 313 ILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKNEDGSTILLTDVR 372
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
E C + ++ G+L + K L +L + +LY LQG + G A
Sbjct: 373 YIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTVLTGKKESTLYFLQGTTLAGEAN 432
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
V +KD T LWH LGH+ KG+ +L +G L K + +SF
Sbjct: 433 VIDK--EKDETSLWHSRLGHIGAKGLQVLVSKGHLDKNIM----------------ISFG 474
Query: 365 TATHRTKGILDYIHSDLWGPSKVT-SYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFK 423
A H TK LDY+HSDLWG + V S G +Y +T IDDF R+ W+YF+R K+E F F
Sbjct: 475 AAKHVTKDKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWIYFIRTKDEAFSKFV 534
Query: 424 KWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIR 483
+W+ +E Q K +K LITDN LEFC+ +F+ FC G+ RH+T PQQNGVAERM R
Sbjct: 535 EWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCAYTPQQNGVAERMNR 594
Query: 484 TLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNL 543
T++ + RCMLS +GL + W EAASTA L+N+SP S+++F +PE+ W+G+ DY L
Sbjct: 595 TIMNKVRCMLSESGL--GKQFWAEAASTAVFLINKSPSSSIEFDIPEEKWTGHPPDYKIL 652
Query: 544 RIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
+ FG AY + GKL PRA + IFL Y K +++W + +K ++SRD+ F E+ +
Sbjct: 653 KKFGSVAYIHSDQGKLNPRAKKGIFLGYPDGVKRFKVWLLE--DRKCVVSRDIVFQENQM 710
>At1g47360 polyprotein, putative
Length = 1182
Score = 300 bits (769), Expect = 1e-81
Identities = 169/419 (40%), Positives = 236/419 (55%), Gaps = 53/419 (12%)
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNESL 262
LDT +FHM P RDLF F+ + G V MGND V IG + + NDG LT+
Sbjct: 277 LDTGCSFHMTPRRDLFKDFKELSSGFVKMGNDTYSPVKGIGSIKIRNNDGTQVILTDVRY 336
Query: 263 V-----------------CKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAAV 305
V C + ++ GVL + KG +LK + ++LY+L+G+ G +
Sbjct: 337 VPNMARNLISLGTLEDKGCWFKSQDGVLKIVKGCSTILKGQKRETLYILEGLAENGESHS 396
Query: 306 SSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFST 365
S+ + KD T LWH LGHM++KGM +L K+ L ++ I +L+FC+ V+ K +VSF+
Sbjct: 397 SAEL--KDETSLWHSRLGHMSQKGMEILVKKDCLQRETIKELKFCEDCVYGKNHRVSFAP 454
Query: 366 ATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKK 424
A H TK L YIHSDLWG P S G +Y ++ +DD+ RK
Sbjct: 455 AQHVTKEKLAYIHSDLWGSPHNPASLGNCQYFISFVDDYSRK------------------ 496
Query: 425 WRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRT 484
VKKL TDN LE+C+ F +FC + GI RHKT PQQNG+AER+ RT
Sbjct: 497 -----------KVKKLRTDNGLEYCNHYFEKFCKDEGIVRHKTCAYTPQQNGIAERLNRT 545
Query: 485 LLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLR 544
++++ R MLS +G+ + W EAASTA +L+NRSP +ALDF +PE+ W+G L D LR
Sbjct: 546 IMDKVRSMLSESGMD--KKFWAEAASTAVYLINRSPSTALDFDLPEEKWTGALPDLKGLR 603
Query: 545 IFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL 603
FGC Y + GKL PRA + +F SY KGY++W + K K ++SR+V F E+ +
Sbjct: 604 KFGCLVYIHADQGKLNPRAKKDVFTSYLEGVKGYKVWVLEEK--KCVISRNVIFREEIM 660
>At3g45520 copia-like polyprotein
Length = 1363
Score = 296 bits (757), Expect = 3e-80
Identities = 169/421 (40%), Positives = 242/421 (57%), Gaps = 26/421 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-- 259
++DT +HM R+ F+ G V MGN +V +G V ++G+ TL N
Sbjct: 317 IMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNKSISRVKGVGTVRIVNDNGLTVTLQNVR 376
Query: 260 ---------------ESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTG-SA 303
E K+ +E G+L + G VLL+ R D+LY+L G T S
Sbjct: 377 YIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGNQVLLEGRRYDTLYILHGKPATDESL 436
Query: 304 AVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSF 363
AV+ + D T LWH L HM++K M LL K+G L K+ + L+ C+ ++ + KK+ F
Sbjct: 437 AVARA---NDDTVLWHRRLCHMSQKNMSLLIKKGFLDKKKVSMLDTCEDCIYGRAKKIGF 493
Query: 364 STATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
+ A H TK L+Y+HSDLWG P+ S G +Y ++ IDD+ RKVWVYFL+ K+E F F
Sbjct: 494 NLAQHDTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYTRKVWVYFLKTKDEAFEKF 553
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
W LVE Q+G+ VK L TDN LEFC+ F+ FC G RH+T PQQNGV ERM
Sbjct: 554 VSWISLVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKGFQRHRTCAYTPQQNGVVERMN 613
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
RT++E+ R ML ++GL + W EA TA L+N++P SA++F+ P+ WSG YS
Sbjct: 614 RTIMEKVRSMLCDSGL--PKRFWAEATHTAVLLINKTPCSAINFEFPDKRWSGKAPIYSY 671
Query: 543 LRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNEDA 602
LR +GC + + GKL RA + + + Y S KGY++W + K K ++SR+V+F E+A
Sbjct: 672 LRRYGCVTFVHTDGGKLNLRAKKGVLIGYPSGVKGYKVWLIEEK--KCVVSRNVSFQENA 729
Query: 603 L 603
+
Sbjct: 730 V 730
>At4g04380 putative polyprotein
Length = 778
Score = 279 bits (713), Expect = 4e-75
Identities = 161/413 (38%), Positives = 230/413 (54%), Gaps = 24/413 (5%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNES 261
++DT ++HM R+ F+ V MGN +V +G V ++G+ TL N
Sbjct: 200 IMDTGCSYHMTHKREWLEDFDEEAGCSVRMGNKTISRVKGVGTVRIANDNGLTVTLQNVR 259
Query: 262 LVC-----------------KYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTG-SA 303
+ K+ +E +L V+ G VLL+ R D+LY+L G T S
Sbjct: 260 YISDMDRIFLSLGTFEKAGRKFESENDMLRVNAGEQVLLEGRRYDTLYILHGKPATDESL 319
Query: 304 AVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSF 363
AV+ + D LWH L HM++K M LL K+G L K+ + L+ C+ ++ K KK+ F
Sbjct: 320 AVAKA---NDDIVLWHRRLCHMSQKNMSLLVKKGFLDKKKVSMLDTCEDCIYGKAKKIGF 376
Query: 364 STATHRTKGILDYIHSDLWG-PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTF 422
+ A H TK L+Y+H DLWG P+ + G +Y ++ IDD RKV VYFL+ K+E F F
Sbjct: 377 NFAQHDTKEKLEYVHYDLWGAPTMPMALGNCQYFISFIDDHTRKVCVYFLKTKDEAFEKF 436
Query: 423 KKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMI 482
W LVE Q+G VK L TDN LEFC+ F+ FC GI RH+T P PQQNGV ERM
Sbjct: 437 VSWISLVENQSGNRVKTLRTDNGLEFCNRMFDGFCEERGIQRHRTCPYTPQQNGVVERMN 496
Query: 483 RTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSN 542
RT++ER R ML ++ L + W EA TA L+N++P SA +F+ P+ WSG YS
Sbjct: 497 RTIMERVRSMLCDSRL--PKRFWAEATHTAVLLINKTPCSANNFEFPDKRWSGKAPIYSY 554
Query: 543 LRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRD 595
LR +GC + +DGKL RA + + + Y S KGY++W + ++ L+ R+
Sbjct: 555 LRRYGCVTFVQTDDGKLNLRAKKGVLIGYPSGVKGYKVWLIEERNSHLVRDRE 607
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 253 bits (646), Expect = 2e-67
Identities = 134/305 (43%), Positives = 184/305 (59%), Gaps = 20/305 (6%)
Query: 299 VTGSAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQ 358
V G + P V L ++ LGHM+ K M++L ++G L + +DKLEFC+ V K
Sbjct: 322 VKGIGKIKILNPDDYVVILTNVRLGHMSLKNMNVLVEEGYLSGKEVDKLEFCESCVLGKS 381
Query: 359 KKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNET 418
K SF TA H TK S GG RY ++ IDDF +KVWVYFL+ K+E
Sbjct: 382 HKQSFPTAKHTTK----------------ESLGGCRYFVSFIDDFSKKVWVYFLKTKDEA 425
Query: 419 FPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVA 478
+ F++W+ VE QTGK +K L TDN LEF ++ F+ C GI RH+T PQQNGV+
Sbjct: 426 YHKFREWKQAVENQTGKKIKYLRTDNGLEFYNTQFDNLCKEDGIKRHRTCTYTPQQNGVS 485
Query: 479 ERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLV 538
ERM +T+++ R ML+ G+ ++ W EA STA +L+NR+P+S + FK+PE++W+G
Sbjct: 486 ERMNKTIMDIVRSMLAETGM--SQEFWAEATSTAVYLINRTPNSFIGFKLPEEVWTGTKP 543
Query: 539 DYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTF 598
D S+LR FGC AY V K +PRAV+ +F+ Y KGYR+W PK K SR+V F
Sbjct: 544 DLSHLRRFGCSAYVHVTQDKTSPRAVKGVFMGYPCGIKGYRVWL--PKEGKCTTSRNVVF 601
Query: 599 NEDAL 603
NE L
Sbjct: 602 NETEL 606
>At4g04280 putative transposon protein
Length = 1104
Score = 228 bits (581), Expect = 8e-60
Identities = 142/403 (35%), Positives = 201/403 (49%), Gaps = 73/403 (18%)
Query: 228 VVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN-----------------ESLVCKYSAEG 270
++L+G+D + G + T+ G++R L N SL K+
Sbjct: 260 MILLGDDHTVESRGCGTIKLNTHGGLIRMLKNVRYVPNLRRNLISTGTLHSLGYKHEGGE 319
Query: 271 GVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAAVSSSMPKKDVTKLWHMHLGHMTEKGM 330
G L K L ++ LY+L G V + S K+ TKL
Sbjct: 320 GKLRFYKNGKTALCGYLMNGLYILDGHTVATETCNAESA--KNSTKLL------------ 365
Query: 331 HLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGILDYIHSDLWGPSKVT-S 389
+ K+ L+FC+HYV K KK+SF+ H T+ IL Y+H+DLWG VT S
Sbjct: 366 --------IEKKEFKDLDFCEHYVMGKSKKLSFNVGKHVTEDILGYVHADLWGSPNVTPS 417
Query: 390 YGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFC 449
G++Y ++IIDD RKVW+ FL+ K+ETF F +W+ +VE GK VK L TDN LEFC
Sbjct: 418 ISGKQYFLSIIDDKSRKVWLMFLKSKDETFDKFCEWKEIVENHVGKKVKTLRTDNGLEFC 477
Query: 450 SSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAA 509
++ FN++C GI RH+T PQQNGV +RM RT++E+
Sbjct: 478 NNRFNDYCAKTGIERHRTCTYTPQQNGVTKRMNRTIMEKV-------------------- 517
Query: 510 STACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAVECIFL 569
RSP SA+D VPE +W Y +LR F AY GKL PR ++ +FL
Sbjct: 518 --------RSPASAVDHNVPEQLWLNREPGYKHLRRFSSIAYVHQEQGKLKPRVLKGVFL 569
Query: 570 SYASESKGYRLWCSDPKSQKLILSRDVTFNEDAL---LSSGKQ 609
Y +KGY++W D +K ++SR+V FNED++ L SG +
Sbjct: 570 GYPQGTKGYKVWLID--EEKCVISRNVVFNEDSVFKDLQSGSK 610
>At1g48710 hypothetical protein
Length = 1352
Score = 224 bits (572), Expect = 8e-59
Identities = 144/428 (33%), Positives = 239/428 (55%), Gaps = 40/428 (9%)
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNESL 262
LD+ + HMC + +F + G V +G++ + +V G + + +G + ++N
Sbjct: 337 LDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYY 396
Query: 263 VCKYSAEGGVL----MVSKGYLVLLKASRI-----------------DSLYVLQGIVVTG 301
+ S + +L ++ KGY + LK + + + ++VL +
Sbjct: 397 IP--SMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLN---IRN 451
Query: 302 SAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKL----EFCKHYVFWK 357
A M K+ + LWH+ GH+ G+ LLS++ + +G+ + + C+ + K
Sbjct: 452 DIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMV--RGLPCINHPNQVCEGCLLGK 509
Query: 358 QKKVSF-STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKN 416
Q K+SF ++ R + L+ IH+D+ GP K S G Y + IDDF RK WVYFL+ K+
Sbjct: 510 QFKMSFPKESSSRAQKSLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKS 569
Query: 417 ETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNG 476
E F FKK++ VE ++G +K + +D EF S +F ++C ++GI R T+PR+PQQNG
Sbjct: 570 EVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNG 629
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
VAER RT+LE AR ML + L ++LW EA + A +L+NRSP ++ K P++ WSG
Sbjct: 630 VAERKNRTILEMARSMLKSKRL--PKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 537 LVDYSNLRIFGCPAYALVND---GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILS 593
S+LR+FG A+A V D KL ++ + IF+ Y + SKGY+L+ +P ++K I+S
Sbjct: 688 KSGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLY--NPDTKKTIIS 745
Query: 594 RDVTFNED 601
R++ F+E+
Sbjct: 746 RNIVFDEE 753
>At1g58140 hypothetical protein
Length = 1320
Score = 224 bits (571), Expect = 1e-58
Identities = 144/428 (33%), Positives = 239/428 (55%), Gaps = 40/428 (9%)
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNESL 262
LD+ + HMC + +F + G V +G++ + +V G + + +G + ++N
Sbjct: 337 LDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYY 396
Query: 263 VCKYSAEGGVL----MVSKGYLVLLKASRI-----------------DSLYVLQGIVVTG 301
+ S + +L ++ KGY + LK + + + ++VL +
Sbjct: 397 IP--SMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLN---IRN 451
Query: 302 SAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKL----EFCKHYVFWK 357
A M K+ + LWH+ GH+ G+ LLS++ + +G+ + + C+ + K
Sbjct: 452 DIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMV--RGLPCINHPNQVCEGCLLGK 509
Query: 358 QKKVSF-STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKN 416
Q K+SF ++ R + L+ IH+D+ GP K S G Y + IDDF RK WVYFL+ K+
Sbjct: 510 QFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKS 569
Query: 417 ETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNG 476
E F FKK++ VE ++G +K + +D EF S +F ++C ++GI R T+PR+PQQNG
Sbjct: 570 EVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNG 629
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
VAER RT+LE AR ML + L ++LW EA + A +L+NRSP ++ K P++ WSG
Sbjct: 630 VAERKNRTILEMARSMLKSKRL--PKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 537 LVDYSNLRIFGCPAYALVND---GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILS 593
S+LR+FG A+A V D KL ++ + IF+ Y + SKGY+L+ +P ++K I+S
Sbjct: 688 KPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLY--NPDTKKTIIS 745
Query: 594 RDVTFNED 601
R++ F+E+
Sbjct: 746 RNIVFDEE 753
>At3g61330 copia-type polyprotein
Length = 1352
Score = 223 bits (567), Expect = 3e-58
Identities = 143/428 (33%), Positives = 238/428 (55%), Gaps = 40/428 (9%)
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNESL 262
LD+ + HMC + +F + G V +G++ + +V G + + +G + ++N
Sbjct: 337 LDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYY 396
Query: 263 VCKYSAEGGVL----MVSKGYLVLLKASRI-----------------DSLYVLQGIVVTG 301
+ S + +L ++ KGY + LK + + + ++VL +
Sbjct: 397 IP--SMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLN---IRN 451
Query: 302 SAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKL----EFCKHYVFWK 357
A M K+ + LWH+ GH+ G+ LLS++ + +G+ + + C+ + K
Sbjct: 452 DIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMV--RGLPCINHPNQVCEGCLLGK 509
Query: 358 QKKVSF-STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKN 416
Q K+SF ++ R + L+ IH+D+ GP K S G Y + IDDF RK WVYFL+ K+
Sbjct: 510 QFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKS 569
Query: 417 ETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNG 476
E F FKK++ VE ++G +K + +D EF S +F ++C ++GI R T+PR+PQQNG
Sbjct: 570 EVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNG 629
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
V ER RT+LE AR ML + L ++LW EA + A +L+NRSP ++ K P++ WSG
Sbjct: 630 VVERKNRTILEMARSMLKSKRL--PKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 537 LVDYSNLRIFGCPAYALVND---GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILS 593
S+LR+FG A+A V D KL ++ + IF+ Y + SKGY+L+ +P ++K I+S
Sbjct: 688 KPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLY--NPDTKKTIIS 745
Query: 594 RDVTFNED 601
R++ F+E+
Sbjct: 746 RNIVFDEE 753
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 223 bits (567), Expect = 3e-58
Identities = 143/428 (33%), Positives = 239/428 (55%), Gaps = 40/428 (9%)
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNESL 262
LD+ + HMC + +F + G V +G++ + +V G + + +G + ++N
Sbjct: 337 LDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYY 396
Query: 263 VCKYSAEGGVL----MVSKGYLVLLKASRI-----------------DSLYVLQGIVVTG 301
+ S + +L ++ KGY + LK + + + ++VL +
Sbjct: 397 IP--SMKTNILSLGQLLEKGYDIRLKDNNLSIRDKESNLITKVPMSKNRMFVLN---IRN 451
Query: 302 SAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKL----EFCKHYVFWK 357
A M K+ + LWH+ GH+ G+ LLS++ + +G+ + + C+ +
Sbjct: 452 DIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMV--RGLPCINHPNQVCEGCLLGN 509
Query: 358 QKKVSF-STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKN 416
Q K+SF ++ R + L+ IH+D+ GP K S G Y + IDDF RK WVYFL+ K+
Sbjct: 510 QFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKS 569
Query: 417 ETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNG 476
E F FKK++ VE ++G +K + +D+ EF S +F ++C ++GI R T+PR+PQQNG
Sbjct: 570 EVFEIFKKFKAHVEKESGLVIKTMRSDSGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNG 629
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
VAER RT+LE AR ML + L ++LW EA + A +L+NRSP ++ K P++ WSG
Sbjct: 630 VAERKNRTILEMARSMLKSKRL--PKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 537 LVDYSNLRIFGCPAYALVND---GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILS 593
S+LR+FG A+A V D KL ++ + IF+ Y + SKGY+L+ +P ++K I+S
Sbjct: 688 KPGVSHLRVFGSIAHAHVPDEKRNKLDDKSEKYIFIGYDNNSKGYKLY--NPDTKKTIIS 745
Query: 594 RDVTFNED 601
R++ F+E+
Sbjct: 746 RNIVFDEE 753
>At3g60170 putative protein
Length = 1339
Score = 213 bits (543), Expect = 2e-55
Identities = 145/428 (33%), Positives = 214/428 (49%), Gaps = 31/428 (7%)
Query: 200 VKLLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRT--- 256
V LD+ + HM ++ F+ E V +GND + V G V K N GV +
Sbjct: 299 VWFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKVN-GVTQVIPE 357
Query: 257 ------LTNESLVCKYSAEGGVLMV-----------SKGYLVLLKASRIDSLYVLQGIVV 299
L N L E G+ ++ SKG ++ S ++L
Sbjct: 358 VYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRMFFLLASKPQ 417
Query: 300 TGSAAVSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKL--EFCKHYVFWK 357
S + + LWH GH+ ++G+ LL+ + + I K E C + K
Sbjct: 418 KNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILKATKEICAICLTGK 477
Query: 358 QKKVSFSTATH-RTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKN 416
Q + S S T ++ L +HSD+ GP S+ G+RY+++ IDDF RK WVYFL K+
Sbjct: 478 QHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYFLHEKS 537
Query: 417 ETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNG 476
E F TFK ++ VE + G + L TD EF S++F EFC +HGI+R T PQQNG
Sbjct: 538 EAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGISRQLTAAFTPQQNG 597
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
VAER RT++ R MLS + + W EA + H+ NRSP +A++ PE+ WSG
Sbjct: 598 VAERKNRTIMNAVRSMLSERQV--PKMFWSEATKWSVHIQNRSPTAAVEGMTPEEAWSGR 655
Query: 537 LVDYSNLRIFGCPAYALVND---GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILS 593
R+FGC Y + D KL ++ +C+FL + ESK +RL+ DP +K+++S
Sbjct: 656 KPVVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLY--DPVMKKIVIS 713
Query: 594 RDVTFNED 601
+DV F+ED
Sbjct: 714 KDVVFDED 721
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 196 bits (497), Expect = 4e-50
Identities = 92/203 (45%), Positives = 139/203 (68%), Gaps = 4/203 (1%)
Query: 401 DDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNH 460
+D+ RKVWVYFL+ K+E F +F +W+ +VETQ+ + +K L TDN LEFC+ F+E C
Sbjct: 319 NDWSRKVWVYFLKTKDEAFASFTEWKKMVETQSERKLKHLRTDNGLEFCNHKFDEVCKKE 378
Query: 461 GIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSP 520
GI RH+T PQQNGVAER+ RT++ + R MLS +GL + W +AASTA +L+NRSP
Sbjct: 379 GIVRHRTCTYTPQQNGVAERLNRTIMNKVRSMLSESGL--DKKFWAKAASTAVYLINRSP 436
Query: 521 HSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRL 580
S+++ K+PE++W+ + ++S L+ FGC Y +GKL PRA + +F+ Y + KG+R+
Sbjct: 437 SSSIENKIPEELWTSAVPNFSGLKRFGCVVYVYSQEGKLDPRAKKGVFVGYPNGVKGFRV 496
Query: 581 WCSDPKSQKLILSRDVTFNEDAL 603
W + ++ +SR+V F ED +
Sbjct: 497 WMIE--EERCSISRNVVFREDVM 517
Score = 32.0 bits (71), Expect = 1.0
Identities = 18/51 (35%), Positives = 25/51 (48%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDG 252
++DT +FHM P ++ F G V M N+ +V IG V F DG
Sbjct: 267 IMDTGCSFHMTPRKEYLMDFVEAKSGKVRMANNSFSEVKGIGKVKFIKKDG 317
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 188 bits (477), Expect = 9e-48
Identities = 108/295 (36%), Positives = 166/295 (55%), Gaps = 16/295 (5%)
Query: 317 LWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEF------CKHYVFWKQKKVSF-STATHR 369
+WH GH+ +G+ L+++ + +G+ K + C + KQ + S + +
Sbjct: 395 MWHKRFGHLNHQGLRSLAEKEMV--KGLPKFDLGEEEAVCDICLKGKQIRESIPKESAWK 452
Query: 370 TKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILV 429
+ +L +H+D+ GP S G+RY++ IDDF RK W Y L K+ETF FK+++ V
Sbjct: 453 STQVLQLVHTDICGPINPASTSGKRYILNFIDDFSRKCWTYLLSEKSETFQFFKEFKAEV 512
Query: 430 ETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERA 489
E ++GK + L +D E+ S +F+E+C GI R T PQQNGVAER R+++
Sbjct: 513 ERESGKKLVCLRSDRGGEYNSREFDEYCKEFGIKRQLTAAYTPQQNGVAERKNRSVMNMT 572
Query: 490 RCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCP 549
RCML + R W EA A +++NRSP AL+ PE+ WS +LRIFG
Sbjct: 573 RCMLMEMSV--PRKFWPEAVQYAVYILNRSPSKALNDITPEEKWSSWKPSVEHLRIFGSL 630
Query: 550 AYALV---NDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNED 601
AYALV KL ++++C+ + ESK YRL+ DP + K+++SRDV F+E+
Sbjct: 631 AYALVPYQKRIKLDEKSIKCVMFGVSKESKAYRLY--DPATGKILISRDVQFDEE 683
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 187 bits (474), Expect = 2e-47
Identities = 111/305 (36%), Positives = 166/305 (54%), Gaps = 14/305 (4%)
Query: 306 SSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEF----CKHYVFWKQKKV 361
++ + K + T+LWH LGH + +L + + G+ K C+ + K +
Sbjct: 148 TAMVAKDEATELWHKRLGHTGHSNLKILQSKEMV--TGLPKFNVEEGKCESCILSKHSRD 205
Query: 362 SFSTATH-RTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFP 420
F + R K L+ IHSD+ GP + +S G RY++T IDD R VWVYFL+ K+E F
Sbjct: 206 PFPKESETRAKHKLELIHSDVCGPMQNSSINGSRYILTFIDDATRMVWVYFLKAKSEVFQ 265
Query: 421 TFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAER 480
TFKK++ LVE +KKL D E+ S +F+EF +GI R T +PQQN V+ER
Sbjct: 266 TFKKFKNLVENNANCRIKKLRIDRGTEYLSKEFSEFLEGNGIERQLTAAYSPQQNEVSER 325
Query: 481 MIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDY 540
R+L+E AR M+ L LW EA A + NR+P L K P + WS +
Sbjct: 326 RNRSLVEMARAMIKAKDL--PLKLWAEAVHVAAYAQNRTPTRTLKNKTPLEAWSDSKPSV 383
Query: 541 SNLRIFGCPAYALVNDGKLA---PRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVT 597
S++++FG Y + D K ++ IF+ Y+S++KGYR++ K K+ +SRDV
Sbjct: 384 SHMKVFGSICYVHIPDEKRRKWDDKSKRAIFVGYSSQTKGYRVYLL--KENKIDISRDVI 441
Query: 598 FNEDA 602
F+ED+
Sbjct: 442 FDEDS 446
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.331 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,945,252
Number of Sequences: 26719
Number of extensions: 533181
Number of successful extensions: 1577
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1227
Number of HSP's gapped (non-prelim): 120
length of query: 610
length of database: 11,318,596
effective HSP length: 105
effective length of query: 505
effective length of database: 8,513,101
effective search space: 4299116005
effective search space used: 4299116005
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149197.1


