

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.7 - phase: 0 /pseudo
(359 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
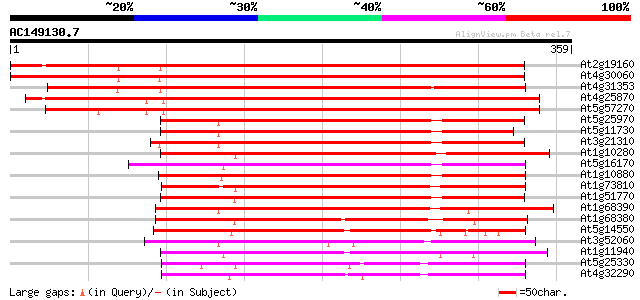
Score E
Sequences producing significant alignments: (bits) Value
At2g19160 unknown protein 380 e-106
At4g30060 unknown protein 366 e-102
At4g31353 unknown protein 364 e-101
At4g25870 unknown protein 358 3e-99
At5g57270 unknown protein 356 1e-98
At5g25970 putative protein 215 3e-56
At5g11730 unknown protein 210 1e-54
At3g21310 unknown protein 207 7e-54
At1g10280 unknown protein 206 2e-53
At5g16170 unknowen protein 204 6e-53
At1g10880 hypothetical protein 203 1e-52
At1g73810 unknown protein (At1g73810) 199 2e-51
At1g51770 hypothetical protein 194 5e-50
At1g68390 hypothetical protein 194 8e-50
At1g68380 unknown protein 177 8e-45
At5g14550 putative protein 176 2e-44
At3g52060 unknown protein 163 2e-40
At1g11940 unknown protein 158 5e-39
At5g25330 putative protein 150 8e-37
At4g32290 unknown protein 141 6e-34
>At2g19160 unknown protein
Length = 394
Score = 380 bits (976), Expect = e-106
Identities = 187/339 (55%), Positives = 244/339 (71%), Gaps = 12/339 (3%)
Query: 1 MKTAKFWRLS-MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSC 59
MK K WRL + M+ L G+ HR +KP+WI+ ++S I +F+ AY++ + +C
Sbjct: 1 MKAIKGWRLGKINYMQSLPGAR--HRAPTRKPIWIIAVLSLIAMFVIGAYMFPHHSKAAC 58
Query: 60 NMFYSKPCI-----IDISVLAVSHIPVSVYIERGITLDMPQ----NPKIAFMFLTPGSLP 110
MF SK C + S+ S ++ + L P+ + KIAFMFLTPG+LP
Sbjct: 59 YMFSSKGCKGLTDWLPPSLREYSDDEIAARVVISEILSSPRVIKKSSKIAFMFLTPGTLP 118
Query: 111 FEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLA 170
FEKLWD FFQGHEGKFSVY+HASK PVH SRYF+NR+IRSD++VWG++S+++AERRLL
Sbjct: 119 FEKLWDLFFQGHEGKFSVYIHASKDTPVHTSRYFLNREIRSDEVVWGRISMIDAERRLLT 178
Query: 171 NALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPE 230
NAL+DP NQ FVLLSDSCVPL +F Y+++Y+M+++ S+VD F DPGP G GR+ +HMLPE
Sbjct: 179 NALRDPENQQFVLLSDSCVPLRSFEYMYNYMMHSNVSYVDCFDDPGPHGTGRHMDHMLPE 238
Query: 231 VEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTI 290
+ +DFR GAQWFS+KRQHAV +AD+LYYSKF+ C KNCI DEHYLPTFF +
Sbjct: 239 IPREDFRKGAQWFSMKRQHAVVTVADNLYYSKFRDYCGPGVEGNKNCIADEHYLPTFFYM 298
Query: 291 VDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
+DP GIA W+VTYVD SE+K HP+ Y +DIT EL+KNI
Sbjct: 299 LDPTGIANWTVTYVDWSERKWHPRKYMPEDITLELIKNI 337
>At4g30060 unknown protein
Length = 401
Score = 366 bits (940), Expect = e-102
Identities = 182/340 (53%), Positives = 241/340 (70%), Gaps = 11/340 (3%)
Query: 1 MKTAKFWRL-SMGDMEILL-GSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTS 58
MK K W + ++ D+ + L G+ P ++ +WI++++S I +F AY+Y + +
Sbjct: 1 MKAVKRWSIGNLADIPVSLPGARYRAPPPGRRRVWIIMVLSLITMFFIMAYMYPHHSKRA 60
Query: 59 CNMFYSKPCI-----IDISVLAVSHIPVSVYIERGITLDMP----QNPKIAFMFLTPGSL 109
C M S+ C + S+ S ++ + L P +N KIAFMFLTPG+L
Sbjct: 61 CYMISSRGCKALADWLPPSLREYSDDEIAARVVIREILSSPPVIRKNSKIAFMFLTPGTL 120
Query: 110 PFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLL 169
PFE+LWD FF GHEGKFSVY+HASK +PVH SRYF+NR+IRSD++VWG++S+V+AERRLL
Sbjct: 121 PFERLWDRFFLGHEGKFSVYIHASKERPVHYSRYFLNREIRSDEVVWGRISMVDAERRLL 180
Query: 170 ANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLP 229
ANAL+D +NQ FVLLSDSCVPL +F YI++YLM+++ S+VD F DPG G GR+ HMLP
Sbjct: 181 ANALRDTSNQQFVLLSDSCVPLRSFEYIYNYLMHSNLSYVDCFDDPGQHGAGRHMNHMLP 240
Query: 230 EVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFT 289
E+ KDFR GAQWF++KRQHAV MAD LYYSKF+ C + KNCI DEHYLPTFF
Sbjct: 241 EIPKKDFRKGAQWFTMKRQHAVATMADSLYYSKFRDYCGPGIENNKNCIADEHYLPTFFH 300
Query: 290 IVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
++DP GIA W+VT VD SE+K HPK+Y +DIT+ELL N+
Sbjct: 301 MLDPGGIANWTVTQVDWSERKWHPKTYMPEDITHELLNNL 340
>At4g31353 unknown protein
Length = 376
Score = 364 bits (935), Expect = e-101
Identities = 170/315 (53%), Positives = 229/315 (71%), Gaps = 10/315 (3%)
Query: 25 RPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMFYSKPC-----IIDISVLAVSHI 79
RP K P WI+ LV + + + A++Y +N+ +C MF C + + ++
Sbjct: 7 RPPFKGPRWIITLVVLVTVVVITAFIYPPRNSVACYMFSGPGCPLYQQFLFVPTRELTDS 66
Query: 80 PVSVYIERGITLDMPQ----NPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA 135
+ + +++PQ NPK+AFMFLTPG+LPFE LW+ FF+GHE KFSVYVHASK
Sbjct: 67 EAAAQVVMNEIMNLPQSKTANPKLAFMFLTPGTLPFEPLWEMFFRGHENKFSVYVHASKK 126
Query: 136 KPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFN 195
PVH S YFV RDI S ++ WG++S+V+AERRLLA+AL DP+NQHF+LLSDSCVPL++FN
Sbjct: 127 SPVHTSSYFVGRDIHSHKVAWGQISMVDAERRLLAHALVDPDNQHFILLSDSCVPLFDFN 186
Query: 196 YIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMA 255
YI+++L++ + SF+D F DPGP G+GRYS+HMLPEVE KDFR G+QWFS+KR+HA+ VMA
Sbjct: 187 YIYNHLIFANLSFIDCFEDPGPHGSGRYSQHMLPEVEKKDFRKGSQWFSMKRRHAIVVMA 246
Query: 256 DHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKS 315
D LYY+KF+ C + ++G+NC DEHY PT F ++DP+GIA WSVT+VD SE K HPK
Sbjct: 247 DSLYYTKFKLYCRPN-MEGRNCYADEHYFPTLFNMIDPDGIANWSVTHVDWSEGKWHPKL 305
Query: 316 YRTQDITYELLKNIK 330
Y +DIT L++ IK
Sbjct: 306 YNARDITPYLIRKIK 320
>At4g25870 unknown protein
Length = 389
Score = 358 bits (919), Expect = 3e-99
Identities = 170/341 (49%), Positives = 242/341 (70%), Gaps = 13/341 (3%)
Query: 11 MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLY-RMQNTTSCNMFYS-KPCI 68
MG+ + +L P H +KKP+W+V+ VS + L C ++Y + ++SC+ YS + C
Sbjct: 1 MGETQKILQGP-RHHTSLKKPLWVVLTVSVTSMLLICTHMYPKHGKSSSCHGLYSTRGCE 59
Query: 69 IDISVLAVSHIPVSVYIE---RGITLDMPQNP-------KIAFMFLTPGSLPFEKLWDNF 118
+S H+ E R + D+ + P KIAF+FLTPG+LPFEKLWD F
Sbjct: 60 DALSKWLPVHVRKFTDEEIAARAVVRDILRTPPFITNNSKIAFLFLTPGTLPFEKLWDEF 119
Query: 119 FQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNN 178
F+GHEGKFS+Y+H SK +PVH+SR+F +R+I SD++ WG++S+V+AE+RLL +AL+DP+N
Sbjct: 120 FKGHEGKFSIYIHPSKERPVHISRHFSDREIHSDEVTWGRISMVDAEKRLLVSALEDPDN 179
Query: 179 QHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRT 238
QHFVL+S+SC+PL+ F+Y + YL+Y++ SF++SF DPGP G GR+ EHMLPE+ +DFR
Sbjct: 180 QHFVLVSESCIPLHTFDYTYRYLLYSNVSFIESFVDPGPHGTGRHMEHMLPEIAKEDFRK 239
Query: 239 GAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAK 298
GAQWF++KRQHA+ VMAD LYYSKF+ C KNCI DEHYLPTFF ++DP GI+
Sbjct: 240 GAQWFTMKRQHAIIVMADGLYYSKFREYCGPGIEADKNCIADEHYLPTFFNMIDPMGISN 299
Query: 299 WSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNFHLFLT 339
WSVT+VD SE++ HPK+Y +I+ E +KN+ + + +T
Sbjct: 300 WSVTFVDWSERRWHPKTYGGNEISLEFMKNVTSEDMSVHVT 340
>At5g57270 unknown protein
Length = 388
Score = 356 bits (913), Expect = 1e-98
Identities = 173/329 (52%), Positives = 232/329 (69%), Gaps = 13/329 (3%)
Query: 24 HRPQMKKPMWIVVLVSFIILFLTCAYLYRMQN---TTSCNMFYSKPCIIDISVLAVSHIP 80
HR +KKP+ IV+LV + L Y+Y N +++C S+ C +S H+
Sbjct: 11 HRSSLKKPLLIVLLVCITSVLLVITYMYPQHNNSKSSACAGLSSRGCQAALSGWLPVHVR 70
Query: 81 VSVYIE---RGITLDMPQNP-------KIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYV 130
E R + D+ + P KIAFMFLTPG+LPFEKLWD FFQG EG+FS+Y+
Sbjct: 71 KFTDEEVAARVVIKDILRLPPALTAKSKIAFMFLTPGTLPFEKLWDKFFQGQEGRFSIYI 130
Query: 131 HASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVP 190
H S+ +PVH+SR+F +R+I SD + WG++S+V+AERRLLANAL+DP+NQHFVLLS+SC+P
Sbjct: 131 HPSRLRPVHISRHFSDREIHSDHVTWGRISMVDAERRLLANALEDPDNQHFVLLSESCIP 190
Query: 191 LYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHA 250
L+ F+Y + YLM+ + SF+DSF D GP G GR+ +HMLPE+ +DFR GAQWF++KRQHA
Sbjct: 191 LHTFDYTYRYLMHANVSFIDSFEDLGPHGTGRHMDHMLPEIPRQDFRKGAQWFTMKRQHA 250
Query: 251 VKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQK 310
V VMAD LYYSKF+ C KNCI DEHYLPTFF ++DP GI+ WSVTYVD SE++
Sbjct: 251 VIVMADGLYYSKFREYCRPGVEANKNCIADEHYLPTFFHMLDPGGISNWSVTYVDWSERR 310
Query: 311 RHPKSYRTQDITYELLKNIKLHNFHLFLT 339
HPK+YR +D++ +LLKNI + + +T
Sbjct: 311 WHPKTYRARDVSLKLLKNITSDDMSVHVT 339
>At5g25970 putative protein
Length = 436
Score = 215 bits (548), Expect = 3e-56
Identities = 110/235 (46%), Positives = 145/235 (60%), Gaps = 8/235 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVNRDIRSDQL 154
PKIAFMFLT G LP LW+ +GHE +SVY+H+ S + S F R I S
Sbjct: 167 PKIAFMFLTMGPLPLAPLWERLLKGHEKLYSVYIHSPVSSSAKFPASSVFYRRHIPSQVA 226
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+M++ +AERRLLANAL D +N+ FVLLS+SC+PL+NF I+ Y+ ++ SF+ SF D
Sbjct: 227 EWGRMTMCDAERRLLANALLDISNEWFVLLSESCIPLFNFTTIYTYMTKSEHSFMGSFDD 286
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
PG G GRY +M PEV I +R G+QWF + R+ AV ++ D LYY KF+ C+ +
Sbjct: 287 PGAYGRGRYHGNMAPEVFIDQWRKGSQWFEINRELAVSIVKDTLYYPKFKEFCQPA---- 342
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
C +DEHY PT TI P +A SVT+VD S HP ++ QDI E I
Sbjct: 343 --CYVDEHYFPTMLTIEKPAALANRSVTWVDWSRGGAHPATFGAQDINEEFFARI 395
>At5g11730 unknown protein
Length = 386
Score = 210 bits (534), Expect = 1e-54
Identities = 106/228 (46%), Positives = 141/228 (61%), Gaps = 8/228 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVNRDIRSDQL 154
PK+AFMFLT G LP LW+ F +GH+G +SVY+H S S F R I S
Sbjct: 117 PKVAFMFLTKGPLPLASLWERFLKGHKGLYSVYLHPHPSFTAKFPASSVFHRRQIPSQVA 176
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+MS+ +AE+RLLANAL D +N+ FVL+S+SC+PLYNF I+ YL + SF+ +F D
Sbjct: 177 EWGRMSMCDAEKRLLANALLDVSNEWFVLVSESCIPLYNFTTIYSYLSRSKHSFMGAFDD 236
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
PGP G GRY+ +M PEV + +R G+QWF + R A ++ D LYY KF+ C +
Sbjct: 237 PGPFGRGRYNGNMEPEVPLTKWRKGSQWFEVNRDLAATIVKDTLYYPKFKEFCRPA---- 292
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDIT 322
C +DEHY PT TI P +A S+T+VD S HP ++ DIT
Sbjct: 293 --CYVDEHYFPTMLTIEKPTVLANRSLTWVDWSRGGPHPATFGRSDIT 338
>At3g21310 unknown protein
Length = 383
Score = 207 bits (527), Expect = 7e-54
Identities = 106/243 (43%), Positives = 151/243 (61%), Gaps = 10/243 (4%)
Query: 91 LDMP--QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVN 146
LD P + PK+AFMFLT G LPF LW+ FF+GHEG +S+YVH + S F
Sbjct: 106 LDYPFKRVPKMAFMFLTKGPLPFAPLWERFFKGHEGFYSIYVHTLPNYRSDFPSSSVFYR 165
Query: 147 RDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDK 206
R I S + WG+MS+ +AERRLLANAL D +N+ FVLLS++C+PL FN+++ Y+ +
Sbjct: 166 RQIPSQHVAWGEMSMCDAERRLLANALLDISNEWFVLLSEACIPLRGFNFVYRYVSRSRY 225
Query: 207 SFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQ 266
SF+ S + GP G GRYS M PEV + ++R G+QWF + R AV ++ D +YY+KF+
Sbjct: 226 SFMGSVDEDGPYGRGRYSYAMGPEVSLNEWRKGSQWFEINRALAVDIVEDMVYYNKFKEF 285
Query: 267 CEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELL 326
C C +DEHY PT +I P+ +A ++T+ D S HP ++ DIT + +
Sbjct: 286 CRPP------CYVDEHYFPTMLSIGYPDFLANRTLTWTDWSRGGAHPATFGKADITEKFI 339
Query: 327 KNI 329
K +
Sbjct: 340 KKL 342
>At1g10280 unknown protein
Length = 412
Score = 206 bits (524), Expect = 2e-53
Identities = 107/251 (42%), Positives = 155/251 (61%), Gaps = 8/251 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRY--FVNRDIRSDQL 154
PK+AFMFLT G LP LW+ FF+G+E SVYVH ++VSR F +R I S ++
Sbjct: 142 PKVAFMFLTRGPLPMLPLWEKFFKGNEKYLSVYVHTPPGYDMNVSRDSPFYDRQIPSQRV 201
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG + +AE+RLLANAL D +N+ FVLLS+SCVP+YNF+ ++ YL+ + SFVDS+ +
Sbjct: 202 EWGSPLLTDAEKRLLANALLDFSNERFVLLSESCVPVYNFSTVYTYLINSAYSFVDSYDE 261
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
P G GRYS MLP++++ +R G+QWF + R+ A+ +++D YYS F+ C +C
Sbjct: 262 PTRYGRGRYSRKMLPDIKLHHWRKGSQWFEVNRKIAIYIISDSKYYSLFKQFCRPACYP- 320
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNF 334
DEHY+PTF + + A SVT+VD S HP +Y +IT L++I+ +
Sbjct: 321 -----DEHYIPTFLNMFHGSMNANRSVTWVDWSIGGPHPATYAAANITEGFLQSIRKNET 375
Query: 335 HLFLTERSTKM 345
E T +
Sbjct: 376 DCLYNEEPTSL 386
>At5g16170 unknowen protein
Length = 411
Score = 204 bits (519), Expect = 6e-53
Identities = 108/256 (42%), Positives = 150/256 (58%), Gaps = 8/256 (3%)
Query: 77 SHIPVSVYIERGITLDMPQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA- 135
S + S ++ R D K+AFMF+T G LP LW+ FF+GHEG +S+YVH + +
Sbjct: 115 SSLSSSSWLGRRHNNDGKMAVKVAFMFMTGGRLPLAGLWEKFFEGHEGFYSIYVHTNPSF 174
Query: 136 -KPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNF 194
+ F +R I S + WG S+V+AE+RLLANAL D +NQ FVLLSDSC+PLYNF
Sbjct: 175 QDSFPETSVFYSRRIPSQPVYWGTSSMVDAEKRLLANALLDESNQRFVLLSDSCIPLYNF 234
Query: 195 NYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVM 254
I+DYL T+ SF+ SF DP G GRY+ M P + I +R G+QWF R+ A+ ++
Sbjct: 235 TTIYDYLTGTNLSFIGSFDDPRKSGRGRYNHTMYPHINITHWRKGSQWFETTRELALHII 294
Query: 255 ADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPK 314
D +YY F C+ C +DEHY+PT ++ A ++T+VD S+ HP
Sbjct: 295 EDTVYYKIFDQHCKPP------CYMDEHYIPTLVHMLHGEMSANRTLTWVDWSKAGPHPG 348
Query: 315 SYRTQDITYELLKNIK 330
+ DIT E L I+
Sbjct: 349 RFIWPDITDEFLNRIR 364
>At1g10880 hypothetical protein
Length = 651
Score = 203 bits (516), Expect = 1e-52
Identities = 100/237 (42%), Positives = 147/237 (61%), Gaps = 8/237 (3%)
Query: 96 NPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASK--AKPVHVSRYFVNRDIRSDQ 153
+PK+AFMFLT +LP LW+ FF+GHEG +S+YVH S + S F + I S
Sbjct: 116 HPKVAFMFLTRWNLPLSPLWEMFFKGHEGFYSIYVHTSPEFTQEPPESSVFYKKRIPSKA 175
Query: 154 LVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFR 213
+ WGK S+++AE+RL+++AL +P+N FVLLS++C+PL+NF I+ YL + +SF+ SF
Sbjct: 176 VEWGKCSMMDAEKRLISHALLEPSNARFVLLSETCIPLFNFTTIYTYLTRSTRSFLGSFD 235
Query: 214 DPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVD 273
DP P+G GRY+ MLP V + D+R G QWF + R+ A ++++D YY+ F+ C
Sbjct: 236 DPRPMGRGRYTPKMLPHVSLSDWRKGNQWFEISRRVAAEIVSDRRYYAVFKDHCRPP--- 292
Query: 274 GKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIK 330
C +DEHYLPT + P + +VT+VD S HP + +DI L I+
Sbjct: 293 ---CYIDEHYLPTLVNKICPEMNSNRTVTWVDWSRGGSHPARFVRKDIRVGFLDRIR 346
>At1g73810 unknown protein (At1g73810)
Length = 418
Score = 199 bits (506), Expect = 2e-51
Identities = 103/239 (43%), Positives = 144/239 (60%), Gaps = 14/239 (5%)
Query: 98 KIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRY------FVNRDIRS 151
K AFMFLT G LP KLW+ FF+GHEG FS+Y+H S P + + F R I S
Sbjct: 147 KAAFMFLTRGKLPLAKLWERFFKGHEGLFSIYIHTSD--PFYFDDHTPETSPFYRRRIPS 204
Query: 152 DQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDS 211
++ WG +S+V AERRLLANAL D N FVLLS+S +PL+NF+ I+ YL+ + S+VD
Sbjct: 205 KEVGWGMVSMVAAERRLLANALLDAGNHRFVLLSESDIPLFNFSTIYSYLINSQHSYVDV 264
Query: 212 FRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSC 271
+ PGP G GRY+ M P + ++R G+QWF + R+ A+ V++D Y+ F+ C
Sbjct: 265 YDLPGPAGRGRYNRRMSPVISRTNWRKGSQWFEIDREVALAVVSDTTYFPVFEKYCLW-- 322
Query: 272 VDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIK 330
NC DEHYL TF + P A S+T+ D S + HP+ Y + +T E L+ ++
Sbjct: 323 ----NCYADEHYLSTFVHAMFPGKNANRSLTWTDWSRRGPHPRKYTRRSVTGEFLRRVR 377
>At1g51770 hypothetical protein
Length = 399
Score = 194 bits (494), Expect = 5e-50
Identities = 99/235 (42%), Positives = 142/235 (60%), Gaps = 8/235 (3%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSR--YFVNRDIRSDQL 154
PK+AFMFL G LPF LW+ F +GHEG +S+YVH+ + SR F R I S +
Sbjct: 116 PKLAFMFLAKGPLPFAPLWEKFCKGHEGLYSIYVHSLPSYKSDFSRSSVFYRRYIPSQAV 175
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+MS+ EAERRLLANAL D +N+ FVLLS+SC+PL F++I+ Y+ + SF+ + +
Sbjct: 176 AWGEMSMGEAERRLLANALLDISNEWFVLLSESCIPLRGFSFIYSYVSESRYSFMGAADE 235
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDG 274
GP G GRY M PE+ + +R G+QWF + R+ AV+++ D YY KF+ C
Sbjct: 236 EGPDGRGRYRTEMEPEITLSQWRKGSQWFEINRKLAVEIVQDTTYYPKFKEFCRPP---- 291
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
C +DEHY PT ++ +A ++T+ D S HP ++ D+T LK +
Sbjct: 292 --CYVDEHYFPTMLSMKHRVLLANRTLTWTDWSRGGAHPATFGKADVTESFLKKL 344
>At1g68390 hypothetical protein
Length = 408
Score = 194 bits (492), Expect = 8e-50
Identities = 99/259 (38%), Positives = 157/259 (60%), Gaps = 10/259 (3%)
Query: 94 PQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVSRYFVNRDIRS 151
P+ PK+AFMF+T G LP +LW+ FF+GHEG F++YVH+ S + F R I S
Sbjct: 134 PRTPKVAFMFMTKGHLPLARLWERFFRGHEGLFTIYVHSYPSYNQSDPEDSVFRGRHIPS 193
Query: 152 DQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDS 211
++ WG +++VEAE+RLLANAL D +N+ FVLLS+SC+PL+NF ++ YL+ + ++ V+S
Sbjct: 194 KRVDWGYVNMVEAEQRLLANALLDISNERFVLLSESCIPLFNFTTVYSYLINSTQTHVES 253
Query: 212 FRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSC 271
+ G VG GRYS M P V+++ +R G+QW + R A+++++D +Y+ F + C
Sbjct: 254 YDQLGGVGRGRYSPLMQPHVQLRHWRKGSQWIEVDRAMALEIISDRIYWPLFYSYCHH-- 311
Query: 272 VDGKNCILDEHYLPTFFTIVD--PNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
C DEHY+PT I + ++T+VD S+ HP + ++T E ++N+
Sbjct: 312 ----GCYADEHYIPTLLNIKSSLKRRNSNRTLTWVDWSKGGPHPNRFIRHEVTAEFMENL 367
Query: 330 KLHNFHLFLTERSTKMDLF 348
+ L+ E + LF
Sbjct: 368 RSGGECLYNGEETNICYLF 386
>At1g68380 unknown protein
Length = 392
Score = 177 bits (449), Expect = 8e-45
Identities = 92/242 (38%), Positives = 147/242 (60%), Gaps = 15/242 (6%)
Query: 94 PQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSR--YFVNRDIRS 151
P+ PK+AFMFLT G LP LW+ FF+GHEG F++YVH + + + + F R I S
Sbjct: 120 PRIPKVAFMFLTWGPLPLAPLWERFFRGHEGLFTIYVHTNSSYDEFMPQDSVFYGRRIPS 179
Query: 152 DQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDS 211
++ WG ++VEAERRLLANAL D NN+ F+LLS+SC+PL+NF+ ++ +L+ + + VDS
Sbjct: 180 KRVDWGNANMVEAERRLLANALLDINNERFILLSESCIPLFNFSTVYSFLIDSTLTHVDS 239
Query: 212 FRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSC 271
+ +G RY M P + + +R G+QWF L R A++V++D Y+ F+A
Sbjct: 240 Y--DLTIGRVRYDRRMYPHIRMHQWRKGSQWFELDRAMALEVVSDTFYWPIFKAYSR--- 294
Query: 272 VDGKNCILDEHYLPTFFTIVDPNGI--AKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
DEHY+PT + G+ A ++T+ D S+++ HP+ + ++ E L+ +
Sbjct: 295 ------CPDEHYIPTLLNMRPSLGLRNANRTLTWTDWSKRRAHPRLFGEWEVNVEFLEWL 348
Query: 330 KL 331
++
Sbjct: 349 RM 350
>At5g14550 putative protein
Length = 364
Score = 176 bits (446), Expect = 2e-44
Identities = 101/256 (39%), Positives = 158/256 (61%), Gaps = 24/256 (9%)
Query: 93 MPQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHV----SRYFVNRD 148
+ Q P+IAF+F+ LP E +WD FF+G +GKFS+YVH+ ++ S+YF++R
Sbjct: 60 LDQRPQIAFLFIARNRLPLEFVWDAFFKGEDGKFSIYVHSRPGFVLNEATTRSKYFLDRQ 119
Query: 149 IR-SDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKS 207
+ S Q+ WG+ +++EAER LL +AL+D N FV LSDSC+PLY+F+Y ++Y+M T S
Sbjct: 120 LNDSIQVDWGESTMIEAERVLLRHALRDSFNHRFVFLSDSCIPLYSFSYTYNYIMSTPTS 179
Query: 208 FVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQC 267
FVDSF D + RY+ M P + ++++R G+QW L R+HA V+ D + FQ C
Sbjct: 180 FVDSFAD---TKDSRYNPRMNPIIPVRNWRKGSQWVVLNRKHAEIVVNDTSVFPMFQQHC 236
Query: 268 EQSCVDG---KNCILDEHYLPTFFTI--VDPNGIAKWSVTY------VDRSEQKR--HPK 314
+ +G NCI DEHY+ T + VD + + + S+T+ +S ++R HP
Sbjct: 237 RPA--EGWKEHNCIPDEHYVQTLLSQKGVD-SELTRRSLTHSAWDLSSSKSNERRGWHPM 293
Query: 315 SYRTQDITYELLKNIK 330
+Y+ D T +L+++IK
Sbjct: 294 TYKFSDATPDLIQSIK 309
>At3g52060 unknown protein
Length = 346
Score = 163 bits (412), Expect = 2e-40
Identities = 93/260 (35%), Positives = 153/260 (58%), Gaps = 14/260 (5%)
Query: 87 RGITLDMPQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA---SKAKPVHVSRY 143
R I+L PKIAF+FLT L F LW++FFQGH+ ++VY+HA S P+ S
Sbjct: 64 RLISLSPNPPPKIAFLFLTNSDLTFLPLWESFFQGHQDLYNVYIHADPTSSVSPLLDSSS 123
Query: 144 FVNRDIRSDQLVWGKMSIVEAERRLLANA-LQDPNNQHFVLLSDSCVPLYNFNYIFDYLM 202
+ I + + +++ AERRLLANA L DPNN +F L+S C+PL++F+YI ++L
Sbjct: 124 INAKFIPARRTARASPTLISAERRLLANAILDDPNNLYFALISQHCIPLHSFSYIHNHLF 183
Query: 203 --YTDKSFVDSFRDPGPV---GNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADH 257
+ +SF++ D + N R + MLPE++ +DFR G+Q+F L ++HA+ V+ +
Sbjct: 184 SDHHQQSFIEILSDEPFLLKRYNARGDDAMLPEIQYQDFRVGSQFFVLAKRHALMVIKER 243
Query: 258 LYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVD-RSEQKRHPKSY 316
+ KF+ C+D ++C +EHY PT ++ DP G + +++T V+ HP +Y
Sbjct: 244 KLWRKFKL----PCLDVESCYPEEHYFPTLLSLEDPQGCSHFTLTRVNWTGSVGGHPHTY 299
Query: 317 RTQDITYELLKNIKLHNFHL 336
+I+ +L+ +++ N L
Sbjct: 300 DASEISPQLIHSLRRSNSSL 319
>At1g11940 unknown protein
Length = 383
Score = 158 bits (399), Expect = 5e-39
Identities = 95/274 (34%), Positives = 148/274 (53%), Gaps = 29/274 (10%)
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHE-GKFSVYVHASKA----KPVHVSRYFVNRDIRS 151
PK+AF+FL LP + +WD FF+G + FS+Y+H+ + S+YF NR + +
Sbjct: 73 PKLAFLFLARRDLPLDFMWDRFFKGVDHANFSIYIHSVPGFVFNEETTRSQYFYNRQLNN 132
Query: 152 D-QLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVD 210
++VWG+ S++EAER LLA+AL+D +NQ FVLLSD C PLY+F YI+ YL+ + +SFVD
Sbjct: 133 SIKVVWGESSMIEAERLLLASALEDHSNQRFVLLSDRCAPLYDFGYIYKYLISSPRSFVD 192
Query: 211 SFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQS 270
SF RYS M P + + +R G+QW +L R HA ++ D + + F+ C++
Sbjct: 193 SFLH---TKETRYSVKMSPVIPEEKWRKGSQWIALIRSHAEVIVNDGIVFPVFKEFCKRC 249
Query: 271 CVDG------------KNCILDEHYLPTFFTIVDPNG--------IAKWSVTYVDRSEQK 310
G +NCI DEHY+ T T+ W+V+ +
Sbjct: 250 PPLGTNEAWLFLKQKRRNCIPDEHYVQTLLTMQGLESEMERRTVTYTVWNVSGTKYEAKS 309
Query: 311 RHPKSYRTQDITYELLKNIKLHNFHLFLTERSTK 344
HP ++ ++ E +K IK + + +E T+
Sbjct: 310 WHPVTFTLENSGPEEIKEIKKIDHVYYESESRTE 343
>At5g25330 putative protein
Length = 366
Score = 150 bits (380), Expect = 8e-37
Identities = 93/246 (37%), Positives = 144/246 (57%), Gaps = 19/246 (7%)
Query: 98 KIAFMFLTPGSLPFEKLWDNFFQG---HEGKFSVYVHASKAKPVHVSRY--FVNRDIRSD 152
K+AFMFLT SLP LW+ FF H+ ++VYVH + + F NR I S
Sbjct: 81 KLAFMFLTTNSLPLAPLWELFFNQSSHHKSLYNVYVHVDPTQKHKPGSHGTFQNRIIPSS 140
Query: 153 QLVWGKM-SIVEAERRLLANAL-QDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVD 210
+ + +++ A RRLLA+AL +DP+N F+LLS SC+PL++FN+ + L+ + KSF++
Sbjct: 141 KPAYRHTPTLISAARRLLAHALLEDPSNYMFILLSPSCIPLHSFNFTYKTLVSSTKSFIE 200
Query: 211 SFRD-PG----PVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQA 265
+D PG G Y+ M PEV ++FR G+Q+++L R HA+ V++D +SKF
Sbjct: 201 ILKDEPGWYERWAARGPYA--MFPEVPPEEFRIGSQFWTLTRAHALMVVSDVEIWSKF-- 256
Query: 266 QCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRS-EQKRHPKSYRTQDITYE 324
+SCV C +EHY PT + DP G +VT+VD S HP++Y+ ++ E
Sbjct: 257 --NKSCVREDICYPEEHYFPTLLNMRDPQGCVSATVTHVDWSVNDHGHPRTYKPLEVRAE 314
Query: 325 LLKNIK 330
L++ ++
Sbjct: 315 LIQKLR 320
>At4g32290 unknown protein
Length = 384
Score = 141 bits (355), Expect = 6e-34
Identities = 88/242 (36%), Positives = 137/242 (56%), Gaps = 14/242 (5%)
Query: 98 KIAFMFLTPGSLPFEKLWDNFFQG-HEGKFSVYVHASKAKPVH--VSRYFVNRDIRSDQL 154
KIAFM+LT LPF LW+ FF G + ++VYVHA + S F+NR I S
Sbjct: 93 KIAFMYLTTSPLPFAPLWEMFFDGISKNLYNVYVHADPTREYDPPFSGVFLNRVIHSKPS 152
Query: 155 VWGKMSIVEAERRLLANAL-QDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFR 213
+ ++ A RRLLA+AL DP N F ++S SCVP+ +F++ + L+ + KSF++ +
Sbjct: 153 LRHTPTLTAAARRLLAHALLDDPLNYMFAVISPSCVPIRSFDFTYKTLVSSRKSFIEILK 212
Query: 214 DPGPVGNGRYS----EHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQ 269
D P R++ MLPEV++++FR G+Q++ LKR+HA V D + KF Q
Sbjct: 213 DE-PWQFDRWTAIGRHAMLPEVKLEEFRIGSQFWVLKRRHARVVARDRRIWVKF----NQ 267
Query: 270 SCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRS-EQKRHPKSYRTQDITYELLKN 328
+CV +C +E Y PT + DP G ++T+VD + HP+ Y +++ EL+
Sbjct: 268 TCVREDSCYPEESYFPTLLNMRDPRGCVPATLTHVDWTVNDGGHPRMYEPEEVVPELVLR 327
Query: 329 IK 330
++
Sbjct: 328 LR 329
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,456,064
Number of Sequences: 26719
Number of extensions: 367664
Number of successful extensions: 978
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 899
Number of HSP's gapped (non-prelim): 32
length of query: 359
length of database: 11,318,596
effective HSP length: 100
effective length of query: 259
effective length of database: 8,646,696
effective search space: 2239494264
effective search space used: 2239494264
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149130.7


