

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.2 + phase: 0
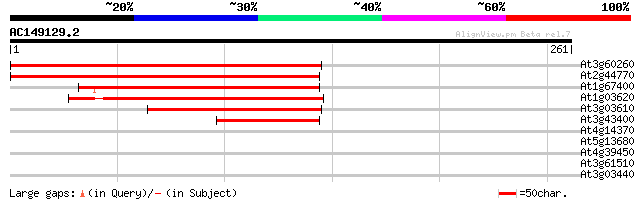
(261 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g60260 unknown protein 248 2e-66
At2g44770 unknown protein 243 1e-64
At1g67400 unknown protein 140 7e-34
At1g03620 hypothetical protein 135 2e-32
At3g03610 unknown protein 115 3e-26
At3g43400 putative protein 75 5e-14
At4g14370 disease resistance N like protein 31 0.59
At5g13680 putative protein 28 3.8
At4g39450 hypothetical protein 28 5.0
At3g61510 1-aminocyclopropane-1-carboxylate synthase-like protein 27 8.5
At3g03440 unknown protein 27 8.5
>At3g60260 unknown protein
Length = 266
Score = 248 bits (634), Expect = 2e-66
Identities = 115/145 (79%), Positives = 129/145 (88%)
Query: 1 MDDRGGSFVAVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFD 60
MDDRGGSFVAVRRISQGL+RG+ YHSSSAE V GS AW+GRGLSCVC Q R+ D R SFD
Sbjct: 1 MDDRGGSFVAVRRISQGLERGSVYHSSSAEVVAGSAAWLGRGLSCVCVQGRDGDPRPSFD 60
Query: 61 LTPYQEECLQRLQSRIDVPYDSSIPEHQASLRALWNAAFPEEELNGLISEQWKDMGWQGK 120
LTP QEECLQRLQSRIDV YDSSIP+HQ +L+ LW AFPEEEL+G++S+QWK+MGWQGK
Sbjct: 61 LTPAQEECLQRLQSRIDVAYDSSIPQHQEALKDLWKLAFPEEELHGIVSDQWKEMGWQGK 120
Query: 121 DPSTDFRGGGYISLENLLFFARNFP 145
DPSTDFRGGG+ISLENLL+FAR FP
Sbjct: 121 DPSTDFRGGGFISLENLLYFARKFP 145
>At2g44770 unknown protein
Length = 266
Score = 243 bits (619), Expect = 1e-64
Identities = 115/144 (79%), Positives = 129/144 (88%)
Query: 1 MDDRGGSFVAVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFD 60
MDDR GSFVAVRRISQGL+RG+ Y+SSSAE V GS AW+GRGLSCVCAQRR+SDA +FD
Sbjct: 1 MDDREGSFVAVRRISQGLERGSVYNSSSAEAVPGSAAWLGRGLSCVCAQRRDSDANSTFD 60
Query: 61 LTPYQEECLQRLQSRIDVPYDSSIPEHQASLRALWNAAFPEEELNGLISEQWKDMGWQGK 120
LTP QEECLQ LQ+RIDV YDS+IP HQ +LR LW +FPEEEL+GLISEQWK+MGWQGK
Sbjct: 61 LTPAQEECLQSLQNRIDVAYDSTIPLHQEALRELWKLSFPEEELHGLISEQWKEMGWQGK 120
Query: 121 DPSTDFRGGGYISLENLLFFARNF 144
DPSTDFRGGG+ISLENLL+FARNF
Sbjct: 121 DPSTDFRGGGFISLENLLYFARNF 144
>At1g67400 unknown protein
Length = 328
Score = 140 bits (353), Expect = 7e-34
Identities = 65/113 (57%), Positives = 88/113 (77%), Gaps = 1/113 (0%)
Query: 33 TGSTAW-IGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSIPEHQASL 91
+ + +W I + L+CVC R+ + R+ +LTP QEE L+RL+ R+ YD+S P+HQ +L
Sbjct: 95 SSTPSWRIKKSLTCVCFNRKRAYERICSNLTPLQEERLKRLRKRMKNYYDASRPDHQDAL 154
Query: 92 RALWNAAFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNF 144
RALW+A +P+E+L LIS+QWK+MGWQGKDPSTDFRG G+ISLENLLFFA+ F
Sbjct: 155 RALWSATYPDEKLQDLISDQWKNMGWQGKDPSTDFRGAGFISLENLLFFAKTF 207
>At1g03620 hypothetical protein
Length = 248
Score = 135 bits (340), Expect = 2e-32
Identities = 63/119 (52%), Positives = 83/119 (68%), Gaps = 3/119 (2%)
Query: 28 SAEFVTGSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSIPEH 87
+A + G+ +WIG R D + + L+P QEE LQRLQ R+ VP+D + P+H
Sbjct: 29 TATGMVGTRSWIG---GLFTRSNRRQDKAVDYTLSPLQEERLQRLQDRMVVPFDETRPDH 85
Query: 88 QASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNFPI 146
Q SL+ALWN AFP L GL++EQWK+MGWQG +PSTDFRG G+I+LENLLF AR +P+
Sbjct: 86 QESLKALWNVAFPNVHLTGLVTEQWKEMGWQGPNPSTDFRGCGFIALENLLFSARTYPV 144
>At3g03610 unknown protein
Length = 204
Score = 115 bits (287), Expect = 3e-26
Identities = 51/81 (62%), Positives = 64/81 (78%)
Query: 65 QEECLQRLQSRIDVPYDSSIPEHQASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPST 124
QEE L+ ++ RI++P+D S EHQ +LR LW A+P+ EL L SE WK+MGWQG DPST
Sbjct: 3 QEERLRNIKRRIEIPFDGSRMEHQDALRQLWRLAYPQRELPPLKSELWKEMGWQGTDPST 62
Query: 125 DFRGGGYISLENLLFFARNFP 145
DFRGGGYISLENL+FFA+ +P
Sbjct: 63 DFRGGGYISLENLIFFAKTYP 83
>At3g43400 putative protein
Length = 213
Score = 74.7 bits (182), Expect = 5e-14
Identities = 32/48 (66%), Positives = 40/48 (82%)
Query: 97 AAFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNF 144
+ + +E+L LIS+QWK+MGWQ KDPSTDFRG G+ISLENL FFA+ F
Sbjct: 71 STYADEKLQDLISDQWKNMGWQRKDPSTDFRGDGFISLENLRFFAKTF 118
>At4g14370 disease resistance N like protein
Length = 1996
Score = 31.2 bits (69), Expect = 0.59
Identities = 29/107 (27%), Positives = 43/107 (40%), Gaps = 16/107 (14%)
Query: 75 RIDVPYDSSIPEHQASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPST-----DFRGG 129
++D+P D P H LR L A+P+ ++ E + WQG P T D
Sbjct: 526 QVDIPEDLEFPPH---LRLLRWEAYPKLDM----KESQLEKLWQGTQPLTNLKKMDLTRS 578
Query: 130 GYI----SLENLLFFARNFPIFCGSRKEIVQCGSTRLRLLVLTLHSC 172
++ L N R +C S EI S +L L +H+C
Sbjct: 579 SHLKELPDLSNATNLERLELSYCKSLVEIPSSFSELRKLETLVIHNC 625
>At5g13680 putative protein
Length = 1319
Score = 28.5 bits (62), Expect = 3.8
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 7/84 (8%)
Query: 26 SSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSIP 85
S + EF G W+ G +R+SD S + ++ L+R RI P D++
Sbjct: 218 SETKEFTQGILEWMPSGAKIAAVYKRKSDDS-SPSIAFFERNGLERSSFRIGEPEDAT-- 274
Query: 86 EHQASLRALWNAAFPEEELNGLIS 109
++ WN+A + L G++S
Sbjct: 275 --ESCENLKWNSA--SDLLAGVVS 294
>At4g39450 hypothetical protein
Length = 1553
Score = 28.1 bits (61), Expect = 5.0
Identities = 23/84 (27%), Positives = 36/84 (42%), Gaps = 6/84 (7%)
Query: 24 YHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSS 83
+ S++A+ V +WI R + +DA LS +PY++ CL +L + D S
Sbjct: 594 FQSNTAKDVNFGASWISR------TAVKAADAVLSACPSPYEKRCLLQLLAATDFGDGGS 647
Query: 84 IPEHQASLRALWNAAFPEEELNGL 107
+ L N A P N L
Sbjct: 648 AATYYRRLYWKVNLAEPSLRENDL 671
>At3g61510 1-aminocyclopropane-1-carboxylate synthase-like protein
Length = 488
Score = 27.3 bits (59), Expect = 8.5
Identities = 11/30 (36%), Positives = 17/30 (56%)
Query: 30 EFVTGSTAWIGRGLSCVCAQRRESDARLSF 59
+FV G A + +C+C +RE+ R SF
Sbjct: 427 DFVVGDRANKNKNCNCICNNKRENKKRKSF 456
>At3g03440 unknown protein
Length = 408
Score = 27.3 bits (59), Expect = 8.5
Identities = 25/88 (28%), Positives = 41/88 (46%), Gaps = 12/88 (13%)
Query: 26 SSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFD-----LTPYQEECLQRLQSRID--- 77
SSS + S+A I R LS + ++ + D+RL LT C + ++
Sbjct: 53 SSSVSVSSSSSASIQRVLSLIRSE--DCDSRLFAAKEIRRLTKTSHRCRRHFSQAVEPLV 110
Query: 78 --VPYDSSIPEHQASLRALWNAAFPEEE 103
+ +DS H+A+L AL N A +E+
Sbjct: 111 SMLRFDSPESHHEAALLALLNLAVKDEK 138
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.145 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,910,896
Number of Sequences: 26719
Number of extensions: 236819
Number of successful extensions: 718
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 710
Number of HSP's gapped (non-prelim): 11
length of query: 261
length of database: 11,318,596
effective HSP length: 97
effective length of query: 164
effective length of database: 8,726,853
effective search space: 1431203892
effective search space used: 1431203892
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149129.2


