

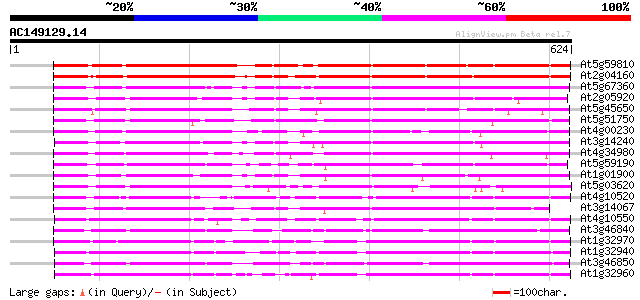
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.14 - phase: 0 /pseudo
(624 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59810 subtilisin-like protease - like protein 495 e-140
At2g04160 subtilisin-like serine protease AIR3 468 e-132
At5g67360 cucumisin-like serine protease (gb|AAC18851.1) 400 e-111
At2g05920 serine protease like protein 397 e-111
At5g45650 subtilisin-like protease 358 5e-99
At5g51750 serine protease-like protein 354 7e-98
At4g00230 subtilisin-type serine endopeptidase XSP1 350 2e-96
At3g14240 unknown protein 348 4e-96
At4g34980 subtilisin proteinase - like 344 1e-94
At5g59190 cucumisin precursor - like 338 4e-93
At1g01900 putative subtilisin-like serine protease 337 1e-92
At5g03620 cucumisin precursor -like protein 334 8e-92
At4g10520 putative subtilisin-like protease 331 6e-91
At3g14067 subtilisin-like serine proteinase, putative, 3' partial 330 2e-90
At4g10550 subtilisin-like protease -like protein 329 3e-90
At3g46840 subtilisin-like proteinase 329 3e-90
At1g32970 hypothetical protein 328 7e-90
At1g32940 unknown protein 327 1e-89
At3g46850 subtilisin-like proteinase 320 2e-87
At1g32960 unknown protein 315 4e-86
>At5g59810 subtilisin-like protease - like protein
Length = 778
Score = 495 bits (1275), Expect = e-140
Identities = 279/580 (48%), Positives = 371/580 (63%), Gaps = 46/580 (7%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA GNFVPGA++FGIGNGT GGSPK+RV YKVCW + A C+ AD+L+AI+ A
Sbjct: 237 TAAGNFVPGANVFGIGNGTASGGSPKARVAAYKVCWPPV----DGAECFDADILAAIEAA 292
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVD++S SVGG + + +D I+IG+F A + +V SAGN GP G+V+NVA
Sbjct: 293 IEDGVDVLSASVGGDAG----DYMSDGIAIGSFHAVKNGVTVVCSAGNSGPKSGTVSNVA 348
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PWV TV AS++DR+F + + + N ++ G SL LP + ++L+ + DA AN DA
Sbjct: 349 PWVITVGASSMDREFQAFVELKNGQSFKGTSLSKPLPEEKMYSLISAADANVANGNVTDA 408
Query: 228 RFCKPGTLDPSKVSGKIVECV-GEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
CK G+LDP KV GKI+ C+ G+ V +G +A +AGA G
Sbjct: 409 LLCKKGSLDPKKVKGKILVCLRGDNAR-------------------VDKGMQAAAAGAAG 449
Query: 287 MILRNQPKFNGKTLLAESNVL--STINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPK 344
M+L N K +G ++++++VL S I+Y D L +S+T K IK P
Sbjct: 450 MVLCND-KASGNEIISDAHVLPASQIDYKDGETLFSY----LSSTKDPKGYIKA----PT 500
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
+ KPAP MASFSSRGPN + P ILKPD+TAPGVNI+AA++ ++L +DNRR P
Sbjct: 501 ATLNTKPAPFMASFSSRGPNTITPGILKPDITAPGVNIIAAFTEATGPTDLDSDNRRT-P 559
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLA 464
FN + GTSMSCPH++G GL+KTLHP+WSPAAI+SAIMTT+ R+N K + D K A
Sbjct: 560 FNTESGTSMSCPHISGVVGLLKTLHPHWSPAAIRSAIMTTSRTRNNRRKPMVDESFKK-A 618
Query: 465 NPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHS 524
NPF+YGSGH+QPN A PGLVYDL+ DYL+FLCA GY+ ++ L + +TC +
Sbjct: 619 NPFSYGSGHVQPNKAAHPGLVYDLTTGDYLDFLCAVGYNNTVVQ-LFAEDPQYTCRQGAN 677
Query: 525 INDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLP-GYNIVVVPDSLTFKKNGE 583
+ D NYPSIT+PNL ++ VTR + NVGPP+TY A+ + P G + V P LTF K GE
Sbjct: 678 LLDFNYPSITVPNL-TGSITVTRKLKNVGPPATYNARFREPLGVRVSVEPKQLTFNKTGE 736
Query: 584 KKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
K FQ+ ++ VTP G Y FGEL WT+ H VRSP+ VQ
Sbjct: 737 VKIFQMTLRPLPVTPSG-YVFGELTWTDSHHYVRSPIVVQ 775
>At2g04160 subtilisin-like serine protease AIR3
Length = 772
Score = 468 bits (1203), Expect = e-132
Identities = 283/582 (48%), Positives = 365/582 (62%), Gaps = 47/582 (8%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G+FVPG SIFG GNGT KGGSP++RV YKVCW GN CY ADVL+A D A
Sbjct: 230 TAAGDFVPGVSIFGQGNGTAKGGSPRARVAAYKVCWPPV--KGNE--CYDADVLAAFDAA 285
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DG D+ISVS+GG +S F D ++IG+F A K I++V SAGN GP +V+NVA
Sbjct: 286 IHDGADVISVSLGGEPTS----FFNDSVAIGSFHAAKKRIVVVCSAGNSGPADSTVSNVA 341
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSF-TLVDSIDAKFANVTNQD 226
PW TV AST+DR+F+S + +GN K G SL P+ F ++ S++AK N + D
Sbjct: 342 PWQITVGASTMDREFASNLVLGNGKHYKGQSLSSTALPHAKFYPIMASVNAKAKNASALD 401
Query: 227 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
A+ CK G+LDP K GKI+ C+ + +GR V +GR G G
Sbjct: 402 AQLCKLGSLDPIKTKGKILVCLRGQ----------NGR--------VEKGRAVALGGGIG 443
Query: 287 MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGIST--TDTIKSVIKIRMSQPK 344
M+L N G LLA+ +VL QLT S +S + T K + I S +
Sbjct: 444 MVLENT-YVTGNDLLADPHVLPAT------QLTSKDSFAVSRYISQTKKPIAHITPS--R 494
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
T KPAPVMASFSS+GP+ V P ILKPD+TAPGV+++AAY+ S +N D RR
Sbjct: 495 TDLGLKPAPVMASFSSKGPSIVAPQILKPDITAPGVSVIAAYTGAVSPTNEQFDPRR-LL 553
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLA 464
FN GTSMSCPH++G AGL+KT +P+WSPAAI+SAIMTTATI D+ I++A + A
Sbjct: 554 FNAISGTSMSCPHISGIAGLLKTRYPSWSPAAIRSAIMTTATIMDDIPGPIQNATNMK-A 612
Query: 465 NPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIH- 523
PF++G+GH+QPN A++PGLVYDL + DYLNFLC+ GY+ IS N FTCS
Sbjct: 613 TPFSFGAGHVQPNLAVNPGLVYDLGIKDYLNFLCSLGYNASQISVFSGNN--FTCSSPKI 670
Query: 524 SINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLP-GYNIVVVPDSLTFKKNG 582
S+ +LNYPSIT+PNL + V V+R V NVG PS Y KV P G + V P SL F K G
Sbjct: 671 SLVNLNYPSITVPNLTSSKVTVSRTVKNVGRPSMYTVKVNNPQGVYVAVKPTSLNFTKVG 730
Query: 583 EKKKFQVI-VQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
E+K F+VI V+++ +G Y FGEL W++ KH VRSP+ V+
Sbjct: 731 EQKTFKVILVKSKGNVAKG-YVFGELVWSDKKHRVRSPIVVK 771
>At5g67360 cucumisin-like serine protease (gb|AAC18851.1)
Length = 757
Score = 400 bits (1027), Expect = e-111
Identities = 242/578 (41%), Positives = 334/578 (56%), Gaps = 46/578 (7%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G+ V GAS+ G +GT +G +P++RV YKVCW C+ +D+L+AID+A
Sbjct: 219 TAAGSVVEGASLLGYASGTARGMAPRARVAVYKVCWL--------GGCFSSDILAAIDKA 270
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I+D V+++S+S+GG S + + D ++IGAF A + IL+ SAGN GP+ S++NVA
Sbjct: 271 IADNVNVLSMSLGGGMS----DYYRDGVAIGAFAAMERGILVSCSAGNAGPSSSSLSNVA 326
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLP-PNQSFTLVDSIDAKFANVTNQD 226
PW+ TV A T+DRDF + +GN K TG SLF P++ + + +A +N TN
Sbjct: 327 PWITTVGAGTLDRDFPALAILGNGKNFTGVSLFKGEALPDKLLPFIYAGNA--SNATN-- 382
Query: 227 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
C GTL P KV GKIV C ++ R V +G +AG G
Sbjct: 383 GNLCMTGTLIPEKVKGKIVMC----------DRGINAR--------VQKGDVVKAAGGVG 424
Query: 287 MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTS 346
MIL N NG+ L+A++++L +K H + TTD + +S T
Sbjct: 425 MILANTAA-NGEELVADAHLLPATTVGEKAGDIIRHYV---TTDPNPTA---SISILGTV 477
Query: 347 YRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFN 406
KP+PV+A+FSSRGPN + P ILKPD+ APGVNILAA++ A + L +D+RR FN
Sbjct: 478 VGVKPSPVVAAFSSRGPNSITPNILKPDLIAPGVNILAAWTGAAGPTGLASDSRR-VEFN 536
Query: 407 IQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANP 466
I GTSMSCPHV+G A L+K++HP WSPAAI+SA+MTTA K + D + P
Sbjct: 537 IISGTSMSCPHVSGLAALLKSVHPEWSPAAIRSALMTTAYKTYKDGKPLLDIATGKPSTP 596
Query: 467 FAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSIN 526
F +G+GH+ P TA +PGL+YDL+ DYL FLCA Y+ I ++ N T S +S+
Sbjct: 597 FDHGAGHVSPTTATNPGLIYDLTTEDYLGFLCALNYTSPQIRSVSRRNYTCDPSKSYSVA 656
Query: 527 DLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKV--QLPGYNIVVVPDSLTFKKNGEK 584
DLNYPS + G+ A TR VT+VG TY KV + G I V P L FK+ EK
Sbjct: 657 DLNYPSFAVNVDGVGAYKYTRTVTSVGGAGTYSVKVTSETTGVKISVEPAVLNFKEANEK 716
Query: 585 KKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
K + V S P G FG ++W++GKH+V SPV +
Sbjct: 717 KSYTVTFTVDSSKPSGSNSFGSIEWSDGKHVVGSPVAI 754
>At2g05920 serine protease like protein
Length = 754
Score = 397 bits (1021), Expect = e-111
Identities = 244/586 (41%), Positives = 337/586 (56%), Gaps = 67/586 (11%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G+ V AS G GT +G + ++RV TYKVCWS C+G+D+L+A+D+A
Sbjct: 215 TAAGSAVRNASFLGYAAGTARGMATRARVATYKVCWSTG--------CFGSDILAAMDRA 266
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVD++S+S+GG S+ + D I+IGAF A + + + SAGN GPT SV NVA
Sbjct: 267 ILDGVDVLSLSLGGGSAPYYR----DTIAIGAFSAMERGVFVSCSAGNSGPTRASVANVA 322
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PWV TV A T+DRDF + +GN K +TG SL+ + ++ + + +
Sbjct: 323 PWVMTVGAGTLDRDFPAFANLGNGKRLTGVSLYSGVGMGTK-----PLELVYNKGNSSSS 377
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
C PG+LD S V GKIV C V+ R V +G AG GM
Sbjct: 378 NLCLPGSLDSSIVRGKIVVC----------DRGVNAR--------VEKGAVVRDAGGLGM 419
Query: 288 ILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPK--- 344
I+ N +G+ L+A+S++L I ++G T D ++ +K S+P
Sbjct: 420 IMANTAA-SGEELVADSHLLPAI------------AVGKKTGDLLREYVKSD-SKPTALL 465
Query: 345 ----TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNR 400
T KP+PV+A+FSSRGPN V P ILKPDV PGVNILA +S + L D+R
Sbjct: 466 VFKGTVLDVKPSPVVAAFSSRGPNTVTPEILKPDVIGPGVNILAGWSDAIGPTGLDKDSR 525
Query: 401 RGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAID 460
R FNI GTSMSCPH++G AGL+K HP WSP+AIKSA+MTTA + DNTN + DA D
Sbjct: 526 RT-QFNIMSGTSMSCPHISGLAGLLKAAHPEWSPSAIKSALMTTAYVLDNTNAPLHDAAD 584
Query: 461 KTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYS-QRLISTLLNPNMTFTC 519
+L+NP+A+GSGH+ P A+ PGLVYD+S +Y+ FLC+ Y+ +++ + P++ C
Sbjct: 585 NSLSNPYAHGSGHVDPQKALSPGLVYDISTEEYIRFLCSLDYTVDHIVAIVKRPSV--NC 642
Query: 520 SGIHS-INDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQL---PGYNIVVVPDS 575
S S LNYPS ++ G V TR VTNVG S+ + KV + P I V P
Sbjct: 643 SKKFSDPGQLNYPSFSVLFGGKRVVRYTREVTNVGAASSVY-KVTVNGAPSVGISVKPSK 701
Query: 576 LTFKKNGEKKKFQV-IVQARSVTPRGRYQFGELQWTNGKHIVRSPV 620
L+FK GEKK++ V V + V+ + +FG + W+N +H VRSPV
Sbjct: 702 LSFKSVGEKKRYTVTFVSKKGVSMTNKAEFGSITWSNPQHEVRSPV 747
>At5g45650 subtilisin-like protease
Length = 791
Score = 358 bits (919), Expect = 5e-99
Identities = 232/594 (39%), Positives = 321/594 (53%), Gaps = 67/594 (11%)
Query: 49 TAGGNFVPGASIFG-IGNGTIKGGSPKSRVVTYKVCWSQTIAD---GNSAVCYGADVLSA 104
TA G V GAS G G+ GG+P +R+ YK CW++ A+ GN +C D+L+A
Sbjct: 242 TAVGRRVLGASALGGFAKGSASGGAPLARLAIYKACWAKPNAEKVEGN--ICLEEDMLAA 299
Query: 105 IDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSV 164
ID AI+DGV +IS+S+G F + D I++GA A +NI++ ASAGN GP PG++
Sbjct: 300 IDDAIADGVHVISISIGTTEPFPFTQ---DGIAMGALHAVKRNIVVAASAGNSGPKPGTL 356
Query: 165 TNVAPWVFTVAASTIDRDFSSTITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTN 224
+N+APW+ TV AST+DR F + +GN ++ LV + + +
Sbjct: 357 SNLAPWIITVGASTLDRAFVGGLVLGNGYTIKTDSITAFKMDKFAPLVYASNVVVPGIAL 416
Query: 225 QDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGA 284
+ C P +L P VSGK+V C L A + + +G E AG
Sbjct: 417 NETSQCLPNSLKPELVSGKVVLC------------------LRGAGSRIGKGMEVKRAGG 458
Query: 285 KGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIR----- 339
GMIL N NG + ++S+ + T + + D I IK
Sbjct: 459 AGMILGNIAA-NGNEVPSDSHFVPTA------------GVTPTVVDKILEYIKTDKNPKA 505
Query: 340 -MSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTD 398
+ KT Y+ + AP M FSSRGPN V P ILKPD+TAPG+ ILAA+S S S + D
Sbjct: 506 FIKPGKTVYKYQAAPSMTGFSSRGPNVVDPNILKPDITAPGLYILAAWSGADSPSKMSVD 565
Query: 399 NRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDA 458
R +NI GTSMSCPHVAG L+K +HP WS AAI+SA+MTTA + ++ K I+D
Sbjct: 566 QRVA-GYNIYSGTSMSCPHVAGAIALLKAIHPKWSSAAIRSALMTTAWMTNDKKKPIQDT 624
Query: 459 IDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFT 518
ANPFA GSGH +P A DPGLVYD S YL + C+ + N + TF
Sbjct: 625 TGLP-ANPFALGSGHFRPTKAADPGLVYDASYRAYLLYGCSV--------NITNIDPTFK 675
Query: 519 C-SGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVG---PPSTYFAKVQLP-GYNIVVVP 573
C S I + NYPSI +PNL V V R VTNVG STY V+ P G ++ +P
Sbjct: 676 CPSKIPPGYNHNYPSIAVPNL-KKTVTVKRTVTNVGTGNSTSTYLFSVKPPSGISVKAIP 734
Query: 574 DSLTFKKNGEKKKFQVIV-----QARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
+ L+F + G+K++F++++ Q + T +G+YQFG WT+ H+VRSP+ V
Sbjct: 735 NILSFNRIGQKQRFKIVIKPLKNQVMNATEKGQYQFGWFSWTDKVHVVRSPIAV 788
>At5g51750 serine protease-like protein
Length = 780
Score = 354 bits (909), Expect = 7e-98
Identities = 230/586 (39%), Positives = 324/586 (55%), Gaps = 57/586 (9%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
T G+ V GA++FG GT +G + K+RV YKVCW C+ +D+LSA+DQA
Sbjct: 234 TVAGSPVKGANLFGFAYGTARGMAQKARVAAYKVCWV--------GGCFSSDILSAVDQA 285
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
++DGV ++S+S+GG S+ D +SI F A + + SAGNGGP P S+TNV+
Sbjct: 286 VADGVQVLSISLGGGVSTYSR----DSLSIATFGAMEMGVFVSCSAGNGGPDPISLTNVS 341
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVN---LPPNQSFTLVDSIDAKFANVTN 224
PW+ TV AST+DRDF +T+ IG +T G SL+ LP N+ + LV N ++
Sbjct: 342 PWITTVGASTMDRDFPATVKIGTMRTFKGVSLYKGRTVLPKNKQYPLV----YLGRNASS 397
Query: 225 QD-ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAG 283
D FC G LD V+GKIV C T V +G+ AG
Sbjct: 398 PDPTSFCLDGALDRRHVAGKIVICDRG------------------VTPRVQKGQVVKRAG 439
Query: 284 AKGMILRNQPKFNGKTLLAESNVLSTINYYDKH-QLTRGHSIGISTTDTIKSVIKIRMSQ 342
GM+L N NG+ L+A+S++L + +K +L + +++ ++ R+
Sbjct: 440 GIGMVLTNTAT-NGEELVADSHMLPAVAVGEKEGKLIKQYAMTSKKATASLEILGTRIGI 498
Query: 343 PKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRG 402
KP+PV+A+FSSRGPN + ILKPD+ APGVNILAA++ + S+L +D RR
Sbjct: 499 -------KPSPVVAAFSSRGPNFLSLEILKPDLLAPGVNILAAWTGDMAPSSLSSDPRR- 550
Query: 403 FPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKT 462
FNI GTSMSCPHV+G A LIK+ HP+WSPAAIKSA+MTTA + DN K + DA
Sbjct: 551 VKFNILSGTSMSCPHVSGVAALIKSRHPDWSPAAIKSALMTTAYVHDNMFKPLTDASGAA 610
Query: 463 LANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLL-NPNMTFTCSG 521
++P+ +G+GHI P A DPGLVYD+ +Y FLC S + + N T +
Sbjct: 611 PSSPYDHGAGHIDPLRATDPGLVYDIGPQEYFEFLCTQDLSPSQLKVFTKHSNRTCKHTL 670
Query: 522 IHSINDLNYPSITL---PNLGLNAVNVTRIVTNVGPP-STYFAKVQ-LPGYNIVVVPDSL 576
+ +LNYP+I+ N + A+ + R VTNVGP S+Y V G ++ V P +L
Sbjct: 671 AKNPGNLNYPAISALFPENTHVKAMTLRRTVTNVGPHISSYKVSVSPFKGASVTVQPKTL 730
Query: 577 TFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
F +K + V + R R +FG L W + H VRSPV +
Sbjct: 731 NFTSKHQKLSYTVTFRTRFRMKRP--EFGGLVWKSTTHKVRSPVII 774
>At4g00230 subtilisin-type serine endopeptidase XSP1
Length = 749
Score = 350 bits (897), Expect = 2e-96
Identities = 228/589 (38%), Positives = 330/589 (55%), Gaps = 77/589 (13%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
T G V AS++GI NGT +G P +R+ YKVCW+++ C D+L+ + A
Sbjct: 217 TVAGVLVANASLYGIANGTARGAVPSARLAMYKVCWARS-------GCADMDILAGFEAA 269
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGV+IIS+S+GG + + +D IS+G+F A K IL VASAGN GP+ G+VTN
Sbjct: 270 IHDGVEIISISIGGPIA----DYSSDSISVGSFHAMRKGILTVASAGNDGPSSGTVTNHE 325
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PW+ TVAAS IDR F S I +GN K+ +G + + P +S+ LV +DA A
Sbjct: 326 PWILTVAASGIDRTFKSKIDLGNGKSFSGMGISMFSPKAKSYPLVSGVDAAKNTDDKYLA 385
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
R+C +LD KV GK++ C R+ G S + GA +
Sbjct: 386 RYCFSDSLDRKKVKGKVMVC----------------RMGGGGVESTIKS----YGGAGAI 425
Query: 288 ILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIG------ISTTDTIKSVIKIRMS 341
I+ +Q N + +A + +++ S+G I++T + +VI+
Sbjct: 426 IVSDQYLDNAQIFMAPATSVNS-------------SVGDIIYRYINSTRSASAVIQ---- 468
Query: 342 QPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRR 401
KT PAP +ASFSSRGPN +LKPD+ APG++ILAA++L S++ L D +
Sbjct: 469 --KTRQVTIPAPFVASFSSRGPNPGSIRLLKPDIAAPGIDILAAFTLKRSLTGLDGDTQF 526
Query: 402 GFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDK 461
F I GTSM+CPHVAG A +K+ HP+W+PAAIKSAI+T+A + + ++ +DA
Sbjct: 527 S-KFTILSGTSMACPHVAGVAAYVKSFHPDWTPAAIKSAIITSA--KPISRRVNKDA--- 580
Query: 462 TLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSG 521
FAYG G I P A PGLVYD+ + Y+ FLC GY+ ++ L+ + +CS
Sbjct: 581 ----EFAYGGGQINPRRAASPGLVYDMDDISYVQFLCGEGYNATTLAPLVG-TRSVSCSS 635
Query: 522 I---HSINDLNYPSI--TLPNLGLNAVNV-TRIVTNVGPPST-YFAKVQLP-GYNIVVVP 573
I + LNYP+I TL + + + V R VTNVGPPS+ Y A V+ P G I V P
Sbjct: 636 IVPGLGHDSLNYPTIQLTLRSAKTSTLAVFRRRVTNVGPPSSVYTATVRAPKGVEITVEP 695
Query: 574 DSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
SL+F K +K+ F+V+V+A+ +TP G+ G L W + +H VRSP+ +
Sbjct: 696 QSLSFSKASQKRSFKVVVKAKQMTP-GKIVSGLLVWKSPRHSVRSPIVI 743
>At3g14240 unknown protein
Length = 775
Score = 348 bits (894), Expect = 4e-96
Identities = 232/600 (38%), Positives = 326/600 (53%), Gaps = 75/600 (12%)
Query: 50 AGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAI 109
+ G +V AS G +G G +PK+R+ YKVCW+ CY +D+L+A D A+
Sbjct: 218 SAGRYVFPASTLGYAHGVAAGMAPKARLAAYKVCWNSG--------CYDSDILAAFDTAV 269
Query: 110 SDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAP 169
+DGVD+IS+SVGG + D I+IGAF A + I + ASAGNGGP +VTNVAP
Sbjct: 270 ADGVDVISLSVGGV----VVPYYLDAIAIGAFGAIDRGIFVSASAGNGGPGALTVTNVAP 325
Query: 170 WVFTVAASTIDRDFSSTITIGN-KTVTGASLF--VNLPPNQSFTLVDSIDAKFANVTNQD 226
W+ TV A TIDRDF + + +GN K ++G S++ L P + + LV
Sbjct: 326 WMTTVGAGTIDRDFPANVKLGNGKMISGVSVYGGPGLDPGRMYPLV--YGGSLLGGDGYS 383
Query: 227 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
+ C G+LDP+ V GKIV C ++ R ++G G G
Sbjct: 384 SSLCLEGSLDPNLVKGKIVLC----------DRGINSR--------ATKGEIVRKNGGLG 425
Query: 287 MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVI----KIRMSQ 342
MI+ N F+G+ L+A+ +VL S+G S D I+ I K R S+
Sbjct: 426 MIIANGV-FDGEGLVADCHVLPAT------------SVGASGGDEIRRYISESSKSRSSK 472
Query: 343 PKTS--------YRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSN 394
T+ +PAPV+ASFS+RGPN P ILKPDV APG+NILAA+ S
Sbjct: 473 HPTATIVFKGTRLGIRPAPVVASFSARGPNPETPEILKPDVIAPGLNILAAWPDRIGPSG 532
Query: 395 LVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKL 454
+ +DNRR FNI GTSM+CPHV+G A L+K HP+WSPAAI+SA++TTA DN+ +
Sbjct: 533 VTSDNRRT-EFNILSGTSMACPHVSGLAALLKAAHPDWSPAAIRSALITTAYTVDNSGEP 591
Query: 455 IRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPN 514
+ D ++ YGSGH+ P AMDPGLVYD++ DY+NFLC + Y++ I T+
Sbjct: 592 MMDESTGNTSSVMDYGSGHVHPTKAMDPGLVYDITSYDYINFLCNSNYTRTNIVTITRRQ 651
Query: 515 MTFTCSGIH---SINDLNYP--SITLPNLGLN--AVNVTRIVTNVG-PPSTYFAKVQLP- 565
C G + +LNYP S+ G + + + R VTNVG S Y K++ P
Sbjct: 652 A--DCDGARRAGHVGNLNYPSFSVVFQQYGESKMSTHFIRTVTNVGDSDSVYEIKIRPPR 709
Query: 566 GYNIVVVPDSLTFKKNGEKKKFQVIVQARSV--TPRG-RYQFGELQWTNGKHIVRSPVTV 622
G + V P+ L+F++ G+K F V V+ V +P + G + W++GK V SP+ V
Sbjct: 710 GTTVTVEPEKLSFRRVGQKLSFVVRVKTTEVKLSPGATNVETGHIVWSDGKRNVTSPLVV 769
>At4g34980 subtilisin proteinase - like
Length = 764
Score = 344 bits (882), Expect = 1e-94
Identities = 225/591 (38%), Positives = 320/591 (54%), Gaps = 61/591 (10%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G AS+ G +G KG +PK+R+ YKVCW + C +D+L+A D A
Sbjct: 212 TAAGRHAFKASMSGYASGVAKGVAPKARIAAYKVCWKDS-------GCLDSDILAAFDAA 264
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
+ DGVD+IS+S+GG + D I+IG++ A +K I + +SAGN GP SVTN+A
Sbjct: 265 VRDGVDVISISIGGGDGIT-SPYYLDPIAIGSYGAASKGIFVSSSAGNEGPNGMSVTNLA 323
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPN-QSFTLVDSIDAKFANVTNQD 226
PWV TV ASTIDR+F + +G+ + G SL+ +P N + F +V + ++
Sbjct: 324 PWVTTVGASTIDRNFPADAILGDGHRLRGVSLYAGVPLNGRMFPVVYPGKSGMSS----- 378
Query: 227 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
A C TLDP +V GKIV C S P V++G AG G
Sbjct: 379 ASLCMENTLDPKQVRGKIVIC-------DRGSSP-----------RVAKGLVVKKAGGVG 420
Query: 287 MILRNQPKFNGKTLLAESNVLSTI----NYYDKHQL-TRGHSIGISTTDTIKSVIKIRMS 341
MIL N NG+ L+ +++++ N D+ + H I++ D +++ I
Sbjct: 421 MILANGAS-NGEGLVGDAHLIPACAVGSNEGDRIKAYASSHPNPIASIDFRGTIVGI--- 476
Query: 342 QPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRR 401
KPAPV+ASFS RGPN + P ILKPD+ APGVNILAA++ + L +D R+
Sbjct: 477 --------KPAPVIASFSGRGPNGLSPEILKPDLIAPGVNILAAWTDAVGPTGLPSDPRK 528
Query: 402 GFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDK 461
FNI GTSM+CPHV+G A L+K+ HP+WSPA I+SA+MTT + DN+N+ + D
Sbjct: 529 T-EFNILSGTSMACPHVSGAAALLKSAHPDWSPAVIRSAMMTTTNLVDNSNRSLIDESTG 587
Query: 462 TLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSG 521
A P+ YGSGH+ AM+PGLVYD++ DY+ FLC+ GY + I + + +
Sbjct: 588 KSATPYDYGSGHLNLGRAMNPGLVYDITNDDYITFLCSIGYGPKTIQVITRTPVRCPTTR 647
Query: 522 IHSINDLNYPSIT----LPNLGLNAVNVTRIVTNVG-PPSTYFAKVQLP-GYNIVVVPDS 575
S +LNYPSIT GL + V R TNVG + Y A+++ P G + V P
Sbjct: 648 KPSPGNLNYPSITAVFPTNRRGLVSKTVIRTATNVGQAEAVYRARIESPRGVTVTVKPPR 707
Query: 576 LTFKKNGEKKKFQVIVQARS---VTPRGRYQFGELQW-TNGKHIVRSPVTV 622
L F +++ + V V + V FG + W GKH+VRSP+ V
Sbjct: 708 LVFTSAVKRRSYAVTVTVNTRNVVLGETGAVFGSVTWFDGGKHVVRSPIVV 758
>At5g59190 cucumisin precursor - like
Length = 693
Score = 338 bits (868), Expect = 4e-93
Identities = 225/585 (38%), Positives = 308/585 (52%), Gaps = 78/585 (13%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA GN V AS +G+ GT +GG P +R+ YKVC+++ C D+L+A D A
Sbjct: 167 TAAGNAVQAASFYGLAQGTARGGVPSARIAAYKVCFNR---------CNDVDILAAFDDA 217
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I+DGVD+IS+S+ SN + ++IG+F A + I+ SAGN GP GSV NV+
Sbjct: 218 IADGVDVISISISADYVSN---LLNASVAIGSFHAMMRGIITAGSAGNNGPDQGSVANVS 274
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PW+ TVAAS DR F + +GN K +TG S+ F +V + N + A
Sbjct: 275 PWMITVAASGTDRQFIDRVVLGNGKALTGISVNTFNLNGTKFPIVYGQNVS-RNCSQAQA 333
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
+C G +D V GKIV C LG+ REA AGA G+
Sbjct: 334 GYCSSGCVDSELVKGKIVLC---------------DDFLGY--------REAYLAGAIGV 370
Query: 288 ILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSY 347
I++N TLL +S + S+G +IKS I+ P+
Sbjct: 371 IVQN-------TLLPDSAFVVPFP---------ASSLGFEDYKSIKSYIE-SAEPPQAEI 413
Query: 348 RR------KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVT-DNR 400
R + AP + SFSSRGP+ V +LKPDV+APG+ ILAA+S AS S+ + +++
Sbjct: 414 LRTEEIVDREAPYVPSFSSRGPSFVIQNLLKPDVSAPGLEILAAFSPVASPSSFLNPEDK 473
Query: 401 RGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAID 460
R +++ GTSM+CPHVAG A +K+ HP+WSP+AIKSAIMTTAT + +
Sbjct: 474 RSVRYSVMSGTSMACPHVAGVAAYVKSFHPDWSPSAIKSAIMTTATPMN---------LK 524
Query: 461 KTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCS 520
K FAYGSG I P A DPGLVY++ DYL LCA G+ ++T N+ TCS
Sbjct: 525 KNPEQEFAYGSGQINPTKASDPGLVYEVETEDYLKMLCAEGFDSTTLTTTSGQNV--TCS 582
Query: 521 GIHSINDLNYPSITLPNLGLNAVNVT--RIVTNVG-PPSTYFAKV--QLPGYNIVVVPDS 575
+ DLNYP++T L+ NVT R VTNVG P STY A V P I + P+
Sbjct: 583 ERTEVKDLNYPTMTTFVSSLDPFNVTFKRTVTNVGFPNSTYKASVVPLQPELQISIEPEI 642
Query: 576 LTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPV 620
L F EKK F V + + + G + + W++G H VRSP+
Sbjct: 643 LRFGFLEEKKSFVVTISGKELKD-GSFVSSSVVWSDGSHSVRSPI 686
>At1g01900 putative subtilisin-like serine protease
Length = 774
Score = 337 bits (864), Expect = 1e-92
Identities = 232/593 (39%), Positives = 316/593 (53%), Gaps = 72/593 (12%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G+ VP A+ FG G G SR+ YK CW+ C DV++AID+A
Sbjct: 232 TAAGDIVPKANYFGQAKGLASGMRFTSRIAAYKACWALG--------CASTDVIAAIDRA 283
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVD+IS+S+GG S + D I+I F A KNI + SAGN GPT +V+N A
Sbjct: 284 ILDGVDVISLSLGGSS----RPFYVDPIAIAGFGAMQKNIFVSCSAGNSGPTASTVSNGA 339
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQD- 226
PW+ TVAAS DR F + + IGN K++ G+SL+ + ++ F ++
Sbjct: 340 PWLMTVAASYTDRTFPAIVRIGNRKSLVGSSLYKGKS-------LKNLPLAFNRTAGEES 392
Query: 227 -ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAK 285
A FC +L V GKIV C+ SGR ++G E +G
Sbjct: 393 GAVFCIRDSLKRELVEGKIVICL----------RGASGR--------TAKGEEVKRSGGA 434
Query: 286 GMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKT 345
M+L + + G+ LLA+ +VL + S+G S T+ + + + +
Sbjct: 435 AMLLVST-EAEGEELLADPHVLPAV------------SLGFSDGKTLLNYLAGAANATAS 481
Query: 346 SYRR-----KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNR 400
R AP++A+FSSRGP+ P I KPD+ APG+NILA +S F+S S L +D R
Sbjct: 482 VRFRGTAYGATAPMVAAFSSRGPSVAGPEIAKPDIAAPGLNILAGWSPFSSPSLLRSDPR 541
Query: 401 RGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRD--- 457
R FNI GTSM+CPH++G A LIK++H +WSPA IKSAIMTTA I DN N+ I D
Sbjct: 542 R-VQFNIISGTSMACPHISGIAALIKSVHGDWSPAMIKSAIMTTARITDNRNRPIGDRGA 600
Query: 458 AIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTF 517
A ++ A FA+G+G++ P A+DPGLVYD S VDYLN+LC+ Y+ I LL +
Sbjct: 601 AGAESAATAFAFGAGNVDPTRAVDPGLVYDTSTVDYLNYLCSLNYTSERI--LLFSGTNY 658
Query: 518 TCSG---IHSINDLNYPS--ITLPN-LGLNAVNVTRIVTNVGPPS-TYFAKVQLP-GYNI 569
TC+ + S DLNYPS + L N L V R VTNVG P+ Y V+ P G +
Sbjct: 659 TCASNAVVLSPGDLNYPSFAVNLVNGANLKTVRYKRTVTNVGSPTCEYMVHVEEPKGVKV 718
Query: 570 VVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
V P L F+K E+ + V A + FG L W K+ VRSP+ V
Sbjct: 719 RVEPKVLKFQKARERLSYTVTYDAEASRNSSSSSFGVLVWICDKYNVRSPIAV 771
>At5g03620 cucumisin precursor -like protein
Length = 766
Score = 334 bits (857), Expect = 8e-92
Identities = 234/606 (38%), Positives = 320/606 (52%), Gaps = 92/606 (15%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
T G V AS+FGI NGT +GG P +R+ YKVCW C D+L+A D+A
Sbjct: 217 TIAGVSVSSASLFGIANGTARGGVPSARIAAYKVCWDSG--------CTDMDMLAAFDEA 268
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
ISDGVDIIS+S+GG S FE D I+IGAF A + IL SAGN GP +V+N+A
Sbjct: 269 ISDGVDIISISIGGASLPFFE----DPIAIGAFHAMKRGILTTCSAGNNGPGLFTVSNLA 324
Query: 169 PWVFTVAASTIDRDFSSTITIGNK-TVTGASLFVNLPPNQSFTLVD-SIDAKFANVTNQD 226
PWV TVAA+++DR F + + +GN T +G SL P + + L S+ + + +
Sbjct: 325 PWVMTVAANSLDRKFETVVKLGNGLTASGISLNGFNPRKKMYPLTSGSLASNLSAGGYGE 384
Query: 227 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
C+PGTL KV GK+V C +GR G + QG++ + KG
Sbjct: 385 PSTCEPGTLGEDKVMGKVVYCE-------------AGREEG---GNGGQGQDHVVRSLKG 428
Query: 287 ----MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQ 342
+ L TL+A S V + D ++T I++T ++VI
Sbjct: 429 AGVIVQLLEPTDMATSTLIAGSYVF----FEDGTKITEY----INSTKNPQAVIF----- 475
Query: 343 PKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRG 402
KT + AP ++SFS+RGP ++ P ILKPD++APG+NILAAYS ASV+ DNRR
Sbjct: 476 -KTKTTKMLAPSISSFSARGPQRISPNILKPDISAPGLNILAAYSKLASVTGYPDDNRRT 534
Query: 403 FPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTAT---IRDNTNKLIRDAI 459
F+I GTSM+CPH A A +K+ HP+WSPAAIKSA+MTTAT I+ N +L
Sbjct: 535 L-FSIMSGTSMACPHAAAAAAYVKSFHPDWSPAAIKSALMTTATPMRIKGNEAEL----- 588
Query: 460 DKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMT--- 516
+YGSG I P A+ PGLVYD++ YL FLC GY+ I L N
Sbjct: 589 --------SYGSGQINPRRAIHPGLVYDITEDAYLRFLCKEGYNSTSIGLLTGDNSNNTT 640
Query: 517 ---FTCSGIH---SINDLNYPSITLPNLGLNAVNVT---------RIVTNVG-PPSTYFA 560
+ C I + LNYPS+ VN T R VTNVG PSTY A
Sbjct: 641 KKEYNCENIKRGLGSDGLNYPSLH------KQVNSTEAKVSEVFYRTVTNVGYGPSTYVA 694
Query: 561 KVQLP-GYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGK-HIVRS 618
+V P G + VVP ++F++ EK+ F+V++ ++W + + H+VRS
Sbjct: 695 RVWAPKGLRVEVVPKVMSFERPKEKRNFKVVIDGVWDETMKGIVSASVEWDDSRGHLVRS 754
Query: 619 PVTVQR 624
P+ + R
Sbjct: 755 PILLFR 760
>At4g10520 putative subtilisin-like protease
Length = 756
Score = 331 bits (849), Expect = 6e-91
Identities = 227/581 (39%), Positives = 314/581 (53%), Gaps = 66/581 (11%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
T GG+F+P S G+G GT +GG+P + YK CWS C GADVL A+D+A
Sbjct: 225 TIGGSFLPNVSYVGLGRGTARGGAPGVHIAVYKACWS--------GYCSGADVLKAMDEA 276
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVDI+S+S+G S F E T+ S+GAF A AK I +V +AGN GPT +++NVA
Sbjct: 277 IHDGVDILSLSLGP-SVPLFPE--TEHTSVGAFHAVAKGIPVVIAAGNAGPTAQTISNVA 333
Query: 169 PWVFTVAASTIDRDFSSTITIGNK-TVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PWV TVAA+T DR F + IT+GN T+ G +++ P F +
Sbjct: 334 PWVLTVAATTQDRSFPTAITLGNNITILGQAIYGG--PELGFVGL--------------- 376
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREA--LSAGAK 285
T S +SG +C EK++ S +L FA ++ S A ++AG
Sbjct: 377 ------TYPESPLSG---DC--EKLSANPNSTMEGKVVLCFAASTPSNAAIAAVINAGGL 425
Query: 286 GMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKT 345
G+I+ P + L + ++ G I T ++KI+ S KT
Sbjct: 426 GLIMAKNPTHS----LTPTRKFPWVSI----DFELGTDILFYIRSTRSPIVKIQAS--KT 475
Query: 346 SYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPF 405
+ + + +A+FSSRGPN V P ILKPD+ APGVNILAA S +S+ N GF
Sbjct: 476 LFGQSVSTKVATFSSRGPNSVSPAILKPDIAAPGVNILAAISPNSSI------NDGGFA- 528
Query: 406 NIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNK-LIRDAIDKTLA 464
+ GTSM+ P V+G L+K+LHP+WSP+AIKSAI+TTA D + + + D + LA
Sbjct: 529 -MMSGTSMATPVVSGVVVLLKSLHPDWSPSAIKSAIVTTAWRTDPSGEPIFADGSSRKLA 587
Query: 465 NPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHS 524
+PF YG G I P A+ PGL+YD++ DY+ ++C+ YS IS +L +T + S
Sbjct: 588 DPFDYGGGLINPEKAVKPGLIYDMTTDDYVMYMCSVDYSDISISRVLG-KITVCPNPKPS 646
Query: 525 INDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLP--GYNIVVVPDSLTFKKNG 582
+ DLN PSIT+PNL V +TR VTNVGP ++ + V P G N+ V P L F
Sbjct: 647 VLDLNLPSITIPNL-RGEVTLTRTVTNVGPVNSVYKVVIDPPTGINVAVTPAELVFDYTT 705
Query: 583 EKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
K+ F V V G Y FG L WT+ H V PV+V+
Sbjct: 706 TKRSFTVRVSTTHKVNTG-YYFGSLTWTDNMHNVAIPVSVR 745
>At3g14067 subtilisin-like serine proteinase, putative, 3' partial
Length = 743
Score = 330 bits (845), Expect = 2e-90
Identities = 221/569 (38%), Positives = 304/569 (52%), Gaps = 70/569 (12%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G+ V AS++ GT G + K+R+ YK+CW+ CY +D+L+A+DQA
Sbjct: 224 TAAGSVVANASLYQYARGTATGMASKARIAAYKICWT--------GGCYDSDILAAMDQA 275
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
++DGV +IS+SVG S+ E TD I+IGAF A I++ SAGN GP P + TN+A
Sbjct: 276 VADGVHVISLSVGASGSA--PEYHTDSIAIGAFGATRHGIVVSCSAGNSGPNPETATNIA 333
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLP-PNQSFTLVDSIDAKFANVTNQD 226
PW+ TV AST+DR+F++ G+ K TG SL+ P+ +LV S D
Sbjct: 334 PWILTVGASTVDREFAANAITGDGKVFTGTSLYAGESLPDSQLSLVYSGDC--------G 385
Query: 227 ARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
+R C PG L+ S V GKIV C V +G AG G
Sbjct: 386 SRLCYPGKLNSSLVEGKIVLCDRG------------------GNARVEKGSAVKLAGGAG 427
Query: 287 MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQP-KT 345
MIL N + +G+ L A+S+++ +G D I+ IK S K
Sbjct: 428 MILANTAE-SGEELTADSHLVPAT------------MVGAKAGDQIRDYIKTSDSPTAKI 474
Query: 346 SYR------RKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDN 399
S+ P+P +A+FSSRGPN + P ILKPDV APGVNILA ++ ++L D
Sbjct: 475 SFLGTLIGPSPPSPRVAAFSSRGPNHLTPVILKPDVIAPGVNILAGWTGMVGPTDLDIDP 534
Query: 400 RRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAI 459
RR FNI GTSMSCPHV+G A L++ HP+WSPAAIKSA++TTA +N+ + I D
Sbjct: 535 RR-VQFNIISGTSMSCPHVSGLAALLRKAHPDWSPAAIKSALVTTAYDVENSGEPIEDLA 593
Query: 460 DKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYS-QRLISTLLNPNMTFT 518
+N F +G+GH+ PN A++PGLVYD+ V +Y+ FLCA GY ++ L +P +
Sbjct: 594 TGKSSNSFIHGAGHVDPNKALNPGLVYDIEVKEYVAFLCAVGYEFPGILVFLQDPTLYDA 653
Query: 519 C--SGIHSINDLNYP--SITLPNLGLNAVNVTRIVTNVGP--PSTYFAKVQLP-GYNIVV 571
C S + + DLNYP S+ + G V R+V NVG + Y V+ P I V
Sbjct: 654 CETSKLRTAGDLNYPSFSVVFASTG-EVVKYKRVVKNVGSNVDAVYEVGVKSPANVEIDV 712
Query: 572 VPDSLTFKKNGEKKKFQVIVQARSVTPRG 600
P L F K EK + V +SV G
Sbjct: 713 SPSKLAFSK--EKSVLEYEVTFKSVVLGG 739
>At4g10550 subtilisin-like protease -like protein
Length = 778
Score = 329 bits (843), Expect = 3e-90
Identities = 215/583 (36%), Positives = 319/583 (53%), Gaps = 54/583 (9%)
Query: 50 AGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAI 109
AGG+FVP S G+ GT++GG+P++ + YK CW + D ++ C AD+L A+D+A+
Sbjct: 230 AGGSFVPNISYKGLAGGTVRGGAPRAHIAMYKACWY--LDDDDTTTCSSADILKAMDEAM 287
Query: 110 SDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAP 169
DGVD++S+S+G E D I+ GAF A K I +V S GN GP +VTN AP
Sbjct: 288 HDGVDVLSISLGSSVPLYGETDIRDGITTGAFHAVLKGITVVCSGGNSGPDSLTVTNTAP 347
Query: 170 WVFTVAASTIDRDFSSTITIG-NKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDAR 228
W+ TVAA+T+DR F++ +T+G NK + G +++ P FT + + N N +
Sbjct: 348 WIITVAATTLDRSFATPLTLGNNKVILGQAMYTG--PGLGFTSLVYPE----NPGNSNES 401
Query: 229 F---CKPGTLDPSK-VSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGA 284
F C+ + ++ + GK+V C T+ P G +L S R AG
Sbjct: 402 FSGTCEELLFNSNRTMEGKVVLCF--------TTSPYGGAVL-------SAARYVKRAGG 446
Query: 285 KGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPK 344
G+I+ P + + L + ++ G I + T + V+KI+ S K
Sbjct: 447 LGVIIARHPGYAIQPCLDDFPCVAV-------DWELGTDILLYTRSSGSPVVKIQPS--K 497
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
T + +A+FSSRGPN + P ILKPD+ APGV+ILAA + T + +G
Sbjct: 498 TLVGQPVGTKVATFSSRGPNSIAPAILKPDIAAPGVSILAA-------TTNTTFSDQG-- 548
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDN-TNKLIRDAIDKTL 463
F + GTSM+ P ++G A L+K LH +WSPAAI+SAI+TTA D ++ + L
Sbjct: 549 FIMLSGTSMAAPAISGVAALLKALHRDWSPAAIRSAIVTTAWKTDPFGEQIFAEGSPPKL 608
Query: 464 ANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIH 523
A+PF YG G + P + +PGLVYD+ + DY+ ++C+ GY++ IS L+ T CS
Sbjct: 609 ADPFDYGGGLVNPEKSANPGLVYDMGLEDYVLYMCSVGYNETSISQLI--GKTTVCSNPK 666
Query: 524 -SINDLNYPSITLPNLGLNAVNVTRIVTNVGP-PSTYFAKVQLP-GYNIVVVPDSLTFKK 580
S+ D N PSIT+PNL + V +TR VTNVGP S Y V+ P G+ + V P++L F
Sbjct: 667 PSVLDFNLPSITIPNL-KDEVTITRTVTNVGPLNSVYRVTVEPPLGFQVTVTPETLVFNS 725
Query: 581 NGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
+K F+V V T G Y FG L W++ H V P++V+
Sbjct: 726 TTKKVYFKVKVSTTHKTNTG-YYFGSLTWSDSLHNVTIPLSVR 767
>At3g46840 subtilisin-like proteinase
Length = 739
Score = 329 bits (843), Expect = 3e-90
Identities = 224/580 (38%), Positives = 314/580 (53%), Gaps = 62/580 (10%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA GN V S +G+GNGT +GG P +R+ YKVC D C +L+A D A
Sbjct: 212 TAAGNAVKHVSFYGLGNGTARGGVPAARIAVYKVC------DPGVDGCTTDGILAAFDDA 265
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I+D VDII++S+GG +SS FEE D I+IGAF A AK IL+V SAGN GP P +V ++A
Sbjct: 266 IADKVDIITISIGGDNSSPFEE---DPIAIGAFHAMAKGILIVNSAGNSGPEPSTVASIA 322
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PW+FTVAAS +R F + + +GN KTV G S+ + + LV A ++ A
Sbjct: 323 PWMFTVAASNTNRAFVTKVVLGNGKTVVGRSVNSFDLNGKKYPLVYGKSAS-SSCGAASA 381
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
FC PG LD +V GKIV C +S EA + GA
Sbjct: 382 GFCSPGCLDSKRVKGKIVLC-----------------------DSPQNPDEAQAMGAIAS 418
Query: 288 ILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSY 347
I+R+ ++V S ++ L ++ +S ++ K+ K + + +T +
Sbjct: 419 IVRSH----------RTDVASIFSFPVSVLLEDDYNTVLSYMNSTKNP-KAAVLKSETIF 467
Query: 348 RRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNI 407
++ APV+AS+ SRGPN + P ILKPD+TAPG I+AAYS A S ++D RR +++
Sbjct: 468 NQR-APVVASYFSRGPNTIIPDILKPDITAPGSEIVAAYSPDAPPS--ISDTRR-VKYSV 523
Query: 408 QQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPF 467
GTSMSCPHVAG A +K+ HP WSP+ I+SAIMTTA + + + + F
Sbjct: 524 DTGTSMSCPHVAGVAAYLKSFHPRWSPSMIQSAIMTTAWPMNASTSPFNELAE------F 577
Query: 468 AYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI-N 526
AYG+GH+ P TA+ PGLVY+ + D++ FLC Y+ + + + + + T S+
Sbjct: 578 AYGAGHVDPITAIHPGLVYEANKSDHIAFLCGLNYTAKNLRLISGDSSSCTKEQTKSLPR 637
Query: 527 DLNYPSITLPNLGLNAVNV--TRIVTNVG-PPSTYFAKVQLPGYNIVVVPDSLTFKKNGE 583
+LNYPS+T V R VTNVG P +TY AKV + VVP L+ K E
Sbjct: 638 NLNYPSMTAQVSAAKPFKVIFRRTVTNVGRPNATYKAKVVGSKLKVKVVPAVLSLKSLYE 697
Query: 584 KKKFQVIVQARSVTPRG-RYQFGELQWTNGKHIVRSPVTV 622
KK F V A P+ +L W++G H VRSP+ V
Sbjct: 698 KKSF--TVTASGAGPKAENLVSAQLIWSDGVHFVRSPIVV 735
>At1g32970 hypothetical protein
Length = 734
Score = 328 bits (840), Expect = 7e-90
Identities = 223/582 (38%), Positives = 316/582 (53%), Gaps = 51/582 (8%)
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G+FVP + G+G GT +GG+P++R+ YK CW + C AD++ AID+A
Sbjct: 186 TAAGSFVPDTNYLGLGRGTARGGAPRARIAMYKACWHLVTG---ATTCSAADLVKAIDEA 242
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFT-DEISIGAFQAFAKNILLVASAGNGGPTPGSVTNV 167
I DGVD++S+S G S F E+ T D +++GAF A AK I +V + GN GP+ +++N
Sbjct: 243 IHDGVDVLSIS-NGFSVPLFPEVDTQDGVAVGAFHAVAKGIPVVCAGGNAGPSSQTISNT 301
Query: 168 APWVFTVAASTIDRDFSSTITIGNK-TVTGASLFVNLPPNQSFT-LVDSIDAKFANVTNQ 225
APW+ TVAA+T DR F + IT+GN TV G +L+ P+ FT LV D+ +N T
Sbjct: 302 APWIITVAATTQDRSFPTFITLGNNVTVVGQALYQG--PDIDFTELVYPEDSGASNETFY 359
Query: 226 DARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVS-QGREALSAGA 284
C+ +P+ + + EKI + T ++ A++ V G + A
Sbjct: 360 GV--CEDLAKNPAHI-------IEEKIVLCFTKSTSYSTMIQAASDVVKLDGYGVIVARN 410
Query: 285 KGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPK 344
G L P F L + + + I +Y + TR I T T+ +
Sbjct: 411 PGHQL--SPCFGFPCLAVDYELGTDILFYIRS--TRSPVAKIQPTRTLVGL--------- 457
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
A +A+FSSRGPN + P ILKPD+ APGVNILAA S D
Sbjct: 458 -----PVATKVATFSSRGPNSISPAILKPDIAAPGVNILAATS--------PNDTFYDKG 504
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNK-LIRDAIDKTL 463
F ++ GTSMS P VAG L+K++HP+WSPAAI+SAI+TTA D + + + D ++ L
Sbjct: 505 FAMKSGTSMSAPVVAGIVALLKSVHPHWSPAAIRSAIVTTAWRTDPSGEPIFADGSNRKL 564
Query: 464 ANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIH 523
A+PF YG G + A +PGLVYD+ V DY+ +LC+ GY+ I+ L++ T +
Sbjct: 565 ADPFDYGGGVVNSEKAANPGLVYDMGVKDYILYLCSVGYTDSSITGLVS-KKTVCANPKP 623
Query: 524 SINDLNYPSITLPNLGLNAVNVTRIVTNVGP-PSTYFAKVQLP-GYNIVVVPDSLTFKKN 581
S+ DLN PSIT+PNL V +TR VTNVGP S Y ++ P G N+ V P +L F
Sbjct: 624 SVLDLNLPSITIPNLA-KEVTITRTVTNVGPVGSVYKPVIEAPMGVNVTVTPSTLVFNAY 682
Query: 582 GEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
K F+V V + G Y FG L WT+ H V PV+V+
Sbjct: 683 TRKLSFKVRVLTNHIVNTG-YYFGSLTWTDSVHNVVIPVSVR 723
>At1g32940 unknown protein
Length = 774
Score = 327 bits (838), Expect = 1e-89
Identities = 214/580 (36%), Positives = 310/580 (52%), Gaps = 47/580 (8%)
Query: 50 AGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAI 109
AGG+FVP S G+ G ++GG+P++R+ YK CW + + C +D+L A+D+++
Sbjct: 225 AGGSFVPNISYKGLAGGNLRGGAPRARIAIYKACWY--VDQLGAVACSSSDILKAMDESM 282
Query: 110 SDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAP 169
DGVD++S+S+G + E D I+ GAF A AK I++V + GN GP +V N AP
Sbjct: 283 HDGVDVLSLSLGAQIPLYPETDLRDRIATGAFHAVAKGIIVVCAGGNSGPAAQTVLNTAP 342
Query: 170 WVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFT-LVDSIDAKFANVTNQDA 227
W+ TVAA+T+DR F + IT+GN K + G +L+ FT LV +A F N T
Sbjct: 343 WIITVAATTLDRSFPTPITLGNRKVILGQALYTG--QELGFTSLVYPENAGFTNETFSGV 400
Query: 228 RFCKPGTLDPSK-VSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKG 286
C+ L+P++ ++GK+V C S S +AG G
Sbjct: 401 --CERLNLNPNRTMAGKVVLCFTTNTLFTAVSRAAS---------------YVKAAGGLG 443
Query: 287 MILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTS 346
+I+ P +N T + I+Y G + + T V+KI+ S +T
Sbjct: 444 VIIARNPGYN-LTPCRDDFPCVAIDY------ELGTDVLLYIRSTRSPVVKIQPS--RTL 494
Query: 347 YRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFN 406
+ +A+FSSRGPN + P ILKPD+ APGV+ILAA S N F+
Sbjct: 495 VGQPVGTKVATFSSRGPNSISPAILKPDIGAPGVSILAATS--------PDSNSSVGGFD 546
Query: 407 IQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDN-TNKLIRDAIDKTLAN 465
I GTSM+ P VAG L+K LHPNWSPAA +SAI+TTA D ++ + + +A+
Sbjct: 547 ILAGTSMAAPVVAGVVALLKALHPNWSPAAFRSAIVTTAWRTDPFGEQIFAEGSSRKVAD 606
Query: 466 PFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI 525
PF YG G + P A DPGL+YD+ DY+ +LC+AGY+ I+ L+ N+T + S+
Sbjct: 607 PFDYGGGIVNPEKAADPGLIYDMGPRDYILYLCSAGYNDSSITQLVG-NVTVCSTPKTSV 665
Query: 526 NDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLP--GYNIVVVPDSLTFKKNGE 583
D+N PSIT+P+L + V +TR VTNVG + + V P G +VV P++L F +
Sbjct: 666 LDVNLPSITIPDL-KDEVTLTRTVTNVGTVDSVYKVVVEPPLGIQVVVAPETLVFNSKTK 724
Query: 584 KKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
F V V G Y FG L WT+ H V PV+V+
Sbjct: 725 NVSFTVRVSTTHKINTGFY-FGNLIWTDSMHNVTIPVSVR 763
>At3g46850 subtilisin-like proteinase
Length = 736
Score = 320 bits (819), Expect = 2e-87
Identities = 218/578 (37%), Positives = 303/578 (51%), Gaps = 60/578 (10%)
Query: 50 AGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAI 109
A GN V S +G+GNGT++GG P +R+ YKVC D C +L+A D AI
Sbjct: 212 AAGNAVKHVSFYGLGNGTVRGGVPAARIAVYKVC------DPGVIRCTSDGILAAFDDAI 265
Query: 110 SDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAP 169
+D VDII+VS+G + FEE D ++IGAF A AK IL V AGN GP ++ ++AP
Sbjct: 266 ADKVDIITVSLGADAVGTFEE---DTLAIGAFHAMAKGILTVNGAGNNGPERRTIVSMAP 322
Query: 170 WVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDAR 228
W+FTVAAS ++R F + + +GN KT+ G S+ + + LV A + A
Sbjct: 323 WLFTVAASNMNRAFITKVVLGNGKTIVGRSVNSFDLNGKKYPLVYGKSAS-SRCDASSAG 381
Query: 229 FCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMI 288
FC PG LD +V GKIV C ++ EA + GA I
Sbjct: 382 FCSPGCLDSKRVKGKIVLC-----------------------DTQRNPGEAQAMGAVASI 418
Query: 289 LRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYR 348
+RN + +VLS +Y +I +S ++ K+ K + + +T +
Sbjct: 419 VRNPYEDAASVFSFPVSVLSEDDY----------NIVLSYVNSTKNP-KAAVLKSETIFN 467
Query: 349 RKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQ 408
+K APV+AS+SSRGPN + ILKPD+TAPG ILAAYS + S + R + +
Sbjct: 468 QK-APVVASYSSRGPNPLIHDILKPDITAPGSEILAAYSPYVPPSE---SDTRHVKYTVI 523
Query: 409 QGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFA 468
GTSMSCPHVAG A IKT HP WSP+ I+SAIMTTA + + + + FA
Sbjct: 524 SGTSMSCPHVAGVAAYIKTFHPLWSPSMIQSAIMTTAWPMNASTSPSNELAE------FA 577
Query: 469 YGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI-ND 527
YG+GH+ P A+ PGLVY+ + D++ FLC Y+ + + + + + T S+ +
Sbjct: 578 YGAGHVDPIAAIHPGLVYEANKSDHITFLCGFNYTGKKLRLISGDSSSCTKEQTKSLTRN 637
Query: 528 LNYPSITLPNLGLNAVNVT--RIVTNVG-PPSTYFAKVQLPGYNIVVVPDSLTFKKNGEK 584
LNYPS++ G VT R VTNVG P +TY AKV + VVP L+ K EK
Sbjct: 638 LNYPSMSAQVSGTKPFKVTFRRTVTNVGRPNATYKAKVVGSKLKVKVVPAVLSLKSLYEK 697
Query: 585 KKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
K F V V +L W++G H VRSP+ V
Sbjct: 698 KSFTVTVSGAGPKAENLVS-AQLIWSDGVHFVRSPIVV 734
>At1g32960 unknown protein
Length = 777
Score = 315 bits (808), Expect = 4e-86
Identities = 213/583 (36%), Positives = 309/583 (52%), Gaps = 53/583 (9%)
Query: 50 AGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAI 109
AGG+FVP S G+ GT++GG+P++R+ YK CW G C +D++ AID+AI
Sbjct: 228 AGGSFVPNVSYKGLAGGTLRGGAPRARIAMYKACWFHEELKG--VTCSDSDIMKAIDEAI 285
Query: 110 SDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAP 169
DGVD++S+S+ G+ N E DE + G F A AK I++V + GN GP +V N+AP
Sbjct: 286 HDGVDVLSISLVGQIPLNSETDIRDEFATGLFHAVAKGIVVVCAGGNDGPAAQTVVNIAP 345
Query: 170 WVFTVAASTIDRDFSSTITIG-NKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDAR 228
W+ TVAA+T+DR F + IT+G NK + G + + P +LV +A+ N T
Sbjct: 346 WILTVAATTLDRSFPTPITLGNNKVILGQATYTG-PELGLTSLVYPENARNNNETFSGV- 403
Query: 229 FCKPGTLDPS-KVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
C+ L+P+ ++ K+V C T T+ +S R F +AG G+
Sbjct: 404 -CESLNLNPNYTMAMKVVLC----FTASRTNAAIS-RAASFVK----------AAGGLGL 447
Query: 288 ILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKS----VIKIRMSQP 343
I+ P + LS N D + + +G I+S V+KI+ S
Sbjct: 448 IISRNPVY----------TLSPCN-DDFPCVAVDYELGTDILSYIRSTRSPVVKIQRS-- 494
Query: 344 KTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGF 403
+T + + +FSSRGPN + P ILKPD+ APGV ILAA S D
Sbjct: 495 RTLSGQPVGTKVVNFSSRGPNSMSPAILKPDIAAPGVRILAATS--------PNDTLNVG 546
Query: 404 PFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDN-TNKLIRDAIDKT 462
F + GTSM+ P ++G L+K LHP WSPAA +SAI+TTA D ++ + +
Sbjct: 547 GFAMLSGTSMATPVISGVIALLKALHPEWSPAAFRSAIVTTAWRTDPFGEQIFAEGSSRK 606
Query: 463 LANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGI 522
+++PF YG G + P A +PGL+YD+ DY+ +LC+AGY+ IS L+ +T +
Sbjct: 607 VSDPFDYGGGIVNPEKAAEPGLIYDMGPQDYILYLCSAGYNDSSISQLVG-QITVCSNPK 665
Query: 523 HSINDLNYPSITLPNLGLNAVNVTRIVTNVG-PPSTYFAKVQLP-GYNIVVVPDSLTFKK 580
S+ D+N PSIT+PNL + V +TR VTNVG S Y V+ P G +VV P++L F
Sbjct: 666 PSVLDVNLPSITIPNL-KDEVTLTRTVTNVGLVDSVYKVSVEPPLGVRVVVTPETLVFNS 724
Query: 581 NGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
F V V G Y FG L WT+ H V P++V+
Sbjct: 725 KTISVSFTVRVSTTHKINTG-YYFGSLTWTDSVHNVVIPLSVR 766
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,492,632
Number of Sequences: 26719
Number of extensions: 571743
Number of successful extensions: 1725
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1246
Number of HSP's gapped (non-prelim): 100
length of query: 624
length of database: 11,318,596
effective HSP length: 105
effective length of query: 519
effective length of database: 8,513,101
effective search space: 4418299419
effective search space used: 4418299419
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149129.14


