

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.4 + phase: 0
(323 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
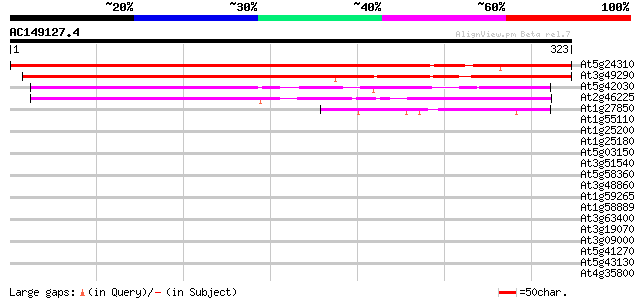
At5g24310 unknown protein 386 e-108
At3g49290 unknown protein 383 e-107
At5g42030 unknown protein 190 1e-48
At2g46225 unknown protein 139 2e-33
At1g27850 transposon protein, putative 48 8e-06
At1g55110 putative zinc finger protein 40 0.002
At1g25200 38 0.007
At1g25180 38 0.007
At5g03150 putative protein 37 0.011
At3g51540 unknown protein 36 0.032
At5g58360 similar to unknown protein (gb|AAB63094.1) 35 0.042
At3g48860 putative protein 35 0.042
At1g59265 polyprotein, putative 35 0.072
At1g58889 polyprotein, putative 35 0.072
At3g63400 cyclophylin (ROC22) 34 0.094
At3g19070 hypothetical protein 34 0.094
At3g09000 unknown protein 34 0.094
At5g41270 unknown protein 34 0.12
At5g43130 unkown protein 33 0.16
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 33 0.16
>At5g24310 unknown protein
Length = 321
Score = 386 bits (992), Expect = e-108
Identities = 206/327 (62%), Positives = 243/327 (73%), Gaps = 10/327 (3%)
Query: 1 MGKIATQPLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQI 60
M AT P+PR ASNYDE+ M Q++LF DSL DLKNLRTQLYSAAEYFELSYTND+QKQI
Sbjct: 1 MSAAATMPMPREASNYDEISMQQSMLFSDSLKDLKNLRTQLYSAAEYFELSYTNDEQKQI 60
Query: 61 LVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMD 120
+VETLKDYA+KAL+NTVDHLGSV YKV+D +DEKV EV G +LR+SCIEQR++ CQ +MD
Sbjct: 61 VVETLKDYAIKALVNTVDHLGSVTYKVNDFVDEKVDEVAGTELRVSCIEQRLRMCQEYMD 120
Query: 121 HEGHTQQSLVISTPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATI 180
HEG +QQSLVI TPK HKRY LP GE G N K K + D EDDW+ FRNAVRATI
Sbjct: 121 HEGRSQQSLVIDTPKFHKRYFLPSGEIKRGGNLAKLKNVEGSFDGEDDWNQFRNAVRATI 180
Query: 181 RETPTSTSSKGNSPSPS-LQPQRVGAFSFTSPNMA-KKDLEKRTVSPHRFPLSRTGSMSS 238
RETP K SPS +PQR FSF+S A KK+ +KR VSPHRFPL R+GS++
Sbjct: 181 RETPPPPVRKPILQSPSQRKPQRSATFSFSSIATAPKKEQDKRAVSPHRFPLLRSGSVAI 240
Query: 239 RSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLS--SDVNNIRDIDQHPSK 296
R ++ R TTP+ S T +P RYPSEPR+SAS+R++ + + Q PSK
Sbjct: 241 RPSS--ISRPTTPSKSRAVTPTPK----RYPSEPRRSASVRVAFEKEAQKEPEHQQQPSK 294
Query: 297 SKRLLKSLLSRRKSKKDDTLYTYLDEY 323
SKRLLK+LLSRRK+KKDDTLYTYLDEY
Sbjct: 295 SKRLLKALLSRRKTKKDDTLYTYLDEY 321
>At3g49290 unknown protein
Length = 312
Score = 383 bits (984), Expect = e-107
Identities = 201/319 (63%), Positives = 240/319 (75%), Gaps = 11/319 (3%)
Query: 8 PLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKD 67
P ASNYDEV M Q++LF D L DLKNLR QLYSAAEYFELSYT DD+KQI+VETLKD
Sbjct: 2 PASHEASNYDEVSMQQSMLFSDGLQDLKNLRAQLYSAAEYFELSYTTDDKKQIVVETLKD 61
Query: 68 YAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQ 127
YAVKAL+NTVDHLGSV YKV+D +DEKV EV +LR+SCIEQR++ CQ +MDHEG +QQ
Sbjct: 62 YAVKALVNTVDHLGSVTYKVNDFIDEKVDEVSETELRVSCIEQRLRMCQEYMDHEGRSQQ 121
Query: 128 SLVISTPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTS- 186
SLVI TPK HKRYILP GE + TN K KY G L+D DDW+ FRNAVRATIRETP
Sbjct: 122 SLVIDTPKFHKRYILPAGEIMTATNLEKLKYFGSSLEDADDWNQFRNAVRATIRETPPPP 181
Query: 187 -TSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKT 245
S S SP PQR FSFTS + KK+ +KR+VSPHRFPL R+GS+++R +
Sbjct: 182 VRKSTSQSSSPRQPPQRSATFSFTS-TIPKKEQDKRSVSPHRFPLLRSGSVATRKSA-SI 239
Query: 246 GRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDID-QHPSKSKRLLKSL 304
R TTP+ S T +RYPSEPR+SAS+R++ + +N ++ + Q PSKSKRLLK+L
Sbjct: 240 SRPTTPSKSRSITP------IRYPSEPRRSASVRVAFEKDNQKETEQQQPSKSKRLLKAL 293
Query: 305 LSRRKSKKDDTLYTYLDEY 323
LSRRK+KKDDTLYT+LDEY
Sbjct: 294 LSRRKTKKDDTLYTFLDEY 312
>At5g42030 unknown protein
Length = 279
Score = 190 bits (482), Expect = 1e-48
Identities = 115/302 (38%), Positives = 168/302 (55%), Gaps = 40/302 (13%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
+SN+DE+FM QTL F ++L DLKNLR QLYSAAEYFE SY + K+ ++ETLK+YA KA
Sbjct: 14 SSNHDELFMKQTLQFSETLKDLKNLRKQLYSAAEYFETSYGKAEHKETVIETLKEYAAKA 73
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
++NTVDHLGSV+ K + L + T LRLS +EQR++ C+ +M G Q L+
Sbjct: 74 VVNTVDHLGSVSDKFNSFLSDNSTHFSTTHLRLSSLEQRMRLCRDYMGKSGTHQHLLLFQ 133
Query: 133 TPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGN 192
P+HHKRY P + GT+ + DD H F +AVR+TI E +T+ K N
Sbjct: 134 YPRHHKRYFFP--QQGRGTSFSAG----------DDSHRFTSAVRSTILENLPNTARKAN 181
Query: 193 SPSPSLQPQRVGAFSF---TSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRST 249
+ G+FSF N+ + KR+ SP RFPL R+GS+ RS++P
Sbjct: 182 ---------KTGSFSFAPIVHNNINNRTPNKRSNSPMRFPLLRSGSLLKRSSSP------ 226
Query: 250 TPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRK 309
+ P + P EP+++ S+ ++++ I+ K + K+L+S K
Sbjct: 227 ---------SQPKKPPLALP-EPQRAISVSRNTEIVEIKQSSSRKGKKILMFKALMSMSK 276
Query: 310 SK 311
S+
Sbjct: 277 SR 278
>At2g46225 unknown protein
Length = 298
Score = 139 bits (351), Expect = 2e-33
Identities = 94/302 (31%), Positives = 156/302 (51%), Gaps = 24/302 (7%)
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
A DEV M + F +L +LKNLR QLYSAA+Y E SY + +QKQ++++ LKDY VKA
Sbjct: 12 AMTLDEVSMERNKSFVKALQELKNLRPQLYSAADYCEKSYLHSEQKQMVLDNLKDYTVKA 71
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L+N VDHLG+VA K++DL D + +++ ++R SC+ Q++ TC+ ++D EG QQ L+
Sbjct: 72 LVNAVDHLGTVASKLTDLFDHQNSDISTMEMRASCVSQQLLTCRTYIDKEGLRQQQLLAV 131
Query: 133 TPKHHKRYILP--VGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSK 190
P HHK YILP V + +H + ++ + +H++ R + P S S
Sbjct: 132 IPLHHKHYILPNSVNKRVHFSPLRRT---------DTRQNHYQAISRLQPSDAPASKSLS 182
Query: 191 GNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTT 250
+ S + + S +P + KD + S+T + + +
Sbjct: 183 WHLGSET--KSTLKGTSTVAP--SSKDSK---------AFSKTSGVFHLLGDDENIANKK 229
Query: 251 PNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRKS 310
P + ++ + P+ + E K + S D N R+I Q P ++K +L + ++K+
Sbjct: 230 PLAGSQVSGVPAASTAHKDLEVPKLLTAHRSLDNNPRREIIQAPVRTKSVLSAFFVKQKT 289
Query: 311 KK 312
K
Sbjct: 290 PK 291
>At1g27850 transposon protein, putative
Length = 1148
Score = 47.8 bits (112), Expect = 8e-06
Identities = 41/143 (28%), Positives = 72/143 (49%), Gaps = 16/143 (11%)
Query: 180 IRETPTSTSSKGNSPSPSLQ--PQRVGAFSFTSPNMAKKDLEKRTVSPHR--FPLSRTG- 234
+R T +SS+GNSPSP ++ + FS +P + L R S R P SR G
Sbjct: 225 VRGTSPVSSSRGNSPSPKIKVWQSNIPGFSLDAPPNLRTSLGDRPASYVRGSSPASRNGR 284
Query: 235 -SMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDI--- 290
++S+RS +S +P++S ++S S+ R R+ S+ + S + D+++++ I
Sbjct: 285 DAVSTRSR-----KSVSPSASRSVSSSHSHERDRFSSQSKGSVASSGDDDLHSLQSIPVG 339
Query: 291 --DQHPSKSKRLLKSLLSRRKSK 311
++ SK L + + R SK
Sbjct: 340 GSERAVSKRASLSPNSRTSRSSK 362
>At1g55110 putative zinc finger protein
Length = 455
Score = 40.0 bits (92), Expect = 0.002
Identities = 32/138 (23%), Positives = 52/138 (37%), Gaps = 10/138 (7%)
Query: 136 HHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPS 195
HH + +G + N + + + E+ HH++N I P + GN
Sbjct: 243 HHHQTQQNIGFSSSSQNIISNSNLHGPMKQEESQHHYQNIPPWLISSNPNPNGNNGNLFP 302
Query: 196 PSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSM-SSRSTTPKTGRSTTPNSS 254
P G SF P+ A L + M S++STTP+ ++ +S
Sbjct: 303 PVASSVNTGRSSFPHPSPAMSATAL---------LQKAAQMGSTKSTTPEEEERSSRSSY 353
Query: 255 NRATTSPSNARVRYPSEP 272
N T+ A + P EP
Sbjct: 354 NNLITTTMAAMMTSPPEP 371
>At1g25200
Length = 248
Score = 38.1 bits (87), Expect = 0.007
Identities = 32/113 (28%), Positives = 53/113 (46%), Gaps = 14/113 (12%)
Query: 199 QPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTP------- 251
+P+ + S +S + A R +SP ++P SR+ S SS + + R +P
Sbjct: 22 RPKYPASRSVSSSSSAHPSQAARQLSPPKYPASRSVSSSSSAHPSQAARQLSPPKYPASR 81
Query: 252 NSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSL 304
+ SN ++ PS A R + P+ AS +SS + HPS++ R L L
Sbjct: 82 SVSNSSSAHPSQA-ARQLTRPKYPASRSVSSSSS------AHPSQADRQLTRL 127
>At1g25180
Length = 258
Score = 38.1 bits (87), Expect = 0.007
Identities = 32/113 (28%), Positives = 53/113 (46%), Gaps = 14/113 (12%)
Query: 199 QPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTP------- 251
+P+ + S +S + A R +SP ++P SR+ S SS + + R +P
Sbjct: 22 RPKYPASRSVSSSSSAHPSQAARQLSPPKYPASRSVSSSSSAHPSQAARQLSPPKYPASR 81
Query: 252 NSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSL 304
+ SN ++ PS A R + P+ AS +SS + HPS++ R L L
Sbjct: 82 SVSNSSSAHPSQA-ARQLTRPKYPASRSVSSSSS------AHPSQADRQLTRL 127
>At5g03150 putative protein
Length = 501
Score = 37.4 bits (85), Expect = 0.011
Identities = 35/130 (26%), Positives = 53/130 (39%), Gaps = 15/130 (11%)
Query: 163 LDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRV------GAFSFTSPNMAKK 216
L D H F+ + +T S+SS S SLQ Q + FS +S N K
Sbjct: 303 LPDFSGHHQFQIPMTSTNPSLTLSSSSTSQQTSASLQHQTLKDSSFSPLFSSSSENKQNK 362
Query: 217 DLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSA 276
L + + ++ GS S S+T + + +S+ AT SP PR S+
Sbjct: 363 PLSPMSATALLQKAAQMGSTRSNSSTAPSFFAGPTMTSSSATASP---------PPRSSS 413
Query: 277 SMRLSSDVNN 286
M + +NN
Sbjct: 414 PMMIQQQLNN 423
>At3g51540 unknown protein
Length = 438
Score = 35.8 bits (81), Expect = 0.032
Identities = 37/160 (23%), Positives = 63/160 (39%), Gaps = 20/160 (12%)
Query: 150 GTNSTKSKYIGCHLDDEDDWHHFR-NAVRATIRETPTSTSSK-------GNSPSPSLQPQ 201
G +S + + H D+D+ H+ N TI E P S+ S S+
Sbjct: 40 GRHSFQQAKVAKHGPDDDNNHNIHANEPLYTIAEVPLGDLSRLVLEARNTTSKLQSITTT 99
Query: 202 RVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT-----------PKTGRSTT 250
RV S + P+ ++ T+ P P R+ S+ ++TT PK S +
Sbjct: 100 RVPLRSESDPS-SRPTRSGSTIRPSNIPTIRSSSVPKKTTTTQIQASASVSSPKRTVSRS 158
Query: 251 PNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDI 290
S+R T SP++ R + + S + + D R +
Sbjct: 159 LTPSSRKTPSPTSTPSRISTTTSTTPSFKTAGDAQRSRSL 198
Score = 31.6 bits (70), Expect = 0.61
Identities = 25/78 (32%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query: 187 TSSKGNSPSPSLQPQRVGAFSFTSPNM--AKKDLEKRTVSPHRFP------LSRTGSMSS 238
T S +PSP+ P R+ + T+P+ A R+++P P SRT SS
Sbjct: 160 TPSSRKTPSPTSTPSRISTTTSTTPSFKTAGDAQRSRSLTPRAKPQIAANASSRTNVRSS 219
Query: 239 RSTTPKTGRSTTPNSSNR 256
S T + RSTT +++ R
Sbjct: 220 -SVTSRPDRSTTVSATPR 236
>At5g58360 similar to unknown protein (gb|AAB63094.1)
Length = 296
Score = 35.4 bits (80), Expect = 0.042
Identities = 28/126 (22%), Positives = 55/126 (43%), Gaps = 13/126 (10%)
Query: 180 IRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTV--SPHRFPLSRTGSMS 237
+R+ SS + P S P + +F K+ ++++TV R LS + S++
Sbjct: 53 LRDPLRRLSSTAHHPQASNSPPKSSSF--------KRKIKRKTVYKPSSRLKLSTSSSLN 104
Query: 238 SRSTTPKTGRSTTPNSSNR---ATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHP 294
RS + + + + ++ +SPS+ + EP S ++ V + D+ + P
Sbjct: 105 HRSKSSSSANAISDSAVGSFLDRVSSPSDQNFVHDPEPHSSIDIKDELSVRKLDDVPEDP 164
Query: 295 SKSKRL 300
S S L
Sbjct: 165 SVSPNL 170
>At3g48860 putative protein
Length = 609
Score = 35.4 bits (80), Expect = 0.042
Identities = 42/164 (25%), Positives = 69/164 (41%), Gaps = 22/164 (13%)
Query: 164 DDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFT----SPNMAK-KDL 218
+DED F +A R P+S SS G++ S + A SF SP+ A ++
Sbjct: 80 EDEDLSLRFASASLKPARHAPSSISSTGSNSSNG--NNNLPAVSFAPRTRSPSPALGRNF 137
Query: 219 EKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYP------SEP 272
++ S R + SMS+RSTTP + PN PS V+ P P
Sbjct: 138 AEQVPSSVRSASAGRPSMSARSTTP----TPIPN-----LMPPSRVSVKTPVSIPPLDPP 188
Query: 273 RKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRKSKKDDTL 316
+S R +DV ++ ++ + L+ L + + ++ L
Sbjct: 189 TRSRDKRFFADVPSVNSKEKGDQREASALRDELDMLQEENENVL 232
>At1g59265 polyprotein, putative
Length = 1466
Score = 34.7 bits (78), Expect = 0.072
Identities = 22/80 (27%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P T+ + N P P+ QP + + +S N ++ + + S LS SS S +
Sbjct: 828 SPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPS 887
Query: 243 PKTGRSTTPNSSNRATTSPS 262
P +T+ +SS+ + T PS
Sbjct: 888 P----TTSASSSSTSPTPPS 903
>At1g58889 polyprotein, putative
Length = 1466
Score = 34.7 bits (78), Expect = 0.072
Identities = 22/80 (27%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P T+ + N P P+ QP + + +S N ++ + + S LS SS S +
Sbjct: 828 SPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPS 887
Query: 243 PKTGRSTTPNSSNRATTSPS 262
P +T+ +SS+ + T PS
Sbjct: 888 P----TTSASSSSTSPTPPS 903
>At3g63400 cyclophylin (ROC22)
Length = 567
Score = 34.3 bits (77), Expect = 0.094
Identities = 43/157 (27%), Positives = 63/157 (39%), Gaps = 17/157 (10%)
Query: 163 LDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAK-KDLEKR 221
+D+ D + ++V++ R + S SPS P RV SP+ + +DL
Sbjct: 325 VDNADQHANLDDSVKSRSRSPIRRRNQNSRSKSPSRSPVRVLGNGNRSPSRSPVRDLGNG 384
Query: 222 TVSPHRFPLSRTGSMSSRSTTP-------KTGRSTTPN-SSNRATTSPSNARVRYPSEPR 273
+ SP P T S RS +P + GR T S R +PS R S PR
Sbjct: 385 SRSPREKPTEETVGKSFRSPSPSGVPKRIRKGRGFTERYSFARKYHTPSPER----SPPR 440
Query: 274 KSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRKS 310
R D R+ D++PS +S R +S
Sbjct: 441 HWPDRRNFQD----RNRDRYPSNRSYSERSPRGRFRS 473
>At3g19070 hypothetical protein
Length = 346
Score = 34.3 bits (77), Expect = 0.094
Identities = 34/124 (27%), Positives = 51/124 (40%), Gaps = 7/124 (5%)
Query: 172 FRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLS 231
FR++V + P S+S+ S SPSLQP + + S P ++ H+ S
Sbjct: 105 FRSSV--SYSHQPPSSSTLATSSSPSLQPPSMSSSSLQPPASLREFFTSSVSYSHQPSSS 162
Query: 232 RTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDID 291
T + SS + P+SS+R + R +PS P S SS R +
Sbjct: 163 STLATSSFFPSSMPYSVRPPDSSDR-----PSLREFFPSSPSSSIQPPESSSSKRARLSN 217
Query: 292 QHPS 295
PS
Sbjct: 218 IFPS 221
>At3g09000 unknown protein
Length = 541
Score = 34.3 bits (77), Expect = 0.094
Identities = 30/90 (33%), Positives = 42/90 (46%), Gaps = 17/90 (18%)
Query: 189 SKGNSPSPSL------QPQRVGAFSFTSPNMAKKDLEKRTVSPHR-------FPLSRTGS 235
S+G SPSP+L +P + FS +P + L R VS R P SR+GS
Sbjct: 280 SRGTSPSPTLNSSRPWKPPEMPGFSLEAPPNLRTTLADRPVSASRGRPGVASAPGSRSGS 339
Query: 236 MSSRSTTPKTGRSTTPNSSNRATTSPSNAR 265
+ R P +G S ++ R + SPS R
Sbjct: 340 I-ERGGGPTSGGS---GNARRQSCSPSRGR 365
Score = 32.0 bits (71), Expect = 0.47
Identities = 28/96 (29%), Positives = 45/96 (46%), Gaps = 6/96 (6%)
Query: 181 RETPTSTSSKGNSPS--PSLQPQRVGAF--SFTSPNMAKKDLEKRTVSPHRFPLSRTGSM 236
+E+ S ++ ++P+ P++ R+G S N K +V+ R P S S
Sbjct: 95 KESHRSVMNQHDAPNSRPTVLKSRLGNCREDIVSGNNNKPQTSSSSVAGLRRPSSSGSSR 154
Query: 237 SSRSTTPKTGRSTTP--NSSNRATTSPSNARVRYPS 270
S+ T RSTTP ++S TT SN+R P+
Sbjct: 155 STSRPATPTRRSTTPTTSTSRPVTTRASNSRSSTPT 190
>At5g41270 unknown protein
Length = 258
Score = 33.9 bits (76), Expect = 0.12
Identities = 23/84 (27%), Positives = 37/84 (43%), Gaps = 1/84 (1%)
Query: 185 TSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKD-LEKRTVSPHRFPLSRTGSMSSRSTTP 243
T+ SS+ PQ + + +SP + KD +E+++V P+ T R P
Sbjct: 147 TARSSRPKIKKEMTMPQEIQSNMLSSPERSVKDQVEEKSVGDTPKPMMLTLERDRRIRKP 206
Query: 244 KTGRSTTPNSSNRATTSPSNARVR 267
K+ + + P S T SN R R
Sbjct: 207 KSKKPSEPQSVPEKTVGGSNKRKR 230
>At5g43130 unkown protein
Length = 689
Score = 33.5 bits (75), Expect = 0.16
Identities = 38/151 (25%), Positives = 61/151 (40%), Gaps = 17/151 (11%)
Query: 171 HFRNAVRATIRETPTSTSSKGNSPSPSLQP--QRVGAFSFTSPNMAKKDLEKRTVSPHRF 228
HF+N+ + P SS + S+ ++ A S TS +DLEK + R
Sbjct: 288 HFQNSSSLPLNSAPGQGSSVSHVKQESVDQSFEKNNAASMTS----NEDLEKES---SRM 340
Query: 229 PLSRTGSMS-----SRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSD 283
LS +M+ S S T + STT NS TS A R P K S+
Sbjct: 341 VLSTPNNMAPASSVSPSMTTQLDASTTMNSRGPLGTSQGGANARMPP---KKPSVGQKKP 397
Query: 284 VNNIRDIDQHPSKSKRLLKSLLSRRKSKKDD 314
+ + PSK +++ + + + + +D
Sbjct: 398 LETLGSSPPPPSKKQKVAGNSMDQSIEQLND 428
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 33.5 bits (75), Expect = 0.16
Identities = 30/78 (38%), Positives = 37/78 (46%), Gaps = 3/78 (3%)
Query: 186 STSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPK 244
S SS G SP SP P G +S TSP + SP P S S +S S +P
Sbjct: 1546 SPSSPGYSPTSPGYSPTSPG-YSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSP- 1603
Query: 245 TGRSTTPNSSNRATTSPS 262
T S +P S + + TSPS
Sbjct: 1604 TSPSYSPTSPSYSPTSPS 1621
Score = 33.5 bits (75), Expect = 0.16
Identities = 29/78 (37%), Positives = 38/78 (48%), Gaps = 3/78 (3%)
Query: 186 STSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPK 244
S +S G SP SP P G +S TSP + SP P S + S +S S +P
Sbjct: 1553 SPTSPGYSPTSPGYSPTSPG-YSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSP- 1610
Query: 245 TGRSTTPNSSNRATTSPS 262
T S +P S + + TSPS
Sbjct: 1611 TSPSYSPTSPSYSPTSPS 1628
Score = 33.1 bits (74), Expect = 0.21
Identities = 30/85 (35%), Positives = 40/85 (46%), Gaps = 7/85 (8%)
Query: 183 TPTSTSSKGNSPS-----PSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMS 237
+PTS S SPS PS P A+S TSP + SP P S + S +
Sbjct: 1616 SPTSPSYSPTSPSYSPTSPSYSPTSP-AYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPT 1674
Query: 238 SRSTTPKTGRSTTPNSSNRATTSPS 262
S S +P T S +P S + + TSP+
Sbjct: 1675 SPSYSP-TSPSYSPTSPSYSPTSPA 1698
Score = 33.1 bits (74), Expect = 0.21
Identities = 28/78 (35%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Query: 186 STSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPK 244
S +S G SP SP+ P G +S TSP + SP P S + S +S S +P
Sbjct: 1567 SPTSPGYSPTSPTYSPSSPG-YSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSP- 1624
Query: 245 TGRSTTPNSSNRATTSPS 262
T S +P S + + TSP+
Sbjct: 1625 TSPSYSPTSPSYSPTSPA 1642
Score = 33.1 bits (74), Expect = 0.21
Identities = 29/85 (34%), Positives = 40/85 (46%), Gaps = 7/85 (8%)
Query: 183 TPTSTSSKGNSP-----SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMS 237
+PTS S SP SP+ P A+S TSP+ + SP P S + S +
Sbjct: 1630 SPTSPSYSPTSPAYSPTSPAYSPTSP-AYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPT 1688
Query: 238 SRSTTPKTGRSTTPNSSNRATTSPS 262
S S +P T + +P S + TSPS
Sbjct: 1689 SPSYSP-TSPAYSPTSPGYSPTSPS 1712
Score = 31.2 bits (69), Expect = 0.79
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 2/81 (2%)
Query: 183 TPT-STSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRST 241
+PT S SS G SP+ ++S TSP+ + SP P S + S +S S
Sbjct: 1577 SPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSY 1636
Query: 242 TPKTGRSTTPNSSNRATTSPS 262
+P T + +P S + TSP+
Sbjct: 1637 SP-TSPAYSPTSPAYSPTSPA 1656
Score = 29.3 bits (64), Expect = 3.0
Identities = 32/104 (30%), Positives = 43/104 (40%), Gaps = 12/104 (11%)
Query: 183 TPTSTSSKGNSPS-----PSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMS 237
+PTS S SPS PS P ++S TSP + SP P S + +
Sbjct: 1665 SPTSPSYSPTSPSYSPTSPSYSPTSP-SYSPTSPAYSPTSPGYSPTSPSYSPTSPSYGPT 1723
Query: 238 SRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLS 281
S S P++ + S SPSNAR+ P+ P S S
Sbjct: 1724 SPSYNPQSAK-----YSPSIAYSPSNARLS-PASPYSPTSPNYS 1761
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,258,744
Number of Sequences: 26719
Number of extensions: 312333
Number of successful extensions: 1195
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 1092
Number of HSP's gapped (non-prelim): 142
length of query: 323
length of database: 11,318,596
effective HSP length: 99
effective length of query: 224
effective length of database: 8,673,415
effective search space: 1942844960
effective search space used: 1942844960
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149127.4


