

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.11 - phase: 0 /pseudo
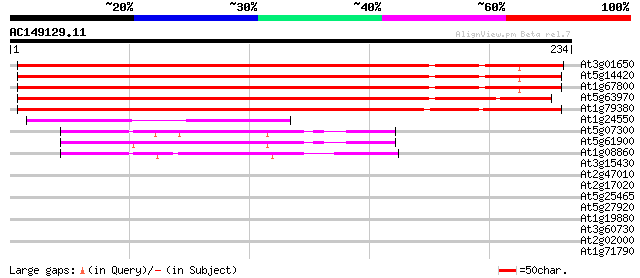
(234 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g01650 unknown protein 267 4e-72
At5g14420 unknown protein 266 7e-72
At1g67800 unknown protein 254 4e-68
At5g63970 unknown protein 223 9e-59
At1g79380 unknown protein 220 6e-58
At1g24550 hypothetical protein 93 1e-19
At5g07300 copine-like protein 61 4e-10
At5g61900 copine - like protein 60 1e-09
At1g08860 hypothetical protein 60 1e-09
At3g15430 unknown protein 30 1.1
At2g47010 unknown protein 30 1.4
At2g17020 unknown protein 29 2.5
At5g25465 unknown protein 28 3.2
At5g27920 unknown protein 28 5.5
At1g19880 unknown protein 28 5.5
At3g60730 pectinesterase - like protein 27 7.1
At2g02000 glutamate decarboxylase like protein 27 9.3
At1g71790 actin capping protein beta-2, putative 27 9.3
>At3g01650 unknown protein
Length = 489
Score = 267 bits (682), Expect = 4e-72
Identities = 142/229 (62%), Positives = 170/229 (74%), Gaps = 5/229 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+SLH + N PN E+AI+IIG+ + F +DNLIPC+GFGDAST D DVFSFY
Sbjct: 170 TGAKSFNRKSLHHLSNTPNPYEQAITIIGRTLAAFDEDNLIPCYGFGDASTHDQDVFSFY 229
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ RFCNG EE+L+RYREI P ++ AG TSFAPIIEMA +V+QS G++HVLVII+DGQV
Sbjct: 230 PEGRFCNGFEEVLARYREIVPQLKLAGPTSFAPIIEMAMTVVEQSSGQYHVLVIIADGQV 289
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D +HG LS Q+ TVDAIV+A PLSI+LVGVGDGPWDM++EF DNI R F
Sbjct: 290 TRSVDTEHGRLSPQEQKTVDAIVKASTLPLSIVLVGVGDGPWDMMQEFDDNI--PARAFD 347
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLGL 231
NF+FVNFTEIMSK N S KE E L A+ EI QYKA EL LLG+
Sbjct: 348 NFQFVNFTEIMSK--NKDQSRKETEFALSALMEIPPQYKATIELNLLGV 394
>At5g14420 unknown protein
Length = 468
Score = 266 bits (680), Expect = 7e-72
Identities = 142/228 (62%), Positives = 173/228 (75%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+SLH I + PN E+AI+IIG+ + F +DNLIPC+GFGDAST D DVFSF
Sbjct: 136 TGARSFNRKSLHFIGSSPNPYEQAITIIGRTLAAFDEDNLIPCYGFGDASTHDQDVFSFN 195
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
++RFCNG EE+LSRY+EI P ++ AG TSFAPII+MA IV+QSGG++HVLVII+DGQV
Sbjct: 196 SEDRFCNGFEEVLSRYKEIVPQLKLAGPTSFAPIIDMAMTIVEQSGGQYHVLVIIADGQV 255
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D ++G LS Q+ TVDAIV+A K PLSI+LVGVGDGPWDM++EF DNI R F
Sbjct: 256 TRSVDTENGQLSPQEQKTVDAIVQASKLPLSIVLVGVGDGPWDMMREFDDNI--PARAFD 313
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFTEIM+K N + SLKE E L A+ EI QYKA EL LLG
Sbjct: 314 NFQFVNFTEIMAK--NKAQSLKETEFALSALMEIPQQYKATIELNLLG 359
>At1g67800 unknown protein
Length = 433
Score = 254 bits (648), Expect = 4e-68
Identities = 134/228 (58%), Positives = 167/228 (72%), Gaps = 5/228 (2%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+SLH I PN ++AISIIGK S F +DNLIPC+GFGDA+T D DVFSF
Sbjct: 107 TGARSFGRKSLHFIGTTPNPYQQAISIIGKTLSVFDEDNLIPCYGFGDATTHDQDVFSFN 166
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
++ +CNG EE+L YREI P ++ +G TSFAPIIE A IV++SGG++HVL+II+DGQV
Sbjct: 167 PNDTYCNGFEEVLMCYREIVPQLRLSGPTSFAPIIERAMTIVEESGGQYHVLLIIADGQV 226
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D +G S Q+ T+DAIV A ++PLSI+LVGVGDGPWD +++F DNI R F
Sbjct: 227 TRSVDTDNGGFSPQEQQTIDAIVRASEYPLSIVLVGVGDGPWDTMRQFDDNI--PARAFD 284
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSK NI P+ KEAE L A+ EI QYKA ELGLLG
Sbjct: 285 NFQFVNFTDIMSK--NIDPARKEAEFALSALMEIPSQYKATLELGLLG 330
>At5g63970 unknown protein
Length = 367
Score = 223 bits (567), Expect = 9e-59
Identities = 118/223 (52%), Positives = 156/223 (69%), Gaps = 3/223 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+YS R+SLH I N EKAISIIG+ SPF +D+LIPCFGFGD +T D VFSFY
Sbjct: 51 TGRYSFNRKSLHAIGKRQNPYEKAISIIGRTLSPFDEDDLIPCFGFGDVTTRDQYVFSFY 110
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ + C+G E + RYREI P+++ +G TSFAP+I+ A IV+Q+ ++HVLVII+DGQV
Sbjct: 111 PENKSCDGLENAVKRYREIVPHLKLSGPTSFAPVIDAAINIVEQNNMQYHVLVIIADGQV 170
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D G LS Q+ T+++I+ A +PLSI+LVGVGDGPWD +K+F DNI R F
Sbjct: 171 TRNPDVPLGRLSPQEEATMNSIMAASHYPLSIVLVGVGDGPWDTMKQFDDNI--PHREFD 228
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKEL 226
NF+FVNFT+IMS+ + + + A L A+ EI FQYKA L
Sbjct: 229 NFQFVNFTKIMSEHKDAAKK-EAAFALAALMEIPFQYKATLSL 270
>At1g79380 unknown protein
Length = 401
Score = 220 bits (560), Expect = 6e-58
Identities = 113/227 (49%), Positives = 157/227 (68%), Gaps = 3/227 (1%)
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S + LH + N EKAI +IG+ +PF +DNLIPCFGFGD++T D +VF F+
Sbjct: 93 TGKTSFDGKCLHALGETSNPYEKAIFVIGQTLAPFDEDNLIPCFGFGDSTTHDEEVFGFH 152
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D C+G EE+L+ Y+ I PN++ +G TS+ P+I+ A IV+++ G+ HVLVI++DGQV
Sbjct: 153 SDNSPCHGFEEVLACYKRIAPNLRLSGPTSYGPLIDAAVDIVEKNNGQFHVLVIVADGQV 212
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D G LS Q+ T+DAIV A + LSI+LVGVGDGPW+ +++F D K+ R F
Sbjct: 213 TRGTDMAEGELSQQEKTTIDAIVNASSYALSIVLVGVGDGPWEDMRKFDD--KIPKREFD 270
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILLAIREILFQYKAAKELGLLG 230
NF+FVNFTEIM++ N+ + + A L A+ EI FQY+AA EL LLG
Sbjct: 271 NFQFVNFTEIMTR-NSPESAKETAFALAALMEIPFQYQAAIELRLLG 316
>At1g24550 hypothetical protein
Length = 177
Score = 93.2 bits (230), Expect = 1e-19
Identities = 52/110 (47%), Positives = 66/110 (59%), Gaps = 22/110 (20%)
Query: 8 SLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFYRDER 67
S RQSLH I PN ++AIS+IGK F +DNLIPC+GFGD
Sbjct: 83 SFNRQSLHYIGTSPNPYQQAISVIGKTLYAFDEDNLIPCYGFGD---------------- 126
Query: 68 FCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVI 117
+++SRYREI P ++ G TSFAPIIE A IV++SGG++HV VI
Sbjct: 127 ------DVVSRYREIVPQLRLVGPTSFAPIIERAMTIVEESGGQYHVHVI 170
>At5g07300 copine-like protein
Length = 586
Score = 61.2 bits (147), Expect = 4e-10
Identities = 50/149 (33%), Positives = 73/149 (48%), Gaps = 22/149 (14%)
Query: 22 NSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDV---FSFYRDERFC--NGHEEIL 76
N+ ++AI +G+ + D P +GFG A D V F+ +C +G + I+
Sbjct: 374 NAYQRAIVEVGEVLQFYDSDKRFPAWGFG-ARPIDIPVSHCFNLNGSSTYCEVDGIQGIM 432
Query: 77 SRYREIRPNIQPAGTTSFAPIIEMATKIVD----QSGGKHHVLVIISDGQVIRRNDAQHG 132
+ Y N+ AG T F P+I A I QS K++VL+II+DG + D Q
Sbjct: 433 NAYNGALFNVSFAGPTLFGPVINAAATIASDSLAQSAKKYYVLLIITDGVI---TDLQE- 488
Query: 133 SLSSQKLHTVDAIVEARKFPLSIILVGVG 161
T D+IV A PLSI++VGVG
Sbjct: 489 --------TRDSIVSASDLPLSILIVGVG 509
>At5g61900 copine - like protein
Length = 578
Score = 59.7 bits (143), Expect = 1e-09
Identities = 47/148 (31%), Positives = 69/148 (45%), Gaps = 20/148 (13%)
Query: 22 NSCEKAISIIGKKFSPFLDDNLIPCFGFG----DASTSDHDVFSFYRDERFCNGHEEILS 77
N+ ++AI +G+ + D P +GFG DA S + +G + I++
Sbjct: 371 NAYQRAIMDVGEVLQFYDSDKRFPAWGFGARPIDAPVSHCFNLNGSSSYSEVDGIQGIMT 430
Query: 78 RYREIRPNIQPAGTTSFAPIIEMATKIVD----QSGGKHHVLVIISDGQVIRRNDAQHGS 133
Y N+ AG T F P+I A I Q K++VL+II+DG + D Q
Sbjct: 431 SYTSALFNVSLAGPTLFGPVINAAAMIASASLAQGSRKYYVLLIITDGVI---TDLQE-- 485
Query: 134 LSSQKLHTVDAIVEARKFPLSIILVGVG 161
T DA+V A PLSI++VGVG
Sbjct: 486 -------TKDALVSASDLPLSILIVGVG 506
>At1g08860 hypothetical protein
Length = 644
Score = 59.7 bits (143), Expect = 1e-09
Identities = 49/152 (32%), Positives = 72/152 (47%), Gaps = 26/152 (17%)
Query: 22 NSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVF-------SFYRDERFCNGHEE 74
NS ++AI +G+ + D P +GFG TSD V + Y DE G E
Sbjct: 434 NSYQQAIMEVGEVIQFYDSDKRFPAWGFG-GRTSDGSVSHAFNLNGASYGDEVV--GVEG 490
Query: 75 ILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQS----GGKHHVLVIISDGQVIRRNDAQ 130
I+ Y N+ AG T F+ +++ A QS K+ VL+II+DG +
Sbjct: 491 IMVAYASALRNVSLAGPTLFSNVVDKAAHTASQSLSQNSPKYFVLLIITDGVL------- 543
Query: 131 HGSLSSQKLHTVDAIVEARKFPLSIILVGVGD 162
+ TVDA+V A PLS+++VGVG+
Sbjct: 544 -----TDMAGTVDALVRASDLPLSVLIVGVGN 570
>At3g15430 unknown protein
Length = 488
Score = 30.0 bits (66), Expect = 1.1
Identities = 23/83 (27%), Positives = 35/83 (41%), Gaps = 10/83 (12%)
Query: 52 ASTSDHDVFSFYRDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGK 111
A T D V+SF FC GH E + E++P + A I+ + S G
Sbjct: 283 AVTQDGSVYSFGSGSNFCLGHGE---QQDELQPRVIQAFKRKGIHILRV-------SAGD 332
Query: 112 HHVLVIISDGQVIRRNDAQHGSL 134
H + + S+G+V G+L
Sbjct: 333 EHAVALDSNGRVYTWGKGYCGAL 355
>At2g47010 unknown protein
Length = 451
Score = 29.6 bits (65), Expect = 1.4
Identities = 16/42 (38%), Positives = 19/42 (45%), Gaps = 2/42 (4%)
Query: 32 GKKFSPFLDDNLIPCFGFG--DASTSDHDVFSFYRDERFCNG 71
GKK PF L PCFG G + T +H RD + G
Sbjct: 164 GKKIPPFNQPGLFPCFGSGCMNQPTLNHGKTELQRDGQTMKG 205
>At2g17020 unknown protein
Length = 656
Score = 28.9 bits (63), Expect = 2.5
Identities = 21/65 (32%), Positives = 27/65 (41%), Gaps = 1/65 (1%)
Query: 2 LVSGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFS 61
L G L + L D+ N+PN + AI + K +P L C GDAS
Sbjct: 475 LFDGSSKLALREL-DLSNLPNLTDAAIFALAKSGAPITKLQLRECRLIGDASVMALASTR 533
Query: 62 FYRDE 66
Y DE
Sbjct: 534 VYEDE 538
>At5g25465 unknown protein
Length = 280
Score = 28.5 bits (62), Expect = 3.2
Identities = 20/69 (28%), Positives = 31/69 (43%), Gaps = 3/69 (4%)
Query: 25 EKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFYRDERFCNGHEEILSRYREIRP 84
EK + +I + + F+DDN + G D HD F + F N EI+ E+ P
Sbjct: 71 EKDLFMIEEDWDEFVDDNHL---GPNDNVFFRHDDKMFLEVQIFKNDGNEIIDAPPEVEP 127
Query: 85 NIQPAGTTS 93
+P T+
Sbjct: 128 ETEPFHPTT 136
>At5g27920 unknown protein
Length = 642
Score = 27.7 bits (60), Expect = 5.5
Identities = 14/30 (46%), Positives = 17/30 (56%)
Query: 22 NSCEKAISIIGKKFSPFLDDNLIPCFGFGD 51
N +K I IG K S L+ +L C GFGD
Sbjct: 440 NISDKGIFHIGSKCSKLLELDLYRCAGFGD 469
>At1g19880 unknown protein
Length = 538
Score = 27.7 bits (60), Expect = 5.5
Identities = 15/55 (27%), Positives = 25/55 (45%)
Query: 103 KIVDQSGGKHHVLVIISDGQVIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIIL 157
KIV + G++H +V+ DGQ + ++G L VE PL ++
Sbjct: 111 KIVKAAAGRNHTVVVSDDGQSLGFGWNKYGQLGLGSAKNGFVSVEVESTPLPCVV 165
>At3g60730 pectinesterase - like protein
Length = 496
Score = 27.3 bits (59), Expect = 7.1
Identities = 11/32 (34%), Positives = 16/32 (49%)
Query: 57 HDVFSFYRDERFCNGHEEILSRYREIRPNIQP 88
H+ +FY+ R GH ++R RPN P
Sbjct: 136 HEALAFYKKSRARQGHGPTRPKHRPTRPNHGP 167
>At2g02000 glutamate decarboxylase like protein
Length = 500
Score = 26.9 bits (58), Expect = 9.3
Identities = 15/65 (23%), Positives = 31/65 (47%), Gaps = 6/65 (9%)
Query: 2 LVSGKYSLIRQSLHDIRNVPNSCEKAISIIGK------KFSPFLDDNLIPCFGFGDASTS 55
+++ Y LIR RNV ++C + + ++ + +F+ +N +P F +S
Sbjct: 325 VIAQYYQLIRLGFEGYRNVMDNCRENMMVLRQGLEKTGRFNIVSKENGVPLVAFSLKDSS 384
Query: 56 DHDVF 60
H+ F
Sbjct: 385 RHNEF 389
>At1g71790 actin capping protein beta-2, putative
Length = 263
Score = 26.9 bits (58), Expect = 9.3
Identities = 12/38 (31%), Positives = 19/38 (49%)
Query: 34 KFSPFLDDNLIPCFGFGDASTSDHDVFSFYRDERFCNG 71
K+ P L+D L P +D+F+ YRD+ + G
Sbjct: 73 KYLPPLEDALYPSSELRKLEVEANDIFAIYRDQYYEGG 110
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,087,858
Number of Sequences: 26719
Number of extensions: 211716
Number of successful extensions: 499
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 473
Number of HSP's gapped (non-prelim): 18
length of query: 234
length of database: 11,318,596
effective HSP length: 96
effective length of query: 138
effective length of database: 8,753,572
effective search space: 1207992936
effective search space used: 1207992936
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149129.11


