

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.3 - phase: 0 /pseudo
(171 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
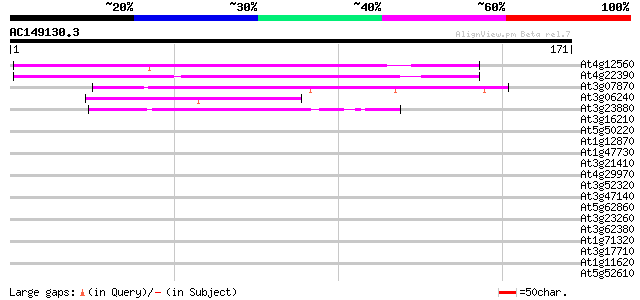
Score E
Sequences producing significant alignments: (bits) Value
At4g12560 putative protein 106 7e-24
At4g22390 unknown protein 72 2e-13
At3g07870 unknown protein 53 7e-08
At3g06240 unknown protein 46 1e-05
At3g23880 unknown protein 43 7e-05
At3g16210 hypothetical protein 38 0.002
At5g50220 putative protein 34 0.034
At1g12870 hypothetical protein 34 0.034
At1g47730 hypothetical protein 33 0.075
At3g21410 hypothetical protein 32 0.13
At4g29970 putative protein 31 0.29
At3g52320 putative protein 30 0.83
At3g47140 putative protein 29 1.4
At5g62860 unknown protein 28 1.9
At3g23260 unknown protein 28 1.9
At3g62380 putative protein 28 2.4
At1g71320 hypothetical protein 28 2.4
At3g17710 unknown protein 28 3.2
At1g11620 hypothetical protein 28 3.2
At5g52610 putative protein 27 5.4
>At4g12560 putative protein
Length = 408
Score = 106 bits (264), Expect = 7e-24
Identities = 56/143 (39%), Positives = 83/143 (57%), Gaps = 8/143 (5%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLP-AELQGETSLLIDVAVLGGC 60
GV NS+ ++ ++ + +LIV F+L LE F V P A G + +D+ VL GC
Sbjct: 212 GVLAGNSLHWVLPRRPGLIAFNLIVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDGC 271
Query: 61 LCITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFEG 120
LC+ NY+ + +DVW+MKEY RDSW K+FT+ K Y P+ YS D K
Sbjct: 272 LCLMCNYDQSYVDVWMMKEYNVRDSWTKVFTVQKPKSVKSFSYMRPLVYSKDKKK----- 326
Query: 121 IEVLLDVHYKKLFWYDLKSERVS 143
VLL+++ KL W+DL+S+++S
Sbjct: 327 --VLLELNNTKLVWFDLESKKMS 347
>At4g22390 unknown protein
Length = 401
Score = 71.6 bits (174), Expect = 2e-13
Identities = 42/142 (29%), Positives = 73/142 (50%), Gaps = 8/142 (5%)
Query: 2 GVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCL 61
GV V N + I+ ++ + + I+ ++L + + P EL E ++ D+ VL GC+
Sbjct: 211 GVVVNNHLHWILPRRQGVIAFNAIIKYDLASDDIGVLSFPQELYIEDNM--DIGVLDGCV 268
Query: 62 CITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFEGI 121
C+ E + +DVWV+KEY SW KL+ + K + + P+ S D SK+L E
Sbjct: 269 CLMCYDEYSHVDVWVLKEYEDYKSWTKLYRVPKPESVESVEFIRPLICSKDRSKILLE-- 326
Query: 122 EVLLDVHYKKLFWYDLKSERVS 143
+ L W+DL+S+ ++
Sbjct: 327 ----INNAANLMWFDLESQSLT 344
>At3g07870 unknown protein
Length = 417
Score = 53.1 bits (126), Expect = 7e-08
Identities = 43/137 (31%), Positives = 63/137 (45%), Gaps = 11/137 (8%)
Query: 26 VAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRDS 85
V+F+L E F E+P P + G + L GCLC V K+D+WVMK YG ++S
Sbjct: 263 VSFDLEDEEFKEIPKP-DCGGLNRTNHRLVNLKGCLCAVVYGNYGKLDIWVMKTYGVKES 321
Query: 86 WCKLF---TLVKSCFTSHLRYSNPIGYSSDGSKV-----LFEGIEVLLDVHYKKLFWYDL 137
W K + T + +L I +++ KV L E E+LL+ + L YD
Sbjct: 322 WGKEYSIGTYLPKGLKQNLDRPMWIWKNAENGKVVRVLCLLENGEILLEYKSRVLVAYDP 381
Query: 138 KSERVS--YVEGIPKLF 152
K + G+P F
Sbjct: 382 KLGKFKDLLFHGLPNWF 398
>At3g06240 unknown protein
Length = 427
Score = 45.8 bits (107), Expect = 1e-05
Identities = 23/68 (33%), Positives = 37/68 (53%), Gaps = 2/68 (2%)
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAV--LGGCLCITVNYETTKIDVWVMKEYG 81
++VAF++ E F E+P+P E + + + V L G LC+ + D+WVM EYG
Sbjct: 288 VVVAFDIQTEEFREMPVPDEAEDCSHRFSNFVVGSLNGRLCVVNSCYDVHDDIWVMSEYG 347
Query: 82 YRDSWCKL 89
SW ++
Sbjct: 348 EAKSWSRI 355
>At3g23880 unknown protein
Length = 364
Score = 43.1 bits (100), Expect = 7e-05
Identities = 31/95 (32%), Positives = 49/95 (50%), Gaps = 7/95 (7%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRD 84
I++++++ + F E+P P G + + L GCL + + DVWVMKE+G
Sbjct: 235 IISYDMSRDEFKELPGPV-CCGRGCFTMTLGDLRGCLSMVCYCKGANADVWVMKEFGEVY 293
Query: 85 SWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFE 119
SW KL ++ T +R P+ + SDG VL E
Sbjct: 294 SWSKLLSI--PGLTDFVR---PL-WISDGLVVLLE 322
>At3g16210 hypothetical protein
Length = 360
Score = 38.1 bits (87), Expect = 0.002
Identities = 30/77 (38%), Positives = 41/77 (52%), Gaps = 8/77 (10%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTK--IDVWVMKEYGY 82
I+ FNL+ + ++PLP QG TS + V LCIT YE K I + VM++ G
Sbjct: 206 ILCFNLSTHEYRKLPLPVYNQGVTSSWL--GVTSQKLCIT-EYEMCKKEIRISVMEKTG- 261
Query: 83 RDSWCKLFTLVKSCFTS 99
SW K+ +L S F S
Sbjct: 262 --SWSKIISLSMSSFIS 276
>At5g50220 putative protein
Length = 357
Score = 34.3 bits (77), Expect = 0.034
Identities = 27/124 (21%), Positives = 54/124 (42%), Gaps = 13/124 (10%)
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYR 83
+++ F++ E F++V P + LI+ G +C ++++WVMK +
Sbjct: 219 VLLCFDVISEKFDQVEAPKTMMDHRYTLINYQGKLGFMCCQ-----NRVEIWVMKNDEKK 273
Query: 84 DSWCKLFTLVKSCFTS-HLRYSNPIGYSSDGSKVLFEGIEVLLDVHYKKLFWYDLKSERV 142
W K+F + F H+ + P G +++F +LL +++Y K +
Sbjct: 274 QEWSKIFFYEMAGFEKWHIARATPSG------EIVFVN-RLLLSCQTLYVYYYGPKRNSM 326
Query: 143 SYVE 146
VE
Sbjct: 327 RRVE 330
>At1g12870 hypothetical protein
Length = 416
Score = 34.3 bits (77), Expect = 0.034
Identities = 18/70 (25%), Positives = 36/70 (50%), Gaps = 4/70 (5%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKI--DVWVMKEYGY 82
++A ++ EIF +P P+ + ID+ ++ LC+ KI ++W +K
Sbjct: 260 VIALDIHTEIFRLLPKPSLIASSEPSHIDMCIIDNSLCMYETEGDKKIIQEIWRLKSS-- 317
Query: 83 RDSWCKLFTL 92
D+W K++T+
Sbjct: 318 EDAWEKIYTI 327
>At1g47730 hypothetical protein
Length = 391
Score = 33.1 bits (74), Expect = 0.075
Identities = 20/93 (21%), Positives = 43/93 (45%), Gaps = 2/93 (2%)
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYR 83
+I++F++ E F+ + LP + L++ ++ G C+ N+E +WV++ R
Sbjct: 239 VILSFDVRSERFDVIELPWD--ENFGLVMMMSYKGRLACLGFNHEKNSRSMWVLENVEQR 296
Query: 84 DSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKV 116
+ C + + Y N G ++DG +
Sbjct: 297 EWSCHTYLPISHYEPGLENYFNLTGITNDGELI 329
>At3g21410 hypothetical protein
Length = 374
Score = 32.3 bits (72), Expect = 0.13
Identities = 27/108 (25%), Positives = 46/108 (42%), Gaps = 6/108 (5%)
Query: 25 IVAFNLTLEIFNEV-PLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEY--G 81
I +FN + E F + LP + + + + ET+KIDVWV + G
Sbjct: 212 IQSFNFSTETFEPLCSLPVRYDVHNVVALSAFKGDNLSLLHQSKETSKIDVWVTNKVKNG 271
Query: 82 YRDSWCKLFTLVKSCFTSHLRYSN---PIGYSSDGSKVLFEGIEVLLD 126
W KLF++ + L + N P+ + ++++ EVL D
Sbjct: 272 VSILWTKLFSVTRPDLPVLLAFENLSYPVHFIDKNNRIVVCCEEVLAD 319
>At4g29970 putative protein
Length = 283
Score = 31.2 bits (69), Expect = 0.29
Identities = 17/64 (26%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRD 84
+++FNL E FN + P +++ S ++ G + +T +D+WVMK+ +
Sbjct: 156 LISFNLRSEEFNVIKFPKDVKHIWSS--NLVNYNGKIALTSYSCNGTLDLWVMKDDASKQ 213
Query: 85 SWCK 88
W K
Sbjct: 214 EWFK 217
>At3g52320 putative protein
Length = 390
Score = 29.6 bits (65), Expect = 0.83
Identities = 31/148 (20%), Positives = 64/148 (42%), Gaps = 11/148 (7%)
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYR 83
++++F+L+ E FN + LP E T + + C + + + +DV V+++ +
Sbjct: 243 VLMSFDLSSEEFNLIELPFENWSRTIHMNYQGKVATCQYMRLASDGF-VDVCVLEDAD-K 300
Query: 84 DSWCKLFTLVKSCFTSHLRYSNP--IGYSSDGSKVLFEGIEVLLDVHYKKLFWYDLKSER 141
W T V + + + +G S D KVL +L + H + F YD++
Sbjct: 301 SQWSNKKTFVLPISQMNFVHGDRLVVGASRDSGKVLMRKANLLRNQH-ARFFLYDMERNE 359
Query: 142 VSYVEGIPKLFFFKKPMISCFKKQMRFL 169
++ + + ++ F K +FL
Sbjct: 360 IA------RRIEIRPSLLGSFNKTNQFL 381
>At3g47140 putative protein
Length = 224
Score = 28.9 bits (63), Expect = 1.4
Identities = 33/140 (23%), Positives = 65/140 (45%), Gaps = 15/140 (10%)
Query: 23 SLIVAFNLTLEIFNEVPLPAELQG---------ETSLLIDVAVLG--GCLCITVNYETTK 71
S+IV+F++ LE FN + +P++L +T + D ++ G + + +
Sbjct: 76 SIIVSFDVRLETFNIINVPSKLLPVDYENMWVVKTLIPTDKTLINYKGKVSVVEHPREGA 135
Query: 72 IDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFEGIEVLLDVHYKK 131
+WV+++ + F L +S + N Y+ + V + ++D H+
Sbjct: 136 FRMWVVEDVEKEEWSMNTFHLHESVVGLDFKVMNTF-YTGEICLVPKVLLTSIVDDHF-C 193
Query: 132 LFWYDL--KSERVSYVEGIP 149
LF+Y+L KS R +EG+P
Sbjct: 194 LFYYNLERKSMRSVTIEGLP 213
>At5g62860 unknown protein
Length = 377
Score = 28.5 bits (62), Expect = 1.9
Identities = 17/79 (21%), Positives = 34/79 (42%), Gaps = 13/79 (16%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSL---------LIDVAVLGGCLCITVNYETTKIDVW 75
+V F++ E F+ + +P LQ L L++ GC + Y ++W
Sbjct: 130 LVRFDVRHESFDRIQMPITLQMNQQLSEVSFDELTLVNYQGKLGC----IRYTKASAEMW 185
Query: 76 VMKEYGYRDSWCKLFTLVK 94
+M+++ + W K+ K
Sbjct: 186 IMEDHIEQQEWSKMMIFEK 204
>At3g23260 unknown protein
Length = 362
Score = 28.5 bits (62), Expect = 1.9
Identities = 23/72 (31%), Positives = 34/72 (46%), Gaps = 8/72 (11%)
Query: 26 VAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGG---CLCITVNYETTKID--VWVMKEY 80
++F+ + E F + LP Q L+ D++V+ CL N+ TT D VWV
Sbjct: 221 LSFDFSTERFQSLSLP---QPFPYLVTDLSVVREEQLCLFGYYNWSTTSEDLNVWVTTSL 277
Query: 81 GYRDSWCKLFTL 92
G SW K T+
Sbjct: 278 GSVVSWSKFLTI 289
>At3g62380 putative protein
Length = 325
Score = 28.1 bits (61), Expect = 2.4
Identities = 17/68 (25%), Positives = 33/68 (48%), Gaps = 1/68 (1%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRD 84
+ A +L E F VPLP+ + + + L LC++ + +DVW +++
Sbjct: 204 LAAIDLHTETFRYVPLPSWYT-KYCKSVYLWSLKDSLCVSDVLQNPSVDVWSLQQEEPSV 262
Query: 85 SWCKLFTL 92
W K+F++
Sbjct: 263 KWEKMFSV 270
>At1g71320 hypothetical protein
Length = 392
Score = 28.1 bits (61), Expect = 2.4
Identities = 22/90 (24%), Positives = 44/90 (48%), Gaps = 2/90 (2%)
Query: 3 VFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLC 62
V+ S+ + KKL L+ A +L E F + LP E + + I++ L LC
Sbjct: 249 VYANGSLFWLTLKKLSQTSYQLL-AIDLHTEEFRWILLP-ECDTKYATNIEMWNLNERLC 306
Query: 63 ITVNYETTKIDVWVMKEYGYRDSWCKLFTL 92
++ E++ + VW + + + W K++++
Sbjct: 307 LSDVLESSNLVVWSLHQEYPTEKWEKIYSI 336
>At3g17710 unknown protein
Length = 368
Score = 27.7 bits (60), Expect = 3.2
Identities = 23/73 (31%), Positives = 33/73 (44%), Gaps = 5/73 (6%)
Query: 25 IVAFNLTLEIFNEVPL-PAELQ-GETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGY 82
I F+ ++EIF L P G L++ V + +ETTKI++WV K
Sbjct: 219 IETFDFSMEIFKPFCLLPCRKDFGSNELVLAVFKEDRFSLLKQCFETTKIEIWVTKMKID 278
Query: 83 RDS---WCKLFTL 92
R+ W K TL
Sbjct: 279 REEEVVWIKFMTL 291
>At1g11620 hypothetical protein
Length = 363
Score = 27.7 bits (60), Expect = 3.2
Identities = 20/72 (27%), Positives = 38/72 (52%), Gaps = 8/72 (11%)
Query: 25 IVAFNLTLEIFNEVPLPAELQGETSLLIDVAV---LGGCLCITVN-YETTKIDVWVMKEY 80
I +F+ + E F + LP + G +L+ +++ G L + + +ET K+ +WVMK
Sbjct: 224 IQSFDFSKERFEPLFLPPQSIGSRNLVKYISLGIFRGDQLSLLLECHETCKLHLWVMK-- 281
Query: 81 GYRDSWCKLFTL 92
+ W +L T+
Sbjct: 282 --KQHWSRLMTV 291
>At5g52610 putative protein
Length = 351
Score = 26.9 bits (58), Expect = 5.4
Identities = 17/84 (20%), Positives = 39/84 (46%), Gaps = 5/84 (5%)
Query: 59 GCLCITVNYETTKIDVWVMKEYGYRDSWCKL-FTLVKSCFTSHLRYSNPIGYSSDGSKVL 117
G L + + D+WV+++ G + W K+ L + F+ L + +G+ + +++
Sbjct: 241 GKLALLAAKSMSMYDLWVLEDAG-KQEWSKVSIVLTREMFSYDLVWLGAVGFVAGSDELI 299
Query: 118 FEGIEVLLDVHYKKLFWYDLKSER 141
+ ++ L + DLK +R
Sbjct: 300 VTAHDRFYQIY---LVYVDLKMKR 320
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.143 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,924,764
Number of Sequences: 26719
Number of extensions: 162815
Number of successful extensions: 463
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 446
Number of HSP's gapped (non-prelim): 29
length of query: 171
length of database: 11,318,596
effective HSP length: 92
effective length of query: 79
effective length of database: 8,860,448
effective search space: 699975392
effective search space used: 699975392
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149130.3


